Abstract
Recently, a positive correlation between basal leukocyte counts and mating system across primates suggested that sexual promiscuity could be an important determinant of the evolution of the immune system. Motivated by this idea, we examined the patterns of molecular evolution of 15 immune defense genes in primates in relation to promiscuity and other variables expected to affect disease risk. We obtained maximum likelihood estimates of the rate of protein evolution for terminal branches of the primate phylogeny at these genes. Using phylogenetically independent contrasts, we found that immunity genes evolve faster in more promiscuous species, but only for a subset of genes that interact closely with pathogens. We also observed a significantly greater proportion of branches under positive selection in the more promiscuous species. Analyses of independent contrasts also showed a positive effect of group size. However, this effect was not restricted to genes that interact closely with pathogens, and no differences were observed in the proportion of branches under positive selection in species with small and large groups. Together, these results suggest that mating system has influenced the evolution of some immunity genes in primates, possibly due to increased risk of acquiring sexually transmitted diseases in species with higher levels of promiscuity.
Keywords: Disease risk, immunity genes, mating system, primates, sexual promiscuity
In recent years, a large body of work has been devoted to understanding the intricate relationship between immunity and reproduction (Schmid-Hempel 2003; Lawniczak et al. 2007). It has become increasingly clear that there are trade-offs between immune and reproductive functions due to the costly nature of both systems (Sheldon and Verhulst 1996; Lochmiller and Deerenberg 2000; Zuk and Stoehr 2002). For example, in flycatchers, infection rates (measured by serological parameters) increase when brood size is experimentally increased, and parasitized females lay smaller clutches than nonparasitized counterparts (Gustafsson et al. 1994). A more direct link between immunity and reproduction has been provided by the discovery that immune molecules are commonly expressed in reproductive tissues of vertebrates (Li et al. 2001; Com et al. 2003; Silphaduang et al. 2006) and invertebrates (Lung et al. 2001). Moreover, there is now evidence that the female immune system can be modulated in response to mating. In Drosophila, for example, a large suite of immune related genes change their expression profiles following mating (Lawniczak and Begun 2004; McGraw et al. 2004).
Another connection between reproduction and immunity comes from the work of Hamilton and Zuk (1982), who first suggested a role for parasites in the context of sexual selection. At the precopulatory level, mate choice could be based on secondary sexual traits that indirectly reflect heritable variation in immune condition (Hamilton and Zuk 1982). At the postcopulatory level, it is possible that male ejaculates interfere with female immunity leading to sexual conflict (Fedorka and Zuk 2005), or that antimicrobial peptides that inhibit sperm motility (Reddy et al. 2004) mediate cryptic female choice (Lawniczak et al. 2007).
Disease transmission during mating provides another connection between reproduction and immunity. Sexually transmitted diseases (STDs) are ubiquitous among animals and differ from infectious diseases in a number of important characteristics (Lockhart et al. 1996). STDs affect fitness mostly by a negative effect on sterility rather than by inducing mortality. STDs also persist longer in their hosts and do not generally exhibit cyclic fluctuations compared with other infectious diseases. For these reasons, it is possible that STDs might impose different selective pressures on their hosts than other types of diseases (Lockhart et al. 1996). Mating system might affect the evolution of immunity because species with higher levels of sexual promiscuity might experience increased risk of STDs. Alternatively mating behavior itself might evolve as a consequence of STDs (Immerman 1986; Loehle 1995; Thrall et al. 1997; Kokko et al. 2002).
Using the baseline number of leukocytes (a common indicator of immunocompetence), Nunn and others (Nunn et al. 2000, 2002b; Anderson et al. 2004) found a positive correlation between levels of white blood cells and several proxies of female sexual promiscuity among species of primates with different mating systems. The lack of associations with several other social, ecological and life-history variables led to the hypothesis that increased levels of transmission of STDs in promiscuous species have resulted in the evolution of a greater investment in immune function (Nunn et al. 2000). An alternative interpretation for a positive correlation between female promiscuity and the strength of the immune system is based on antagonistic coevolution between male ejaculate and female immunity. Support for this idea comes from studies in crickets of the genus Allonemobious, in which multiple mating with diverse males results in suppression of female immunity (Fedorka and Zuk 2005). If such immunodepression is costly to the female, over evolutionary time, coevolutionary processes caused by conflicting male and female interests might result in the evolution of stronger immunity.
Here, we sought to extend the disease risk/promiscuity hypothesis (Nunn et al. 2000) to the molecular level, by exploring the relationship between sexual promiscuity and the evolution of immunity genes in primates. If the increase in leukocyte levels in promiscuous species reported by Nunn and others truly reflects differences in disease risk among species, we might expect that natural selection will have shaped other aspects of the immune system in a similar way. In particular, natural selection on genes involved in immunity might be stronger or more frequent in species in which females routinely mate with multiple males than in species in which females mate with one male. We thus predict an acceleration of the rate of molecular evolution at immunity genes (particularly those that participate directly in host-pathogen interactions) in more promiscuous species. Primates constitute a good study-system for testing this hypothesis for several reasons. First, the original observations made by Nunn and colleagues were done on primates. Second, primates exhibit a diversity of social and mating systems, and several socioecological variables have been recorded. Some of these variables are also expected to affect disease risk. Thus, we use this information and also explore the effect of group size, density, diet, and habitat on the rate of molecular evolution of immunity genes.
Using phylogenetically independent comparisons for a set of 15 genes related to immune defense in primates we found that both female promiscuity and group size show a weak but significant positive correlation with the rate of protein evolution. The effect of mating system (female promiscuity) was stronger for a subset of genes that interact directly with pathogens, and this seems to be driven by positive selection. Mating system and group size, however, explain only a small fraction of the variation in the rate of protein evolution, emphasizing that other factors related to the particular biology of individual species play a major role in the evolution of immune defense genes.
Methods
SAMPLES
DNA samples from 14 primate species were obtained from the following sources: Cercopithecus mona, Theropithecus gelada, and Mandrillus sphinx from William Switzer; Papio anubis, Callithrix jacchus, and Macaca fascicularis from the Southwest National Primate Research Center; Chiropotes satanas and Saguinus midas from Smithsonian Institution; Ateles geoffroyi, Allenopithecus nigrovirdis, Pithecia pithecia, Cercocebus agilis, Symphalangus syndactylus, and Colobus guereza from Coriell Cell Repositories.
MOLECULAR DATA
We gathered published sequence data on 15 genes related to immune defense in several primate species. These genes were originally sequenced as part of molecular evolutionary studies that did not focus on the effects of mating system. We made an effort to include genes for which there was previous evidence of positive selection in the patterns of protein evolution (Filip and Mundy 2004; Sawyer et al. 2004, 2005; OhAinle et al. 2006; Zelezetsky et al. 2006; Osorio et al. 2007; Sawyer et al. 2007; Kerns et al. 2008; Zhang et al. 2008; Elde et al. 2009; Wlasiuk et al. 2009). By looking at a set of genes that in most cases have a recognized history of positive selection we sought to maximize the chances of uncovering a positive relationship between promiscuity and molecular evolution, if such a relationship exists. The sample includes several innate immunity genes [including three pattern recognition receptors (TLR1, TLR4, TLR5), one antimicrobial peptide (CAMP), a virus-activated protein kinase (PKR), and two chemokine receptors (CCR5, DUFFY)], a series of intrinsic immunity genes with antiviral function (APOBEC3G, APOBEC3H, TRIM5, TRIM22, ZAP), two adaptive immunity genes (CD4, CD45), and a gene with putative immune function (ANG). Some of these genes (CAMP, APOBEC3G, APOBEC3H, PKR, TLR1, TLR4, TLR5, TRIM5, TRIM22, ZAP) interact directly with pathogens as part of their normal function, whereas others do not. We will refer to these two classes of genes as “pathogen-interacting genes” and “non–pathogen-interacting genes,” respectively. More information about the function of these genes is provided in the Supporting information. An average of 16 species (9–29) was used per gene, although there was only partial overlap of species among genes. The complete list of species and accession numbers is presented in the Supporting information.
We also generated sequence data for the DUFFY gene for the 14 species listed above. Special interest in this gene comes from its recently reported association with differences in white-blood cell counts within and between human populations (Nalls et al. 2008; Reich et al. 2009). A fragment of ~1800 bp containing the entire coding region was sequenced. Together with eight additional sequences from GenBank (Supporting information), the complete dataset for DUFFY consists of 22 primate species. PCR was performed in 50 µl reactions using Platinum Taq High Fidelity DNA Polymerase (Invitrogen, San Diego, CA), with primers F1-CTTTCTGGTCCCCACCTTTT and R1-TAAGAAACCACCCGCYTCAC. PCR products were purified using the Qiagen PCR purification kit (Qiagen, Valencia, CA) and sequenced using an ABI 3700 automated sequencer (Applied Biosystems, Foster City, CA), using the following sequencing primers: F2-TAGTCCCRACCAGYCAAATC, F5-ATCGGCTTCCCCAGGA and R2-CGCTTCACAAARGCAKTGTA. Sequences were deposited in GenBank under the following accession numbers: GU219517-GU219530. Sequence editing and assembly were performed using SEQUENCHER (Gene Codes, Ann Arbor, MI).
HYPOTHESES AND PREDICTIONS FOR OTHER SOCIOECOLOGICAL VARIABLES
Many host traits and ecological factors have been proposed to influence disease risk in primates (reviewed in Nunn and Altizer, 2006), and these hypotheses generate testable predictions. Aside from sexual promiscuity, we investigated the effect of group size, density, diet, and habitat. Disease risk is expected to increase with group size and density, because more contacts among individuals should promote transmission of infectious diseases (Altizer et al. 2003). Disease risk is also expected to be higher in species that consume leaves (because folivorous primates consume larger volumes of food, and potentially more parasites) (Moore 2002) or insect prey (because insects can be intermediate hosts for trophically transmitted diseases) (Dunn 1968) than in frugivorous primates. Finally, disease risk is expected to be higher in terrestrial primates than in arboreal primates, because terrestrial species should be exposed to fecal contamination more than arboreal species (Nunn et al. 2000).
PRIMATE VARIABLES
Data on mating system, testis size, group size, density, diet, and habitat were obtained from several published compilations (Kenagy and Trombulak 1986; Harcourt 1991; Harcourt et al. 1995; Rowe 1996; Lindenfors and Tullberg 1998; Nunn 2002b; Semple et al. 2002; Nunn et al. 2003; Anderson et al. 2004). The values for these variables are presented in the Supporting information.
Mating system was further categorized as unimale (UM-monogamous or polygynous) or multimale (MM-polyandrous or promiscuous). To deal with the problem of ambiguities in mating system we grouped mating system three ways. First, we assigned all ambiguous cases as UM (mating system partition 1, MS1). Second, we assigned all ambiguous cases as MM (mating system partition 2, MS2), and third, we excluded all species with ambiguous mating system (mating system partition 3, MS3). A categorization separating monogamous species from the rest was not possible due to the very low number of monogamous primate species. In primates, large testes are likely the result of selection for high sperm production due to sperm competition, in species in which females mate multiply (Harcourt 1991). Thus, the residuals of the regression of log testis size versus log body size (residual testis size-RTS) were also used as a proxy for female promiscuity. Information on testis size was not available for all species, so in several instances we imputed values from other species in the same genus. This is shown in the Supporting information. The amount of imputed data for RTS constitutes a small fraction of the total on a gene-by-gene basis.
Group size and density were log transformed to approach normality. In the linear regression models described below, continuous variables were used. For the other analyses described below, RTS, group size, and density were transformed into discrete variables. For RTS, we coded as 0 the negative values and as 1 the positive values. For group size and density, taking an interval of one standard deviation centered on the mean, we coded as 1 all the values above this range, and as 0 all the values below it. Below we refer to the discrete categories of small group size (SG) and large group size (LG). Habitat and diet were treated as discrete variables with three states each. For habitat we used: strictly arboreal, terrestrial in wooded environments and terrestrial in open environments. For diet we used: insectivores, folivores, and frugivores.
PHYLOGENETIC RECONSTRUCTION
Sequences were aligned in Revtrans (http://cbs.dtu.dk/services/RevTrans/) with manual adjustment of small indels. Sites with indels were removed from the alignments. To assess the concordance between individual gene trees and the published species phylogenies (Purvis 1995; Bininda-Emonds et al. 2007), we reconstructed phylogenetic trees for each gene in PAUP version 4.0b10 (Swofford 2000) using parsimony, distance, and maximum likelihood methods (ML). In all cases there was very close agreement between the gene tree and the species trees (Purvis 1995; Bininda-Emonds et al. 2007), with only a few branches in slightly different positions. The Purvis (1995) and Bininda-Emonds et al. (2007) phylogenies differ in many respects (e.g., topologically and in the number of species included), but for the species analyzed here, there is almost perfect concordance between the two topologies. Because the methods for detecting selection described below are relatively robust to minor changes in the phylogeny and the inferences of trait evolution are based on the species relationships, for all the subsequent analyses we used the species phylogeny (Purvis 1995). We used the Purvis (1995) phylogeny as the species tree because it has fewer unresolved polytomies than the Bininda-Emonds et al. (2007) phylogeny (Fig. 1) (Purvis 1995). Moreover, the results of the CAIC analyses with and without branch length information were similar. Thus, our results appear to be relatively insensitive to differences in branch lengths.
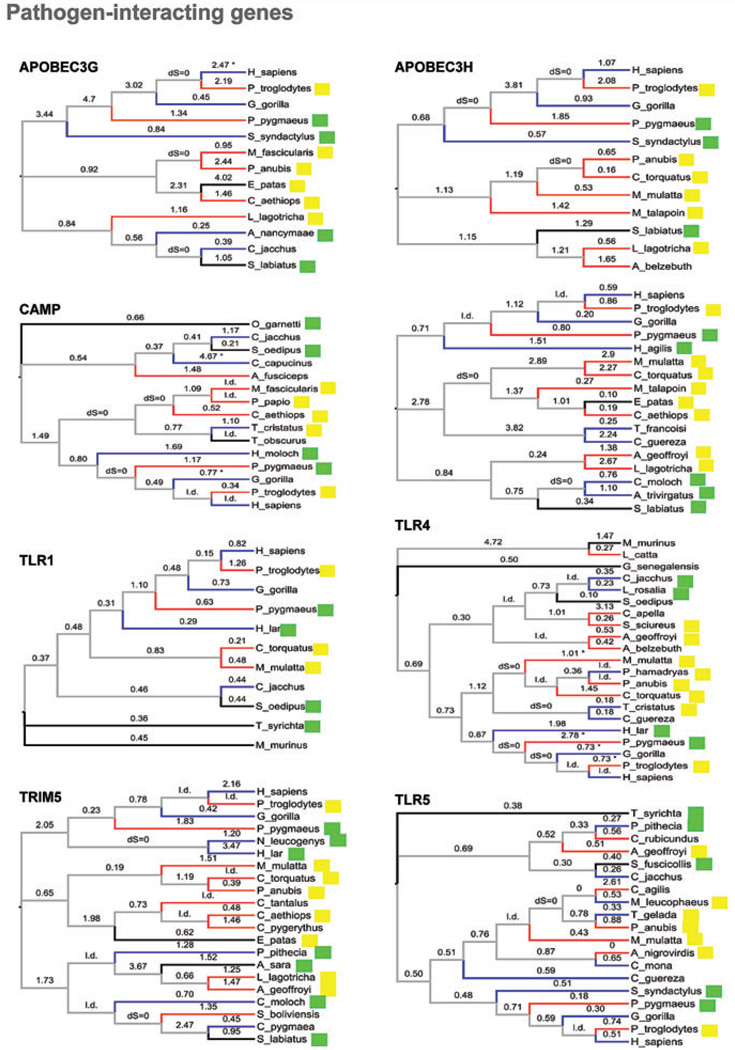
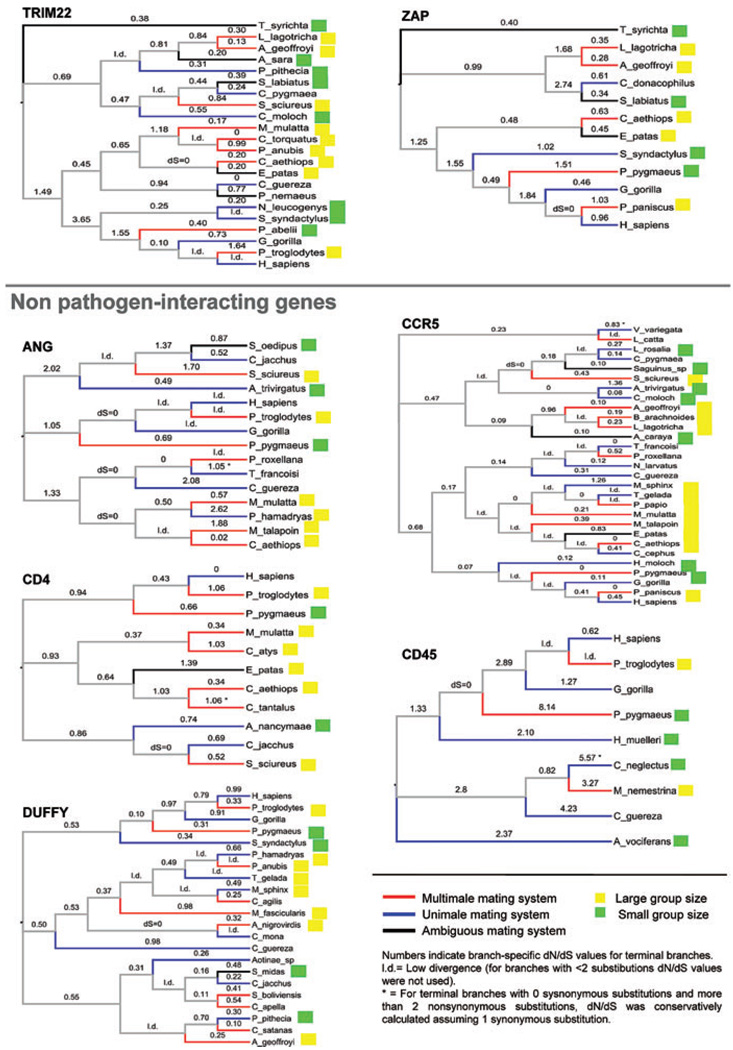
Figure 1.
Phylogenies of the 15 genes [species trees according to Purvis (1995)] showing dN/dS values (from free-ratio ML models), mating system, and group size (discretized). Branches are not to scale. Red branches indicate multimale species, blue branches indicate unimale species, and black branches represent species with ambiguous mating system. Yellow squares indicate species with large groups and green squares indicate species with small groups. l., low divergence; “dS = 0,” no synonynonymous substitutions. For the terminal branches with 0 sysnonymous substitutions and more than 2 nonsynonymous substitutions, dN/dS was conservatively calculated assuming 1 synonymous substitution. These cases are indicated with an asterix.
MAXIMUM LIKELIHOOD ESTIMATE OF EVOLUTIONARY RATES
We estimated dN/dS, the number of nonsynonymous substitutions per nonsynonymous site (dN) divided by the number of synonymous substitutions per synonymous site (dS) in a maximum likelihood (ML) framework using Codeml, in the PAML ver 4.2 package (Yang 1997, 2007). A dN/dS ratio > 1 represents unambiguous evidence of positive selection whereas a value < 1 indicates purifying or negative selection.
We ran a free-ratio model, in which dN/dS is estimated independently for each branch in the phylogeny (Yang 1998). These estimates of the rate of protein evolution on terminal branches of the phylogeny were used to test for correlations with socioecological variables. For some genes, a few branches lacked synonymous substitutions, preventing the calculation of dN/dS. This is a common problem with short branches. However, in some of these cases, the number of nonsynonymous substitutions was high, arguing against low divergence. To minimize the amount of missing data in subsequent analyses that were based on these values, for the branches with dS = 0 and more than two nonsynonymous substitutions, we calculated the dN/dS ratio assuming one synonymous substitution. This assumption is conservative with respect to detecting positive selection. To check for convergence, the free-ratio models were run twice, using initial ω values of 0.5 and 1.5. In all cases we used the F3×4 model of codon frequencies.
STATISTICAL ANALYSES
To minimize the problem of uncertainty in mating system reconstruction along long branches or uncertainty due to incomplete phylogenetic sampling, we restricted the analyses to the terminal branches of the phylogeny. In all the analyses described below the rate of protein evolution, dN/dS, was treated as the dependent variable, whereas mating system, relative testis size, group size, density, diet, and habitat were treated as independent variables. For the first analysis (independent contrasts; see below) we included all variables, but for additional analyses we used only female promiscuity and group size because these were the only two variables correlated with dN/dS in the first analysis. We conducted four major sets of analyses, outlined with their specific predictions in Figure 2.
Figure 2.
Summary of tests using sexual promiscuity (residual testis size or mating system) and group size as independent variables. Red branches indicate multimale species or species with large groups, whereas blue branches represent unimale species or species with small groups. Gray branches indicate internal branches, whose dN/dS values were not used in the analyses. In panel 3 log group size was used as a continuous variable whereas in the rest of the analyses group size was discretized (see the Methods section for details). Each of the analyses is shown in detail in Tables 1–4, corresponding to panels 1–4 in this figure. MS, mating system; GS, group size; UM, unimale mating system; MM, multimale mating system; SG, small group size; LG, large group size; n.s., not significant; Panel 1. Sign tests of the number of positive contrasts [in which an increase in dN/dS is accompanied by an increase in promiscuity (MS3) or group size]. Panel 2. T-tests of differences in mean dN/dS between unimale and multimale species (or species with small groups and large groups). The right half of the figure (under OBSERVATIONS) shows the number of genes with the predicted pattern, followed by the number that are significant or marginally significant in parentheses. For example, for pathogen-interacting genes, 8/10 had higher dN/dS in MM branches than in UM branches, and 3/10 of these were marginally significant at P < 0.10. Panel 3. Multiple regressions of dN/dS as dependent variable, with promiscuity (RTS or MS3) and log group size as independent variables. Only the effects of individual variables are shown. Panel 4. Z-test of differences in the proportion of branches with dN/dS>1 between unimale and multimale species (MS3), or species with small groups and large groups.
First, we performed an analysis based on phylogenetically independent contrasts (Felsenstein 1985), in which variation in the set of independent variables was examined (separately) in relation to variation in dN/dS. Although dN/dS was estimated independently for each branch of the phylogeny, mating system (or other traits) might be the same in closely related species due to shared ancestry, leading to nonindependence. Phylogentically independent contrasts take this potential problem into account. Because branches of the phylogeny are used only once, these contrasts represent independent transitions in the predictor variables given a certain topology. An excess of positive contrasts (i.e., the independent variable and dN/dS vary in the direction predicted by the hypothesis) can be taken as evidence of correlated evolution (Fig. 2, panel 1). Phylogenetically independent contrasts were obtained using the BRUNCH algorithm implemented in the CAIC software (Purvis and Rambaut 1995). The number of contrasts per gene was generally low resulting in little statistical power. Thus, we summed the number of positive contrasts across the 15 genes. To test for deviations from a null expectation of equal number of positive and negative contrasts, we performed sign tests on the number of positive contrasts. We used the False Discovery Rate (FDR) to correct for multiple tests (Verhoeven et al. 2005).
Because of the inferred effect of sexual promiscuity and group size (but not other variables) from the analyses of phylogentically independent contrasts, we focused on these two variables for all subsequent analyses. These analyses do not explicitly correct for phylogenetic effects. However, because for each gene only one species per genus was usually included, we do not expect a high degree of phylogenetic correlation. In the second set of analyses, the mean and variance in dN/dS of UM and MM species were compared with a t-test and a Z-test, respectively (Fig. 2, panel 2). Similarly, means and variances were compared between SG and LG species. Third, we investigated multiple regression models including RTS or mating system and group size as predictor variables (Fig. 2, panel 3). Fourth, differences in the proportion of branches with dN/dS>1 between UM and MM, and between SG and LG species, were evaluated using a Z-test (Fig. 2, panel 4).
Results
We used a combination of approaches to evaluate the effect of sexual promiscuity, group size, density, habitat, and diet on the rate of molecular evolution of immunity genes. We began implementing free-ratio models, in which the dN/dS ratio can vary only among branches. The values of dN/dS of the terminal branches of the phylogeny (Fig. 1) were used in the analyses described below. Panels 1–4 in Figure 2 summarize the results obtained in these analyses, which are presented in detail in Tables 1–4.
Table 1.
Phylogenetically independent contrasts of dN/dS and sexual, social and ecological variables across genes.
| Independent variable |
Positive contrasts1 |
Total contrasts |
Sign-test2 | Significance after correction for multiple tests3 |
|
|---|---|---|---|---|---|
| All genes | Habitat | 20 | 44 | P>0.50 | |
| Diet | 23 | 51 | P>0.50 | ||
| Mating system (1) | 41 | 73 | P=0.17 | ||
| Mating system (2) | 43 | 74 | P=0.10# | ||
| Mating system (3) | 39 | 67 | P=0.11 | ||
| Residual testis size | 19 | 36 | P=0.43 | ||
| Group size | 24 | 38 | P=0.07# | ||
| Density | 13 | 27 | P>0.50 | ||
| Non pathogen interacting genes |
Habitat | 8 | 15 | P>0.50 | |
| Diet | 6 | 14 | P>0.50 | ||
| Mating system (1) | 10 | 25 | P>0.50 | ||
| Mating system (2) | 11 | 27 | P>0.50 | ||
| Mating system (3) | 10 | 25 | P>0.50 | ||
| Residual testis size | 3 | 11 | P>0.50 | ||
| Group size | 8 | 10 | P=0.05* | ||
| Density | 7 | 13 | P>0.50 | ||
| Pathogen interacting genes |
Habitat | 12 | 29 | P>0.50 | |
| Diet | 17 | 37 | P>0.50 | ||
| Mating system (1) | 31 | 48 | P=0.03* | ||
| Mating system (2) | 32 | 47 | P=0.01* | * | |
| Mating system (3) | 29 | 42 | P=0.01* | * | |
| Residual testis size | 16 | 25 | P=0.11 | ||
| Group size | 18 | 27 | P=0.06# | ||
| Density | 6 | 14 | P>0.50 |
P<0.1,
P<0.05.
A contrast is positive when both variables vary in the direction predicted by the hypothesis.
One-tailed test.
Using the false discovery rate (Verhoeven et al. 2005) within each class (all genes, non pathogen-interacting genes, pathogen-interacting genes).
Table 4.
Adaptive evolution at immunity genes along lineages of multimale species or species with large groups.
| Mating system partition 1 | Mating system partition 2 | Mating system partition 3 | Group Size | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Gene | UM1 branches |
MM2 branches |
UM1 branches |
MM2 branches |
UM1 branches |
MM2 branches |
SG3 branches |
LG4 branches |
||||||||
| ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ω5>1 | Total | ||
| Non– pathogen- interacting genes |
ANG | 3 | 6 | 2 | 5 | 3 | 5 | 2 | 6 | 2 | 5 | 2 | 5 | 0 | 3 | 3 | 5 |
| CCR5 | 2 | 15 | 0 | 10 | 2 | 13 | 0 | 12 | 2 | 13 | 0 | 10 | 1 | 7 | 1 | 10 | |
| CD4 | 1 | 4 | 3 | 7 | 0 | 3 | 4 | 8 | 0 | 3 | 3 | 7 | 0 | 2 | 3 | 6 | |
| CD45 | 5 | 6 | 2 | 2 | 5 | 6 | 2 | 2 | 5 | 6 | 2 | 2 | 4 | 4 | 1 | 1 | |
| DUFFY | 0 | 10 | 0 | 9 | 0 | 9 | 0 | 10 | 0 | 9 | 0 | 9 | 0 | 5 | 0 | 6 | |
| Pathogen- interacting genes |
APOBEC3G | 3 | 7 | 5 | 6 | 1 | 5 | 7 | 8 | 1 | 5 | 5 | 6 | 2 | 4 | 5 | 6 |
| APOBEC3H | 2 | 4 | 4 | 8 | 1 | 3 | 5 | 9 | 1 | 3 | 4 | 8 | 2 | 3 | 2 | 6 | |
| CAMP | 4 | 7 | 2 | 4 | 4 | 5 | 2 | 6 | 4 | 5 | 2 | 4 | 2 | 4 | 1 | 3 | |
| PKR | 3 | 9 | 4 | 8 | 3 | 7 | 4 | 10 | 3 | 7 | 4 | 8 | 1 | 5 | 4 | 8 | |
| TLR1 | 0 | 7 | 1 | 4 | 0 | 4 | 1 | 7 | 0 | 4 | 1 | 4 | 0 | 4 | 1 | 3 | |
| TLR4 | 2 | 9 | 4 | 9 | 1 | 7 | 5 | 11 | 1 | 7 | 4 | 9 | 2 | 4 | 2 | 6 | |
| TLR5 | 0 | 11 | 1 | 8 | 0 | 9 | 1 | 10 | 0 | 9 | 1 | 8 | 0 | 5 | 0 | 7 | |
| TRIM5 | 5 | 10 | 6 | 8 | 4 | 7 | 7 | 11 | 4 | 7 | 6 | 8 | 5 | 7 | 3 | 6 | |
| TRIM22 | 0 | 11 | 1 | 9 | 0 | 6 | 1 | 14 | 0 | 5 | 1 | 9 | 0 | 7 | 1 | 9 | |
| ZAP | 1 | 7 | 2 | 5 | 1 | 4 | 2 | 8 | 1 | 4 | 2 | 5 | 2 | 4 | 1 | 5 | |
| ALL GENES | 31 | 123 | 37 | 102 | 25 | 93 | 43 | 132 | 24 | 92 | 37 | 102 | 21 | 68 | 28 | 87 | |
| P-value6 | P=0.05* | n.s. | P=0.09# | n.s. | |||||||||||||
| PATHOGEN INTERACTING |
20 | 82 | 30 | 69 | 15 | 57 | 35 | 94 | 15 | 56 | 30 | 69 | 16 | 47 | 20 | 59 | |
| P-value6 | P=0.01* | P=0.12 | P=0.04* | n.s. | |||||||||||||
| Non PATHOGEN INTERACTING |
11 | 41 | 7 | 33 | 10 | 36 | 8 | 38 | 9 | 36 | 7 | 33 | 5 | 21 | 8 | 2 | |
| P-value6 | n.s | n.s | n.s | n.s. | |||||||||||||
P<0.05,
P<0.1.
Unimale.
Multimale.
Below one standard deviation around the mean log group size.
Above one standard deviation around the mean log group size.
ω=dN/dS.
one-tailed Z-test of the difference between the proportions of branches with dN/dS>1 between UM and MM classes or (SG and LG), across the 15 genes.
We first examined the direction of the change in dN/dS in relation to sexual promiscuity using phylogenetically independent contrasts. We used three mating system partitions and RTS as proxies for sexual promiscuity. When summed across genes, the number of positive contrasts in which an increase in promiscuity was accompanied by an increase in dN/dS showed a very slight trend in the predicted direction but was not significant (Sign test, MS1 = P = 0.17, MS2 P = 0.10, MS3 P = 0.11) (Table 1). When we separated the pathogen-interacting (PI) genes (APOBEC3G, APOBEC3H, CAMP, PKR, TLR1, TLR4, TLR5, TRIM5, TRIM22 and ZAP) from the rest (non PI genes), for the three mating system partitions the number of positive contrasts significantly exceeded the null expectation of 50% for the PI genes (Sign test P = 0.03, 0.01 and 0.01, respectively). Of these, mating system partitions 2 and 3 remained significant after correction for multiple tests (FDR). In contrast, none of the measures of sexual promiscuity deviated from the null expectation for the nonpathogen interacting genes (Table 1).
We repeated these analyses with group size, density, diet, and habitat as independent variables. Only group size showed a significant or marginally significant excess of positive contrasts (Table 1), but did not remain significant after FDR correction. These results, summarized in the first panel of Figure 2, suggest that both promiscuity and group size might influence the rate of evolution of immunity genes. Interestingly, mating system was significant only for genes that interact directly with pathogens whereas group size showed a similar trend for both classes of genes.
Next, for each gene, we compared the mean and variance in dN/dS between UM and MM branches and between SG and LG branches. In 10 of the 15 genes (8/10 PI, 2/5 non-PI) we observed a higher mean dN/dS in MM branches than in UM branches in at least one of the three mating system partitions. Similarly, in 9 of the 15 genes (5/10 PI, 4/5 non-PI), LG branches had a higher mean dN/dS than SG branches. Only in a few cases, however, were these differences significant, with four genes showing a weak effect of promiscuity (t-test APOBEC3G P = 0.06, CD45 P = 0.07, PKR P = 0.08, TLR4 P = 0.09) and only one gene showing a significant effect of group size (t-test APOBEC3G P = 0.04) (Table 2, Fig. 2 panel 2).
Table 2.
Mean and variance in dN/dS of immunity genes in relation to mating system and group size.
| Mating system partition 1 | Mating system partition 2 | Mating system partition 3 | Group size | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Gene | UM1 | MM2 | UM1 | MM2 | UM1 | UM1 | SG3 | LG4 | ||||||||
| mean5 | var6 | mean5 | var6 | mean5 | var6 | mean5 | var6 | mean5 | var6 | mean5 | var6 | mean5 | var6 | mean5 | var6 | ||
| Non– | ANG | 1.39 | 1.09 | 0.97 | 0.63 | 1.50 | 1.28 | 0.96 | 0.50 | 1.50 | 1.28 | 0.97 | 0.63 | 0.68 | 0.04 | 1.36 | 1.10* |
| pathogen- | CCR5 | 0.38 | 0.19 | 0.21 | 0.04 | 0.37 | 0.19 | 0.25 | 0.06 | 0.37 | 0.19 | 0.21 | 0.04 | 0.29 | 0.23 | 0.36 | 0.16 |
| interacting | CD4 | 0.71 | 0.32 | 0.72 | 0.11 | 0.48 | 0.17 | 0.80 | 0.15 | 0.48 | 0.17 | 0.72 | 0.11 | 0.70 | 0.00 | 0.78 | 0.19# |
| genes | CD45 | 2.69 | 3.48 | 5.71# | 11.86 | 2.69 | 3.48 | 5.71# | 11.86 | 2.69 | 3.48 | 5.71# | 11.86 | 4.55 | 8.23 | 3.27 | - |
| DUFFY | 0.56 | 0.09 | 0.39 | 0.06 | 0.57 | 0.10 | 0.40 | 0.06 | 0.57 | 0.10 | 0.39 | 0.06 | 0.34 | 0.01 | 0.45 | 0.07* | |
| Pathogen- | APOBEC3G | 1.35 | 1.95 | 1.59 | 0.35 | 0.88 | 0.84 | 1.83# | 1.07 | 0.88 | 0.84 | 1.59 # | 0.35 | 0.87 | 0.21 | 2.04* | 1.28# |
| interacting | APOBEC3H | 0.97 | 0.09 | 1.11 | 0.52# | 0.86 | 0.07 | 1.13 | 0.46 | 0.86 | 0.07 | 1.11 | 0.52 | 1.24 | 0.41 | 0.90 | 0.51 |
| genes | CAMP | 1.47 | 2.21 | 0.88 | 0.29 | 1.88 | 2.54 | 0.73 | 0.20 | 1.88 | 2.54 | 0.88 | 0.29 | 0.93 | 0.41 | 0.65 | 0.16 |
| PKR | 0.79 | 0.51 | 1.42# | 1.14 | 0.95 | 0.54 | 1.18 | 1.15 | 0.95 | 0.54 | 1.42 | 1.14 | 0.90 | 0.19 | 1.33 | 1.33* | |
| TLR1 | 0.50 | 0.04 | 0.65 | 0.20* | 0.57 | 0.06 | 0.55 | 0.11 | 0.57 | 0.06 | 0.65 | 0.20 | 0.43 | 0.02 | 0.65 | 0.30* | |
| TLR4 | 0.64 | 0.43 | 1.18# | 1.17# | 0.54 | 0.45 | 1.14# | 0.99 | 0.54 | 0.45 | 1.11 | 1.08 | 1.27 | 1.74 | 0.69 | 0.23 | |
| TLR5 | 0.43 | 0.02 | 0.74 | 0.65* | 0.44 | 0.02 | 0.67 | 0.53* | 0.44 | 0.02 | 0.74 | 0.65* | 0.35 | 0.02 | 0.49 | 0.08# | |
| TRIM5 | 1.28 | 0.88 | 1.22 | 0.26 | 1.38 | 1.21 | 1.17 | 0.23 | 1.38 | 1.21 | 1.22 | 0.26 | 1.56 | 0.84 | 0.95 | 0.26 | |
| TRIM22 | 0.36 | 0.06 | 0.52 | 0.29 | 0.34 | 0.07 | 0.47 | 0.20 | 0.34 | 0.07 | 0.52 | 0.29 | 0.35 | 0.02 | 0.50 | 0.30* | |
| ZAP | 0.61 | 0.08 | 0.76 | 0.26 | 0.76 | 0.07 | 0.62 | 0.19 | 0.76 | 0.07 | 0.76 | 0.26 | 0.82 | 0.31 | 0.55 | 0.09 | |
P<0.05.
P<0.1.
Shaded cells indicate cases in which the mean or variance in dN/dS are higher in MM or LG species.
Unimale species.
Multimale species.
Small group size.
Large group size.
t-test between average dN/dS of MM branches and average dN/dS of UM branches (1 tail test).
z-test of differences in variance of dN/dS between MM and UM branches (1 tail test).
The analyses based on independent contrasts (Fig. 2 panel 1, Table 1) strongly suggest a link between promiscuity, group size, and molecular evolution of immune genes. In these analyses, however, all the variables were analyzed separately, precluding teasing apart potential correlations among them. In an attempt to disentangle the potentially confounding effects of sexual and social factors on dN/dS, for each gene we fit multiple regression models using dN/dS as the dependent variable and promiscuity (RTS or Mating system partition 3) and group size as independent factors. Table 3 shows the multiple regression models. In five of the 15 genes, variation in promiscuity (RTS or MS3), group size, or both, explain a significant (or close to significant) proportion of the variance in dN/dS (Table 3, Fig. 2 panel 3). In most of these cases group size showed a stronger effect than promiscuity. Also, in some cases the effect (slope) was negative, indicating, contrary to expectations, that for a given level of promiscuity species with smaller group sizes have higher dN/dS.
Table 3.
Relative effects of promiscuity and group size on the rate of evolution of immunity genes.
| Class | Gene | Model 11 fit | Individual factor effects2 | Model 23 fit | Individual factor effects4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 |
P- value4 |
RTS5 | log GS6 |
RTS*log GS7 |
R2 |
P- value4 |
MS38 | log GS6 |
M3*log GS9 |
||
| Non–pathogen- interacting genes |
ANG | 0.52 | 0.07 (#) | − (*) | + (*) | 0.36 | 0.42 | − | + | + | |
| CCR5 | 0.16 | 0.33 | − | + | + | 0.12 | 0.49 | − | + | − | |
| CD4 | 0.24 | 0.67 | − | + | − | 0.02 | 0.99 | + | − | + | |
| CD45 | 0.4 | 0.51 | + | − | + | 0.63 | 0.08 (#) | + | −(#) | ||
| DUFFY | 0.24 | 0.29 | − | + | − | 0.16 | 0.49 | − | + | − | |
| Pathogen- interacting genes |
APOBEC3G | 0.34 | 0.04 (*) | + (*) | 0.58 | 0.03 (*) | + | +(*) | |||
| APOBEC3H | 0.07 | 0.88 | − | + | + | 0.13 | 0.8 | + | − | − | |
| CAMP | 0.53 | 0.34 | − | − | + | 0.23 | 0.75 | − | − | ||
| PKR | 0.16 | 0.53 | + | − | + | 0.11 | 0.73 | + | − | + | |
| TRL1 | 0.35 | 0.42 | − | + | + | 0.22 | 0.77 | + | + | − | |
| TLR4 | 0.23 | 0.35 | + | − | + | 0.46 | 0.03 (*) | + (*) | − (*) | ||
| TLR5 | 0.14 | 0.56 | + | + | + | 0.15 | 0.72 | + | + | + | |
| TRIM5 | 0.06 | 0.85 | + | − | − | 0.09 | 0.82 | − | − | − | |
| TRIM22 | 0.18 | 0.64 | + | − | + | 0.29 | 0.52 | + | − | − | |
| ZAP | 0.31 | 0.05 (*) | + (*) | 0.59 | 0.03 (*) | + | +(*) | ||||
P<0.05,
P<0.10.
Model includes residual testis size and group size.
Contribution of individual factors to the model based on the direction of the slope.
Model includes mating system partition 3 and group size.
Significance of the model.
Residual testis size.
Group size.
Interaction term between RTS and log GS.
Mating system partition 3 (excludes species with ambiguous mating systems).
Interaction term between MS3 and log GS.
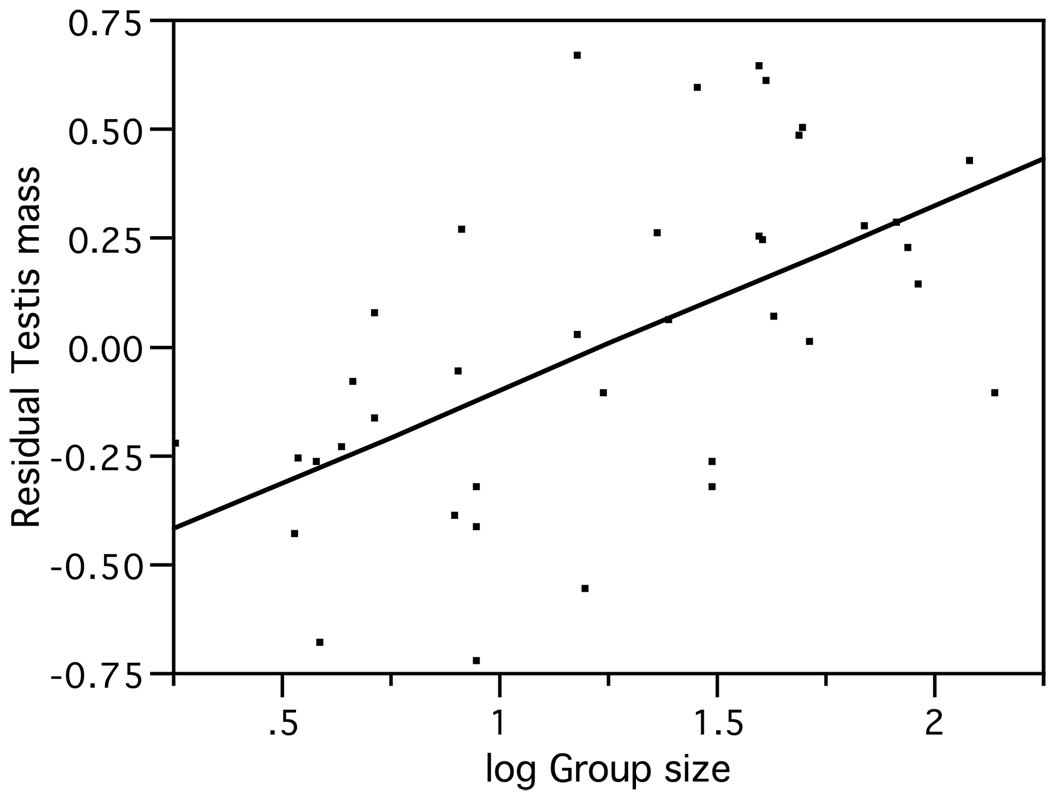
Because RTS and group size show a significant positive correlation across species (P < 0.01) (Fig. 3), we also used the first axis derived from a principal component analysis of RTS and log group size (which captured ~78% of the variation in both variables) as a combined index of promiscuity and sociality. With a couple of exceptions, this resulted in a loss of significance and poorer fit with respect to the multiple regression models (data not shown).
Figure 3.
Positive correlation between residual testis size and log group size (R2 = 0.33, P = 0.0001).
The previous analyses, particularly the independent contrasts, suggest that increases in dN/dS are associated with increases in promiscuity and increases in group size. An increase in dN/dS is suggestive of adaptive evolution, but only a dN/dS>1 constitutes unambiguous evidence of positive selection. Therefore, we compared the relative proportion of branches with dN/dS>1 between UM and MM species and between SG and LG species. When summed across genes, we found a significantly or marginally significantly greater proportion of branches with dN/dS>1 among MM species, than among UM species for two of the three mating system partitions (Table 4). This pattern is driven by the pathogen-interacting genes (Table 4, Fig. 2 panel 4). This indicates that MM species have, on average, more instances of positive selection than UM species and argues for a role of sexual promiscuity in the evolution of immunity genes. On the other hand, the proportion of branches with dN/dS>1 was the same among SG and LG species for the entire dataset as well as for the two groups of genes considered separately (Table 4, Fig. 2 panel 4).
Discussion
The main determinants of rates of protein evolution at immune loci other than adaptive immune receptors are largely unknown. Coevolution between host and pathogens is frequently invoked to explain the rapid evolution of immune loci (Holmes 2004), but certain host features such as promiscuous behavior or sociality might also have an effect by influencing disease risk. Here we investigated whether female mating promiscuity and other social and ecological variables have had a major effect on the rate of molecular evolution in functionally diverse immune defense genes. The underlying hypothesis is that the risk of STD (or more generally infectious diseases) should be higher in species with multiple mating, increasing pathogen exposure and/or diversity, and thus exerting stronger selective pressures on the host.
Using a comparative approach, we found a positive correlation between promiscuity and the rate of protein evolution at these genes across primates (Tables 1 and 4, Fig. 2). We also found a positive correlation between group size and the rate of evolution (Table 1, Fig. 2). The effect is weak, and was only significant when data were combined across genes. However, there are many sources of variation in these analyses, including the noise introduced by trait measurement errors, unknown trait variation within species or over time, incomplete phylogenetic sampling, and the fact that disease risk is likely influenced by other unmeasured variables. Given all the potential sources of variation, it is in fact remarkable to find a signal, suggesting that promiscuity, group size, or some other correlated variable, genuinely affects immune protein evolution.
By controlling for phylogeny, we first found that transitions to higher promiscuity and larger group size were associated with increases in dN/dS (Table 1). In spite of the low statistical power to conduct tests on a gene-by-gene basis, most genes showed the same trend that emerged when we combined genes (data not shown), ruling out the possibility that one or a few outliers are driving the general pattern. Interestingly, we observed more transitions to higher dN/dS associated with higher promiscuity in the group of genes that directly interact with pathogens, such as the antiretroviral genes and the pattern recognition receptors. Genes that lie at the host-pathogen interface might exhibit more evidence of selection due to coevolutionary arms races with pathogens. The increase in dN/dS associated with large groups, on the other hand, was not restricted to pathogen-interacting genes, but instead was distributed across the entire set of immunity genes.
The trends that emerged when comparing the mean dN/dS among species with low and high promiscuity or small and large groups, or when regressing dN/dS against the range of promiscuity and group size, were generally consistent with the independent contrasts but were largely not significant (Tables 2 and 3). Higher mean and variance in dN/dS were usually associated with more promiscuous mating systems or species with larger groups, as expected if more contacts increase the opportunities for disease transmission. However, the results of the multiple regression models showed that, at least in some cases, promiscuity and group size might have different effects on the rate of molecular evolution (Table 3). Consistent with this, dN/dS was not significantly correlated with the principal component that represents a combined index of promiscuity and sociality. Thus, in spite of being positively correlated at a large taxonomic scale, mating system and group size might influence dN/dS independently and sometimes in opposite ways.
An increase in dN/dS is suggestive of positive selection but might also reflect relaxed constraint. A more stringent analysis based on branches with unambiguous evidence of selection (dN/dS>1), also revealed more adaptive evolution in more promiscuous species, but not in species with larger groups (Table 4).
In spite of the overall pattern reported, a high degree of heterogeneity in dN/dS is evident from the branch-based analysis (Fig. 1). This heterogeneity is not restricted to the promiscuous or large group branches but instead is distributed across the different gene phylogenies, and indicates that other lineage-specific factors might have similar importance. In light of such a high degree of heterogeneity, the comparison that focused on the proportion of branches with dN/dS>1 was more informative about the relative potential for natural selection. Similarly, the sign-test of positive contrasts (which focuses on the direction of the change but not the magnitude) resulted in more statistical power to expose the relationship between dN/dS, promiscuity and group size. Taken together, these two analyses suggest that higher levels of promiscuity and larger group size might underlie an increase in the rate of protein evolution. Nevertheless, only for promiscuity does this seem to be due to positive selection.
Interestingly, in spite of underlying differences in leukocyte levels in humans, the DUFFY gene did not exhibit patterns of substitution between species consistent with differences in promiscuity. At least three explanations can account for this result. First, it is possible that regulatory rather than coding variation at DUFFY is responsible for leukocyte differences between primates. In fact, in humans, that seems to be the case (Nalls et al. 2008; Reich et al. 2009). Second, leukocyte levels might be under the genetic control of loci other than DUFFY. Third, the pattern originally reported by Nunn et al. (2000) might not reflect evolved differences in leukocyte levels between species but instead might be caused by some other difference among promiscuous and monogamous primates in captivity, such as density, sex ratio, or stress, as pointed out by Read and Allen (2000).
A potential problem with the interpretation of differences in evolutionary rates among species in the context of adaptation is that relaxation of purifying selection due to reduction in population size can also affect dN/dS (Ohta 1993b). In smaller populations, selection is less efficient at removing deleterious mutations, which should result in an increase in the rate of fixation of nonsynonymous changes, and a concomitant increase in dN/dS (Ohta 1993b). For example, primates have a higher dN/dS and lower effective population size than rodents (Ohta 1993a; Hughes and Friedman 2009). If promiscuity or group size is correlated with effective population size in primates, this might result in a spurious correlation between these variables and dN/dS.
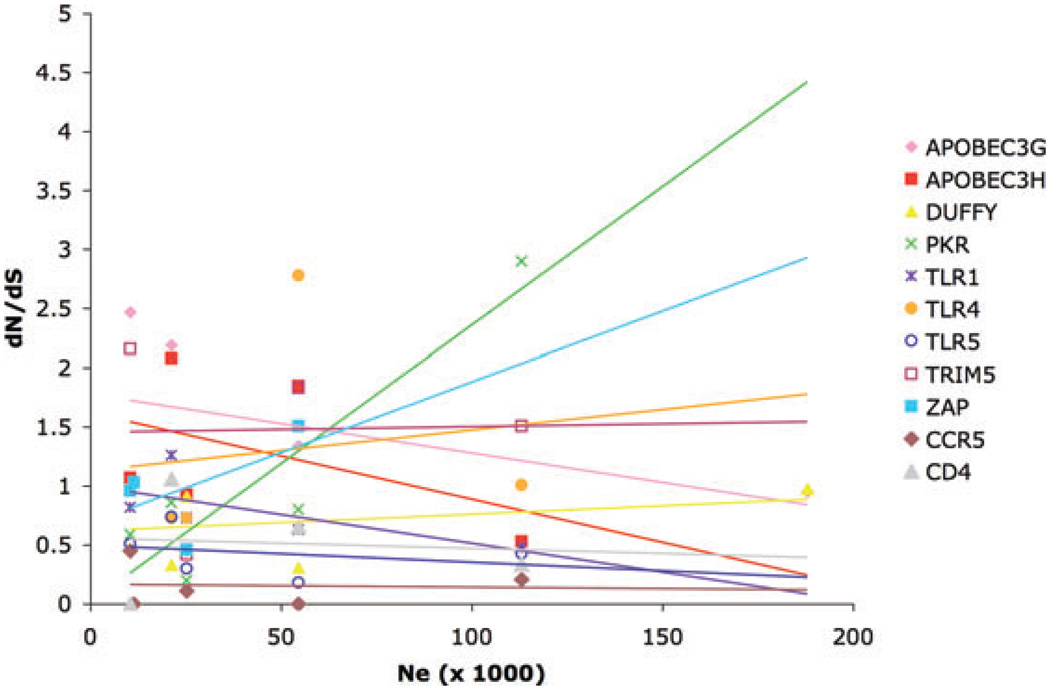
A few lines of evidence seem to argue against this possibility for the data presented here. For a few species of primates, estimates of effective population sizes (Ne) are available (human: Yu et al. 2004; chimpanzee: Yu et al. 2004; bonobo: Yu et al. 2004; Won and Hey 2005; gorilla: Yu et al. 2004; rhesus and cynomolgus macaques: Stevison and Kohn 2009). Additionally, for the orangutan, we estimated Ne based on available estimates of polymorphism and divergence (Fischer et al. 2006). We calculated the neutral mutation rate as μ = Da/2t (Kimura 1983) where Da is the net sequence divergence and t is the divergence time between the two species compared. We used a net sequence divergence of 2.96% between orangutans and humans, obtained by subtracting the average of the human and orangutan nucleotide diversity (π) from the raw sequence divergence between the species, and a divergence time of 13.5 MY (Goodman et al. 1998). Assuming a generation time of 15 years we obtained a mutation rate per site per generation of 1.65 × 10−8. Then, using the nucleotide diversity estimated by Fischer et al. (2006) of 0.36%, we calculated Ne as π/4μ and obtained an effective population size of 54,545. These few species of apes, human, and macaques do not show any consistent relation between population size and dN/dS in the 11 genes for which at least four of these species were included (Fig. 4). Similarly, no consistent pattern has been found in the rate of molecular evolution of social and nonsocial insects (Schmitz and Moritz 1998; Bromham and Leys 2005). Finally, as mentioned above, dN/dS values greater than one are only expected under positive selection. Our analyses based on branches with dN/dS>1 should therefore reflect patterns of adaptation and not simply relaxation of constraint.
Figure 4.
Relationship between population size and dN/dS for a sample of primates that includes human, apes, and macaques. Effective population sizes were taken from the literature or calculated based on multilocus polymorphism and divergence estimates (see the section “Discussion”). Only genes with a minimum of four species with available dN/dS values were included. Regression lines are shown. None of these were significant in the expected direction (P > 0.05 for all genes except for PKR). PKR showed a positive correlation between Ne and dN/dS (R2 = 0.84, P = 0.03).
However, it is still possible that at the larger scale of the primate radiation the accumulation of slightly deleterious substitutions in species with smaller population sizes has contributed to some extent to the pattern. If more social primates tend to have on average lower effective population sizes (as has been proposed for social insects; Crozier 1979), this might offer an explanation for the weaker effect of group size. The fact that in the analysis of independent contrasts, species with larger groups (a proxy for more social species) had higher dN/dS at both the pathogen-interacting and nonpathogen-interacting genes is consistent with this idea, because population size is expected to affect all genes equally. Also in line with this hypothesis, none of the genes or groups of genes showed an excess of branches with dN/dS>1 among the large group species, indicating that positive selection is not necessarily more prevalent in large group species. Thus, it is plausible that overall acceleration in dN/dS in large group species is due to relaxed purifying selection along these branches.
Many theoretical and empirical studies have suggested strong connections between social organization and the spread of horizontally transmitted parasites (e.g., Cote and Poulin 1995; reviewed in Altizer et al. 2003). In primates, somewhat contradictory results have been obtained when correlating direct and indirect measures of disease risk and sociality defined in a broad sense (components of mating and social systems). For example, in spite of the positive relationship between white blood cells and sexual promiscuity, spleen mass, another surrogate measure of disease risk, was not associated with measures of sociality or promiscuity (Nunn 2002a). Similarly, Nunn (2003) did not find support for the hypothesis that behaviors expected to reduce STD transmission are correlated with promiscuity. On the other hand, sociality measured as group size accounts for helminth diversity (Vitone et al. 2004), but population density (another measure of social contact) is the main predictor of parasite species richness in primates, including all the main classes of parasites (Nunn et al. 2003). Neither the previously mentioned studies nor ours found a strong effect of population density, although in some cases, the incorporation of density in our multiple regression models significantly improved the fit (data not shown). The integration of all these results, however, is not straightforward because different aspects of immune defense might be characterized by different trade-offs and constraints.
Using the rate of molecular evolution at immunity genes as a surrogate of disease risk, our comparative data on 15 primate defense genes provide support for the idea that female promiscuity increases the potential for natural selection to act on the immune system. The detected effect of promiscuity, to the exclusion of group size and density, is consistent with the idea that STDs might be important drivers of this pattern. This is an intriguing result, because even if they are expected to interact with sexually transmitted pathogens or participate in pathways that lead to their clearance, the genes included in this study are not specifically involved in immunity against STDs. Recently compiled information of primate parasites show that STDs are common in nonhuman primates and the documented STDs appear to be more frequent in promiscuous species (Nunn and Altizer 2006). Moreover, most of the known sexually transmitted pathogens in nonhuman primates are viruses, and among viruses, those transmitted by close contact (sexual or nonsexual) exhibit higher levels of host specificity (Pedersen et al. 2005). In light of the close relationship with their hosts, it is possible that sexually transmitted pathogens engage more often in arms races with their hosts than pathogens with other transmission modes.
The hypotheses tested here are not mutually exclusive, and the variables studied as well as other potentially confounding variables could interact in complicated ways. Importantly, focusing on the opportunities for disease transmission facilitated by social structure is only one of the possible theoretical frameworks in which to cast this problem. Another equally valid approach would be to study how social behavior is shaped by disease risk over evolutionary time or as a plastic response. Our results provide another interesting piece of information linking promiscuity, STDs and the evolution of the immune system, but this complex relationship is far from being understood. Even if sexual promiscuity causally underlies the pattern of evolution of some immunity genes, a large portion of the variance in dN/dS remains unexplained and suggests that the biological details of host-pathogen interactions in particular lineages play a large role in determining rates of evolution of immunity genes.
Supplementary Material
The following supporting information is available for this article:
Appendix S1. Sequence accession numbers, gene information and primate variables used in this study.
Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
ACKNOWLEDGMENTS
We are grateful to A. Geraldes, M. Carneiro, M. Dean, J. Good, T. Salcedo, M. Sans-Fuentes, P. Campbell, M. Phifer-Rixey, A. Bjork, M. Hammer, N. Moran, D. Vercelli and M. Worobey for discussions and comments. We especially thank Dr. W. Switzer, The Southwest National Primate Research Center, and The Smithsonian Institution for providing DNA or tissue samples. This work was supported by an NIH grant to MWN.
LITERATURE CITED
- Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, Cunningham AA, Dobson AP, Ezenwa V, Jones KE, Pedersen AB, et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Ann. Rev. Ecol. Evol. Syst. 2003;34:517–547. [Google Scholar]
- Anderson MJ, Hessel JK, Dixson AF. Primate mating systems and the evolution of immune response. J. Reprod. Immunol. 2004;61:31–38. doi: 10.1016/j.jri.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Bromham L, Leys R. Sociality and the rate of molecular evolution. Mol. Biol. Evol. 2005;22:1393–1402. doi: 10.1093/molbev/msi133. [DOI] [PubMed] [Google Scholar]
- Com E, Bourgeon F, Evrard B, Ganz T, Colleu D, Jegou B, Pineau C. Expression of antimicrobial defensins in the male reproductive tract of rats, mice, and humans. Biol. Reprod. 2003;68:95–104. doi: 10.1095/biolreprod.102.005389. [DOI] [PubMed] [Google Scholar]
- Cote IM, Poulin R. Parasitism and group size in social animals—a metaanalysis. Behav. Ecol. 1995;6:159–165. [Google Scholar]
- Crozier R. Genetics of Sociality. In: Hermann HR, editor. Social Insects. New York: Academic Press; 1979. pp. 223–286. [Google Scholar]
- Dunn FL. The parasites of Saimiri: in the context of platyrrhine parasitism. In: Rosenblum LA, Cooper RW, editors. The Squirrel Monkey. New York: Academic Press; 1968. pp. 31–68. [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorka KM, Zuk M. Sexual conflict and female immune suppression in the cricket, Allonemobious socius. J. Evol. Biol. 2005;18:1515–1522. doi: 10.1111/j.1420-9101.2005.00942.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. [Google Scholar]
- Filip LC, Mundy NL. Rapid evolution by positive Darwinian selection in the extracellular domain of the abundant lymphocyte protein CD45 in primates. Mol. Biol. Evol. 2004;21:1504–1511. doi: 10.1093/molbev/msh111. [DOI] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Gustafsson L, Nordling D, Andersson MS, Sheldon BC, Qvarnstrom A. Infectious diseases, reproductive effort and the cost of reproduction in birds. Philos. Trans. R. Soc. B-Biol. Sci. 1994;346:323–331. doi: 10.1098/rstb.1994.0149. [DOI] [PubMed] [Google Scholar]
- Hamilton WD, Zuk M. Heritable true fitness and bright birds—a role for parasites. Science. 1982;218:384–387. doi: 10.1126/science.7123238. [DOI] [PubMed] [Google Scholar]
- Harcourt AH. Sperm competition and the evolution of nonfertilizing sperm in mammals. Evolution. 1991;45:314–328. doi: 10.1111/j.1558-5646.1991.tb04406.x. [DOI] [PubMed] [Google Scholar]
- Harcourt AH, Purvis A, Liles L. Sperm competition—mating system, not breeding season, affects testes of primates. Funct. Ecol. 1995;9:468–476. [Google Scholar]
- Holmes EC. Adaptation and immunity. PLoS Biol. 2004;2:e307. doi: 10.1371/journal.pbio.0020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. More radical amino acid replacements in primates than in rodents: support for the evolutionary role of effective population size. Gene. 2009;440:50–56. doi: 10.1016/j.gene.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immerman RS. Sexually transmitted disease and human evolution: survival of the ugliest? Human Ethology Newsletter. 1986;4:6–7. [Google Scholar]
- Kenagy GJ, Trombulak SC. Size and function of mammalian testes in relation to body size. J. Mammal. 1986;67:1–22. [Google Scholar]
- Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, United Kingdom: Cambridge Univ. Press; 1983. [Google Scholar]
- Kokko H, Ranta E, Ruxton G, Lundberg P. Sexually transmitted disease and the evolution of mating systems. Evolution. 2002;56:1091–1100. doi: 10.1111/j.0014-3820.2002.tb01423.x. [DOI] [PubMed] [Google Scholar]
- Lawniczak MKN, Begun DJ. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. [DOI] [PubMed] [Google Scholar]
- Lawniczak MKN, Barnes AI, Linklater JR, Boone JM, Wigby S, Chapman T. Mating and immunity in invertebrates. Trends Ecol. Evol. 2007;22:48–55. doi: 10.1016/j.tree.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Li P, Chan HC, He B, So SC, Chung YW, Shang Q, Zhang YD, Zhang YL. An antimicrobial peptide gene found in the male reproductive system of rats. Science. 2001;291:1783–1785. doi: 10.1126/science.1056545. [DOI] [PubMed] [Google Scholar]
- Lindenfors P, Tullberg BS. Phylogenetic analyses of primate size evolution: the consequences of sexual selection. Biol. J. Linn. Soc. 1998;64:413–447. [Google Scholar]
- Lochmiller RL, Deerenberg C. Trade-offs in evolutionary immunology: just what is the cost of immunity? Oikos. 2000;88:87–98. [Google Scholar]
- Lockhart AB, Thrall PH, Antonovics J. Sexually transmitted diseases in animals: ecological and evolutionary implications. Biol. Rev. Camb. Philos. Soc. 1996;71:415–471. doi: 10.1111/j.1469-185x.1996.tb01281.x. [DOI] [PubMed] [Google Scholar]
- Loehle C. Social barriers to pathogen transmission in wild animal populations. Ecology. 1995;76:326–335. [Google Scholar]
- Lung O, Kuo L, Wolfner MF. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J. Insect Physiol. 2001;47:617–622. doi: 10.1016/s0022-1910(00)00151-7. [DOI] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Moore J. Parasites and the Behavior of Animals. Oxford: Oxford Univ. Press; 2002. [Google Scholar]
- Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, Huntsman S, Garcia M, Hu DL, Li RL, Beamer BA, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the health ABC and Jackson Heart Studies. Am. J. Hum. Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn CL. A comparative study of leukocyte counts and disease risk in primates. Evolution. 2002a;56:177–190. doi: 10.1111/j.0014-3820.2002.tb00859.x. [DOI] [PubMed] [Google Scholar]
- Nunn CL. Spleen size, disease risk and sexual selection: a comparative study in primates. Evol. Ecol. Res. 2002b;4:91–107. [Google Scholar]
- Nunn CL. Behavioural defenses against sexually transmitted diseases in primates. Anim. Behav. 2003;66:37–48. [Google Scholar]
- Nunn CL, Altizer SM. Behavior Ecology and Evolution. New York: Oxford Univ. Press; 2006. Infectious Diseases in Primates. [Google Scholar]
- Nunn CL, Gittleman JL, Antonovics J. Promiscuity and the primate immune system. Science. 2000;290:1168–1170. doi: 10.1126/science.290.5494.1168. [DOI] [PubMed] [Google Scholar]
- Nunn CL, Altizer S, Jones KE, Sechrest W. Comparative tests of parasite species richness in primates. Am. Nat. 2003;162:597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Amino-acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc. Natl Acad. Sci USA. 1993a;90:4548–4551. doi: 10.1073/pnas.90.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. An examination of the generation-time effect on molecular evolution. Proc. Natl. Acad. Sci. USA. 1993b;90:10676–10680. doi: 10.1073/pnas.90.22.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio DS, Antunes A, Ramos MJ. Structural and functional implications of positive selection at the primate angiogenin gene. BMC Evol. Biol. 2007;7:167. doi: 10.1186/1471-2148-7-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AB, Altizer S, Poss M, Cunningham A, Nunn CL. Petterns of host specificity and transmission among parasites of wild primates. Int. J. Parasitol. 2005;35:647–657. doi: 10.1016/j.ijpara.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Purvis A. A Composite Estimate of Primate Phylogeny. Phil. Trans. R. Soc. B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrasts (CAIC)—an Apple–Macintosh application for analyzing comparative data. Comput. Appl. Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Read AF, Allen JE. Evolution and immunology—the economics of immunity. Science. 2000;290:1104–1105. doi: 10.1126/science.290.5494.1104. [DOI] [PubMed] [Google Scholar]
- Reddy KVR, Yedery RD, Aranha C. Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Reich D, Nalls MA, Kao WHL, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, et al. Reduced Neutrophil Count in People of African Descent Is Due To a Regulatory Variant in the Duffy Antigen Receptor for Chemokines Gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe N. The Pictorial Guide of Living Primates. East Hampton, NY: Pogonias Press; 1996. [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Path. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5 alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. Variation in immune defence as a question of evolutionary ecology. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:357–366. doi: 10.1098/rspb.2002.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Moritz RFA. Sociality and the rate of rDNA sequence evolution in wasps (Vespidae) and honeybees (Apis) J. Mol. Evol. 1998;47:606–612. doi: 10.1007/pl00006417. [DOI] [PubMed] [Google Scholar]
- Semple S, Cowlishaw G, Bennett PM. Immune system evolution among anthropoid primates: parasites, injuries and predators. Proc. R. Soc. Lond. B. 2002;269:1031–1037. doi: 10.1098/rspb.2001.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon BC, Verhulst S. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 1996;11:317–321. doi: 10.1016/0169-5347(96)10039-2. [DOI] [PubMed] [Google Scholar]
- Silphaduang U, Hincke MT, Nys Y, Mine Y. Antimicrobial proteins in chicken reproductive system. Biochem. Biophys. Res. Commun. 2006;340:648–655. doi: 10.1016/j.bbrc.2005.12.054. [DOI] [PubMed] [Google Scholar]
- Stevison LS, Kohn MH. Divergence population genetic analysis of hybridization between rhesus and cynomolgus macaques. Mol. Ecol. 2009;18:2457–2475. doi: 10.1111/j.1365-294X.2009.04212.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer; 2000. [Google Scholar]
- Thrall PH, Antonovics J, Bever JD. Sexual transmission of disease and host mating systems: within-season reproductive success. Am. Nat. 1997;149:485–506. [Google Scholar]
- Verhoeven KJF, Simonsen KL, McIntyre LM. Implementing false discovery rate control: increasing your power. Oikos. 2005;108:643–647. [Google Scholar]
- Vitone ND, Altizer S, Nunn CL. Body size, diet and sociality influence the species richness of parasitic worms in anthropoid primates. Evol. Ecol. Res. 2004;6:183–199. [Google Scholar]
- Wlasiuk G, Khan S, Switzer WM, Nachman MW. A history of recurrent positive selection at the toll-like receptor 5 in primates. Mol. Biol. Evol. 2009;26:937–949. doi: 10.1093/molbev/msp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol. Biol. Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang ZH. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH. Nucleotide diversity in gorillas. Genetics. 2004;166:1375–1383. doi: 10.1534/genetics.166.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezetsky I, Pontillo A, Puzzi L, Antcheva N, Segat L, Pacor S, Crovella S, Tossi A. Evolution of the primate cathelicidin—correlation between structural variations and antimicrobial activity. J. Biol. Chem. 2006;281:19861–19871. doi: 10.1074/jbc.M511108200. [DOI] [PubMed] [Google Scholar]
- Zhang ZD, Weinstock G, Gerstein M. Rapid evolution by positive Darwinian selection in t-cell antigen CD4 in primates. J. Mol. Evol. 2008;66:446–456. doi: 10.1007/s00239-008-9097-1. [DOI] [PubMed] [Google Scholar]
- Zuk M, Stoehr AM. Immune defense and host life history. Am. Nat. 2002;160:S9–S22. doi: 10.1086/342131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supporting information is available for this article:
Appendix S1. Sequence accession numbers, gene information and primate variables used in this study.
Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.