Abstract
We report a chemical lipidation model for the study of protein lipidations in vitro and in live mammalian cells based on a bioorthogonal, photoinduced tetrazole-alkene cycloaddition reaction.
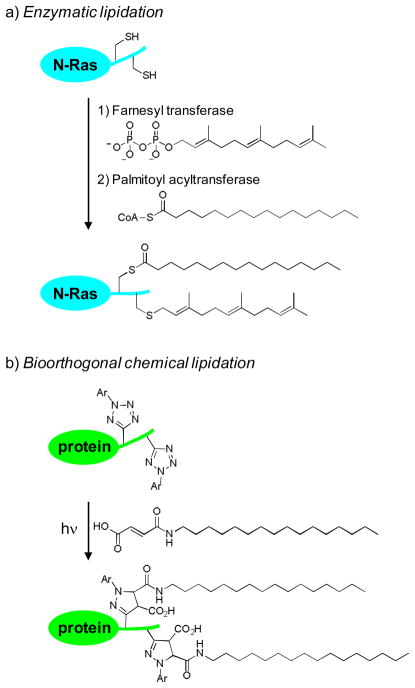
Protein lipidation—the covalent attachment of lipid anchors to a protein—regulates many cellular processes by controlling subcellular protein localization.1 A prominent example is N-Ras, which controls cell growth, differentiation, and apoptosis by acting as a molecular switch in the signal transduction pathway, cycling between the inactive GDP-bound form and the active GTP-bound form.2 As a prerequisite for its biological activity, N-Ras requires two sequential lipidations —a palmitoylation at Cys-181 and a farnesylation at Cys-186—in order to partition into proper compartments (Fig. 1a).3 While the affinities of various lipid anchors toward membrane structures have been studied in vitro using the lipid bilayer model,4 the dynamics of spatiotemporal segregation of proteins upon lipidation in vivo remains less well understood.5
Fig. 1.
(a) Scheme for the enzyme-mediated protein lipidation. N-Ras is used as an example. (b) Scheme for the bioorthogonal chemical lipidation based on the photoinduced tetrazole-alkene cycloaddition reaction.
To probe the effect of lipidation on protein localization dynamics in vivo, two strategies have been successfully developed. One involved the fusion of fluorescent proteins to the target protein to facilitate its monitoring by confocal fluorescent microscopy.6 Another involved the microinjection of the dye-labeled, semisynthesized proteins with the pre-determined lipidation patterns into living cells followed by examination of its subcellular localizations by fluorescent microscopy.7 While these two approaches have elucidated the effect of lipidation on protein localization, the spatiotemporal resolution of intracellular trafficking upon lipidation was limited. We have recently reported a photoinduced tetrazole-alkene cycloaddition reaction for selective functionalization of either tetrazole or alkene-containing proteins both in vitro and in vivo.8 We envisioned that this photoinduced bioorthogonal reaction may provide a chemical strategy for probing protein lipidation in live cells with an improved spatiotemporal resolution. A two-step procedure will be employed in this approach; the photoreactive tetrazoles are first incorporated at the target lipidation sites, which then react with exogenous lipid dipolarophiles to form the lipidated products via the photoinduced cycloaddition reaction (Fig. 1b). Compared to enzymatic lipidation, this chemical strategy offers three unique advantages: 1) while enzymatic lipidations such as palmitoylation,9 are reversible, chemical lipidations are irreversible which simplify the data analysis; 2) the extent of chemical lipidations is tunable through controlling the duration of photoillumination as well as the concentration of exogenous lipid reagents; and 3) exogenous lipid dipolarophiles with either natural lipid chains or unnatural ones can be used in the reactions such that intricate structural effect can be probed in vivo. To validate this strategy, here we report the bioorthogonal chemical lipidations of enhanced green fluorescent protein (EGFP) carrying photoreactive tetrazoles in vitro, and the effect of photoinduced chemical lipidations on EGFP localization both in an organic/PBS buffer mixture and in live HeLa cells.
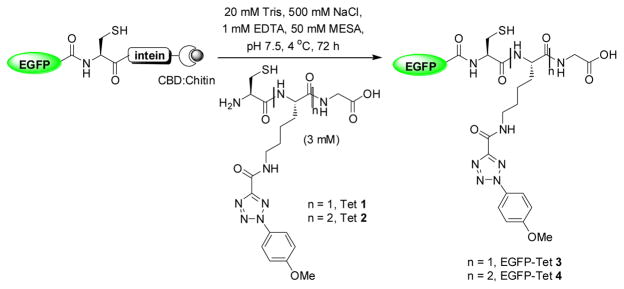
Since successive lipidations are typically required for stable membrane association, we decided to examine the effect of lipid numbers by preparing two semi-synthetic EGFP proteins carrying varying number of photoreactive tetrazole moieties using the intein-mediated protein ligation strategy (Scheme 1). Thus, two short peptides containing either one (Tet 1) or two (Tet 2) tetrazole sidechains were prepared via solid-phase peptide synthesis.10 In parallel, we cloned EGFP(1-239) into the ligation vector, pTXB1, and overexpressed EGFP-intein-CBD in E. coli. The ligation products, EGFP-Tet 3 and 4, were obtained by incubating the immobilized fusion protein with 3 mM Tet 1 or 2 in the ligation buffer followed by FPLC purification (Scheme 1).10 ESI-MS analysis revealed an intact mass of 27547.7 ± 8.7 Da for EGFP-Tet 3, which matches closely with the expected mass of 27536.9 Da. However, repeated attempts in obtaining the intact mass of EGFP-Tet 4 were not successful, presumably due to poor ionization.
Scheme 1.
Preparation of the tetrazole-containing EGFP proteins via native chemical ligation. CBD = chitin binding domain.
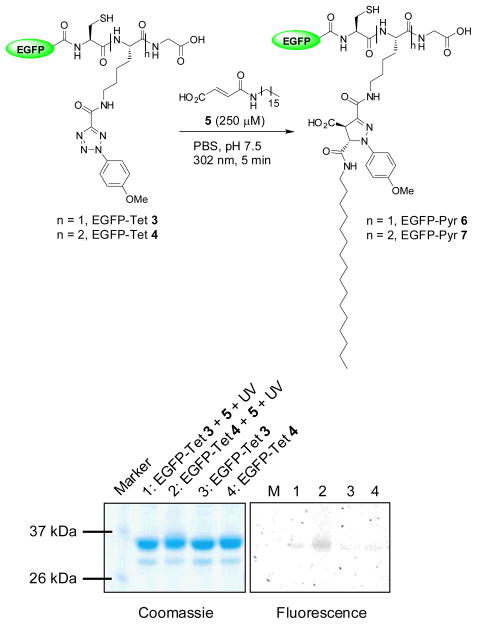
To assess the reactivity of the tetrazole-modified EGFP, we incubated EGFP-Tet 3 and 4, respectively, with N-palmityl fumaric acid (5) and subjected the mixture to 5-min 302-nm photoirradiation with a handheld UV lamp (UVM-57, 0.16 Amps). Both EGFP-Tet 3 and 4 showed the fluorescent adduct formations upon photoirradiation during the in-gel fluorescence analysis (Fig. 2), confirming the photoreactivity of tetrazoles toward a lipid dipolarophile in vitro.11 Unexpectedly, the fluorescence intensity of EGFP-Pyr 7 was about 6-fold greater than that of EGFP-Pyr 6 (compare lane 2 to lane 1 in fluorescence panel), more than two-fold anticipated due to the multiplicity (two tetrazoles in 4 vs. one in 3). This could be attributed to higher effective concentration of the lipid dipolarophile 5 due to the presence of significantly more hydrophobic Tet 2. However, the difference was reduced to 5-fold when 1 mM lipid dipolarophile 5 was used in the reactions, indicating the concentration effect was attenuated.
Fig. 2.
Photoinduced chemical lipidations of the tetrazole-containing EGFP by N-palmityl fumaric acid: scheme for the lipidation reaction was shown in the top and the analysis of the lipidation products by SDS-PAGE was shown in the bottom. Left panel, Coomassie blue staining; right panel, fluorescence imaging. Duration of 5 min of 302-nm UV irradiation was applied to the reaction mixtures.
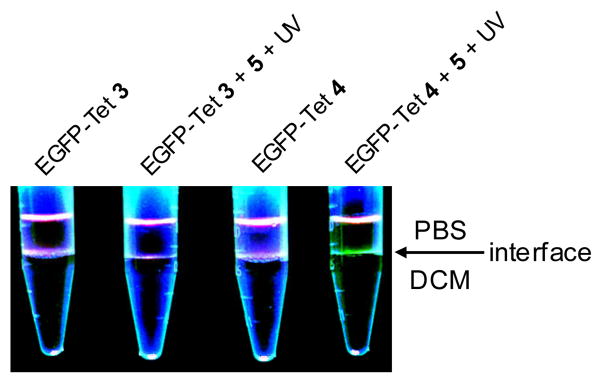
To examine how chemical lipidation affects the phase-transfer behaviour of EGFP, we performed the photoinduced lipidation reaction and added the product mixture to a DCM/PBS buffer (~2:1) mixture. We expected that EGFP lipidation will drive the fluorescent proteins from the aqueous phase to the interface of PBS/DCM as reported previously.12 To our surprise, only EGFP-Tet 4 reaction product showed visible, distinct fluorescence at the solvent interface while EGFP-Tet 3 reaction product did not (Fig. 3). This result could be due to either the high lipidation yield (5–6-fold higher for EGFP-Tet4) or the formation of the bis-lipidated EGFP-Pyr 7.
Fig. 3.
Photoinduced partitioning of EGFP-Tet 3 and 4 in the organic and aqueous phases after the photoinduced cycloaddition reaction: Ten μL of EGFP-Tet 3 (or 4; 20 μM) or the reaction product was added into 400 μL DPBS and 750 μL dichloromethane in a 1.5-mL tube. After vortexing, the EGFP localization was monitored via fluorescence by illuminating the samples with a handheld 365-nm UV lamp from the top.
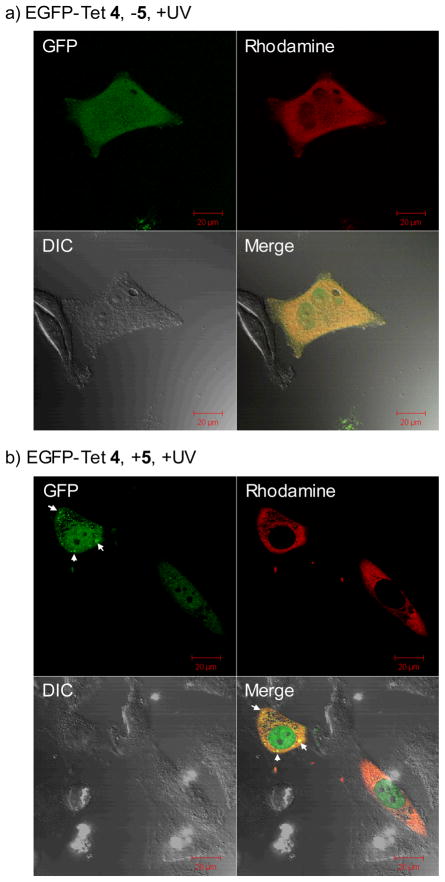
To probe whether this photoinduced chemical lipidation alters protein localization in vivo, we injected EGFP-Tet 4 into HeLa cells together with Dextran-tetramethylrhodamine (MW ~70 KDa). After adding 100 μM N-palmityl fumaric acid (5) to the culture medium, the cells were subjected to 1-min 302-nm UV irradiation before immediate image acquisitions by confocal fluorescent microscopy (Fig. 4). In the GFP channel (ex 488 nm, em 500–550 nm), punctate fluorescent pattern was observed, which was absent in HeLa cells lacking the lipid dipolarophile (compare Fig. 4b to 4a). This was not due to dye-induced non-specific aggregation of EGFP-Tet 4 as the punctate green fluorescence was visible when the GFP channel image was merged with the rhodamine channel (ex 561 nm, em 576–615 nm) image (compare the merged images of Fig. 4b to 4a). Importantly, in the presence of lipid dipolarophile and before the photoirradiation, no punctate fluorescence pattern was observed (see ESI). Therefore, these intense fluorescent spots are most likely the result of EGFP translocation from the cytosol to the vesicles upon its chemical lipidation,13 which is consistent with the tendency of the lipid-containing EGFP-Pyr 7 to localize at the organic/water interface in vitro (Fig. 3).
Fig. 4.
Representative cofoncal micrographs illustrating a bioorthogonal chemical control of the subcellular localization of EGFP in live HeLa cells in the absence (a) and presence (b) of lipid dipolarophile 5 in the culture medium: HeLa cells were microinjected with 25 μM EGFP-Tet 4 together with 2 μM Dextran-TMR, which was used as a red fluorescent marker that is excluded from the nucleus when injected in the cytoplasm. The images were acquired on a confocal microscope with GFP (green), Differential Interference Contrast (DIC), and rhodamine (red) channels following 1 min 302-nm UV irradiation. The merged cell images were shown as the last panels. The scale bars denote 20 μm.
In summary, we have demonstrated a bioorthogonal chemistry strategy in probing protein lipidation in vitro and in vivo without the use of lipidation enzymes. This chemical strategy recapitulated some aspects of protein lipidation in vivo, e.g., the effect of lipid numbers on membrane association stability and the lipidation induced translocation into vesicles inside cells. While we have used the native protein ligation method in preparing the photoreactive lipidation substrates, it is possible in the future that photoreactive tetrazole amino acids14 can be genetically incorporated into target protein site-specifically in mammalian cells. With further development, this photoinduced bioortho-gonal strategy should provide a spatiotemporally controlled tool for dissection of protein lipidation dynamics, specificity, and function in vivo.
Supplementary Material
Acknowledgments
This work is supported by US National Institute of General Medical Sciences, the NIH (GM 085092). We thank Prof. John Kolega at Department of Pathology and Anatomical Sciences for access of his microinjection setup, and Dr. Wade Sigurdson at the Confocal Microscopy Facility for his expert assistance in fluorescent microscopy.
Footnotes
This article is part of 2010 Molecular BioSystems Emerging Investigators Issue highlighting the work of outstanding young scientists at the chemical- and system-biology interfaces.
Electronic supplemental information (ESI) available: Full experimental details and compound characterization data. See DOI: 10.1039/b000000x
References
- 1.(a) Casey PJ. Science. 1995;268:221. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]; (b) Resh MD. Nat Chem Biol. 2006;2:584. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 2.(a) Ehrhardt A, Ehrhardt GR, Guo X, Schrader JW. Exp Hematol. 2002;30:1089. doi: 10.1016/s0301-472x(02)00904-9. [DOI] [PubMed] [Google Scholar]; (b) Vetter IR, Wittinghofer A. Science. 2001;294:1299. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]; (c) Rodriguez-Viciana P, Sabatier C, McCormick F. Mol Cell Biol. 2004;24:4943. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Silvius JR. J Membr Biol. 2002;190:83. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]; (b) Hancock JF. Nat Rev Mol Cell Biol. 2003;4:373. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]; (c) Wright LP, Philips MR. J Lipid Res. 2006;47:883. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.(a) Silvius JR, I’Heureux F. Biochemistry. 1994;33:3014. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]; (b) Buser CA, Sigal CT, Resh MD, McLaughlin S. Biochemistry. 1994;33:13093. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- 5.Rocks O, Peyker A, Bastiaens PI. Curr Opin Cell Biol. 2006;18:351. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 6.(a) Zacharias DA, Violin JD, Newton AC, Tsien RY. Science. 2002;296:913. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]; (b) Chiu VK, Bivona T, Hach A, Sajous JB, Wiener J, Johnson RL, Cox AD, Philips MR. Nat Cell Biol. 2002;4:343. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 7.(a) Reents R, Wagner M, Kuhlmann J, Waldmann H. Angew Chem Int Ed. 2004;43:2711. doi: 10.1002/anie.200353265. [DOI] [PubMed] [Google Scholar]; (b) Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PIH. Science. 2005;307:1746. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]; (c) Brunsveld L, Kuhlmann J, Alexandrov K, Wittinghofer A, Goody RS, Waldmann H. Angew Chem Int Ed. 2006;45:6622. doi: 10.1002/anie.200600855. [DOI] [PubMed] [Google Scholar]
- 8.(a) Song W, Wang Y, Qu J, Madden MM, Lin Q. Angew Chem, Int Ed. 2008;47:2832. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]; (b) Song W, Wang Y, Qu J, Lin Q. J Am Chem Soc. 2008;130:9654. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]; (c) Wang Y, Song W, Hu WJ, Lin Q. Angew Chem Int Ed. 2009;48:5330. doi: 10.1002/anie.200901220. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lim RKV, Lin Q. Chem Commun. 2010:46. doi: 10.1039/B925931G.. [DOI] [Google Scholar]
- 9.Mumby SM. Curr Opin Cell Biol. 1997;9:148. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 10.See Electronic Supplementary Information (ESI) for details.
- 11.Madden MM, Rivera Vera CI, Song W, Lin Q. Chem Commun. 2009:5588. doi: 10.1039/b912094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellois JP, Muir TW. Angew Chem Int Ed. 2005;44:5713. doi: 10.1002/anie.200501244. [DOI] [PubMed] [Google Scholar]
- 13.Matallanas D, Sanz-Moreno V, Arozarena I, Calvo F, Agudo-Ibanez L, Santos E, Berciano MT, Crespo P. Mol Cell Biol. 2006;26:100. doi: 10.1128/MCB.26.1.100-116.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Lin Q. Org Lett. 2009;11:3570. doi: 10.1021/ol901300h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.