Abstract
The plant hormone abscisic acid (ABA) regulates many key processes in plants, including seed germination and development and abiotic stress tolerance, particularly drought resistance. Understanding early events in ABA signal transduction has been a major goal of plant research. The recent identification of the PYRABACTIN (4-bromo-N-[pyridin-2-yl methyl]naphthalene-1-sulfonamide) RESISTANCE (PYR)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) family of ABA receptors and their biochemical mode of action represents a major breakthrough in the field. The solving of PYR/RCAR structures provides a context for resolving mechanisms mediating ABA control of protein–protein interactions for downstream signaling. Recent studies show that a pathway based on PYR/RCAR ABA receptors, PROTEIN PHOSPHATASE 2Cs (PP2Cs), and SNF1-RELATED PROTEIN KINASE 2s (SnRK2s) forms the primary basis of an early ABA signaling module. This pathway interfaces with ion channels, transcription factors, and other targets, thus providing a mechanistic connection between the phytohormone and ABA-induced responses. This emerging PYR/RCAR–PP2C–SnRK2 model of ABA signal transduction is reviewed here, and provides an opportunity for testing novel hypotheses concerning ABA signaling. We address newly emerging questions, including the potential roles of different PYR/RCAR isoforms, and the significance of ABA-induced versus constitutive PYR/RCAR–PP2C interactions. We also consider how the PYR/RCAR–PP2C–SnRK2 pathway interfaces with ABA-dependent gene expression, ion channel regulation, and control of small molecule signaling. These exciting developments provide researchers with a framework through which early ABA signaling can be understood, and allow novel questions about the hormone response pathway and possible applications in stress resistance engineering of plants to be addressed.
Keywords: Signal transduction, PYR1, RCAR, stomata, ABA receptor, phytohormone
Drought is one of the major abiotic stresses affecting plants; >50% of the Earth's surface area, including the vast majority of agricultural lands, is vulnerable to drought (Kogan 1997). Drought-induced crop losses have a significant economic impact, which is predicted to increase with global climate change (Marris 2008; Battisti and Naylor 2009). The phytohormone abscisic acid (ABA) is the central regulator of abiotic stress resistance in plants, and coordinates a complex regulatory network enabling plants to cope with decreased water availability (Cutler et al. 2010; Kim et al. 2010). Plant ABA content significantly increases under drought or salinity stress conditions, stimulating stomatal closure, changes in gene expression, and the accumulation of osmo-compatible solutes, thus increasing the plant's capacity to cope with stress conditions (Seki et al. 2007; Cutler et al. 2010; Kim et al. 2010). ABA also plays important roles during plant development, including embryo and seed development, and the promotion of seed dormancy (Finkelstein et al. 2008). Given the importance of ABA to plant physiology and development, understanding the signal transduction processes linking the hormone to target responses is a focus of abiotic stress research.
While many intermediate signaling components have been extensively characterized, our understanding of ABA signaling has been hampered by the lack of knowledge regarding the ABA receptor(s). The recent identification of PYRABACTIN (4-bromo-N-[pyridin-2-yl methyl]naphthalene-1-sulfonamide) RESISTANCE (PYR)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) proteins (Ma et al. 2009; Park et al. 2009) provided the field with exciting new avenues of research into ABA perception, allowing existing hypotheses to be tested and novel ideas concerning ABA signaling to be generated. This review summarizes the many new findings in early ABA signaling, and also highlights and discusses emerging questions about the signaling network.
The central ABA signaling module
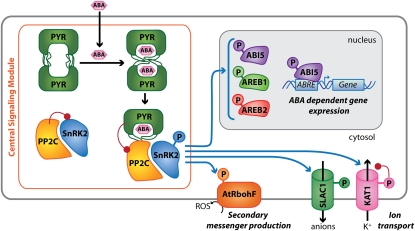
Much recent progress into ABA signal transduction indicates that the earliest events of the signaling pathway occur through a central signaling module made up of three protein classes: PYR/RCARs, Protein Phosphatase 2Cs (PP2Cs), and SNF1-related protein kinase 2s (SnRK2s) (Fig. 1). In this model, the PYR/RCARs act as ABA receptors, the PP2Cs act as negative regulators of the pathway, and SnRK2s act as positive regulators of downstream signaling (Ma et al. 2009; Park et al. 2009). A double-negative regulatory pathway is established, whereby ABA-bound PYR/RCARs inhibit PP2C activity, and PP2Cs inactivate SnRK2s (Park et al. 2009; Umezawa et al. 2009; Vlad et al. 2009). Thus, in the absence of ABA, the PP2Cs are active and repress SnRK2 activity and downstream signaling. In the presence of ABA, PYR/RCARs interact with PP2Cs and inhibit phosphatase activity, allowing SnRK2 activation and phosphorylation of target proteins (Fig. 1). This newly developed signaling model forms the basis for addressing key discussion questions below, but should continue to be actively investigated.
Figure 1.
The core ABA signaling pathway. Recent progress in understanding early ABA signal transduction has led to the construction of a PYR/RCAR–PP2C–SnRK2 signal transduction model. In the absence of ABA, PP2Cs inhibit protein kinase (SnRK2) activity through removal of activating phosphates. ABA is bound by intracellular PYR/PYL dimers, which dissociate to form ABA receptor–PP2C complexes. Complex formation therefore inhibits the activity of the PP2C in an ABA-dependent manner, allowing activation of SnRK2s. Several SnRK2 targets have been identified both at the plasma membrane and in the nucleus, resulting in control of ion channels, secondary messenger production, and gene expression. Red connections on left indicate an inhibitory interaction.
Identification of ABA receptors
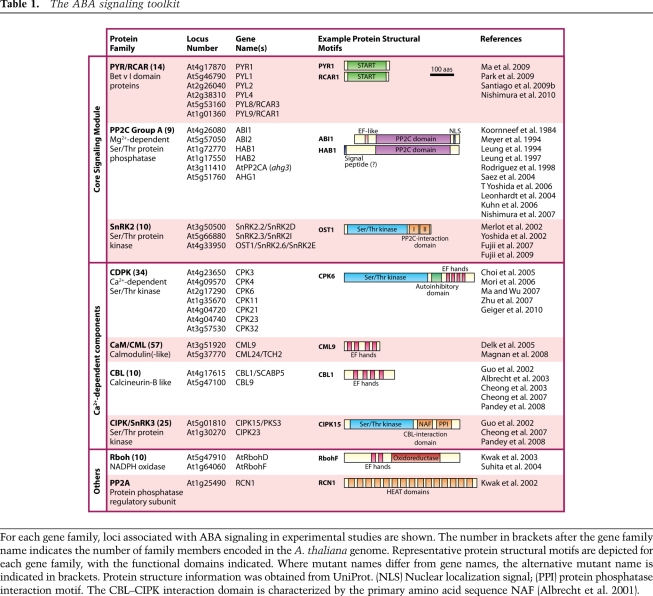
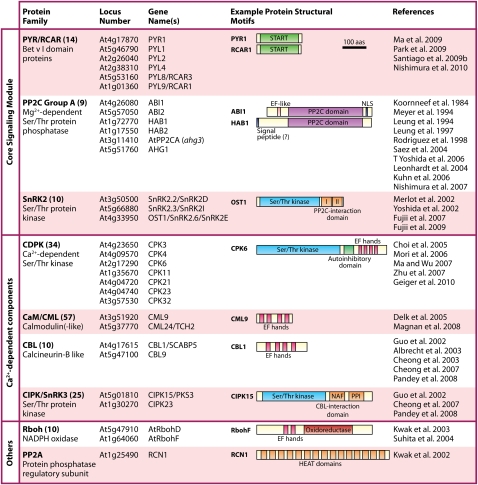
The first stage of hormone signaling must be the specific recognition of the hormone by a receptor. Recent studies identified and confirmed the PYR/RCAR proteins as ABA-binding proteins that interact with the PP2Cs, which function as negative regulators of ABA signaling (Ma et al. 2009; Park et al. 2009). Six of the nine PP2C proteins in the clade containing ABI1 have been shown to be negative regulators of ABA responses (see Table 1; Koornneef et al. 1984; Leung et al. 1994, 1997; Meyer et al. 1994; Rodriguez et al. 1998; Leonhardt et al. 2004; Saez et al. 2004; Kuhn et al. 2006; T Yoshida et al. 2006; Nishimura et al. 2007). Combined cell signaling and genetic analyses provided early evidence that the PP2C ABA-INSENSITIVE 1 (ABI1) functions very early in the ABA signaling pathway, upstream of all known rapid signaling responses—including anion channel activation (Pei et al. 1997), increases in cytosolic free calcium concentration ([Ca2+]cyt) (Allen et al. 1999), activation of the SnRK2 OPEN STOMATA 1 (OST1) (Mustilli et al. 2002), Ca2+ channel activation, and reactive oxygen species (ROS) production (Murata et al. 2001). Therefore, identifying proteins that interact with PP2Cs was one strategy used to isolate novel components of early ABA signaling and identify the PYR/RCARs (Ma et al. 2009; Santiago et al. 2009b; Nishimura et al. 2010). RCAR1/PYR1-LIKE 9 (PYL9) was identified in a yeast two-hybrid screen using the PP2C ABI2 as bait (Ma et al. 2009), and a similar strategy using HOMOLOGY TO ABI1 (HAB1) as bait identified PYL5, PYL6, and PYL8 (Santiago et al. 2009b). Independently, an in vivo strategy of ABI1 complex purification from Arabidopsis plants led to the identification of nine of the 14 PYR/RCARs as the major in planta interactors of ABI1 (Nishimura et al. 2010). In an alternative approach, chemical genetics identified mutations in the PYR1 gene based on insensitivity to the synthetic ABA agonist pyrabactin (Park et al. 2009). These multiple independent lines of evidence indicated that the previously uncharacterized PYR/RCAR proteins are major early ABA signaling components. The Arabidopsis thaliana genome encodes 14 PYR/RCAR proteins that are highly conserved at the amino acid sequence level (Table 1). PYR/RCARs are small soluble proteins belonging to the START/Bet v I superfamily that contain a central hydrophobic ligand-binding pocket (Iyer et al. 2001). The identification of this new class of ABA signaling proteins has resulted in great excitement within the plant hormone signaling field, providing new avenues of research into ABA signal transduction.
Table 1.
The ABA signaling toolkit
For each gene family, loci associated with ABA signaling in experimental studies are shown. The number in brackets after the gene family name indicates the number of family members encoded in the A. thaliana genome. Representative protein structural motifs are depicted for each gene family, with the functional domains indicated. Where mutant names differ from gene names, the alternative mutant name is indicated in brackets. Protein structure information was obtained from UniProt. (NLS) Nuclear localization signal; (PPI) protein phosphatase interaction motif. The CBL–CIPK interaction domain is characterized by the primary amino acid sequence NAF (Albrecht et al. 2001).
While the case for PYR/RCARs acting in ABA signaling is strong, this does not strictly exclude the possibility that other ABA receptors exist (for detailed discussion, see Klingler et al. 2010). Two other unrelated ABA receptors have been proposed:ChlH/GUN5 and GTG1/GTG2. ChlH/GUN5 was identified through homology with an ABA-binding protein from Vicia faba (Zhang et al. 2002; Shen et al. 2006), and overexpression of either the full-length protein (Shen et al. 2006) or the C-terminal half of the protein was reported to confer ABA hypersensitivity (Wu et al. 2009). However, the homologous ChlH/GUN5 protein in barley did not bind ABA (Müller and Hansson 2009), and further analyses are required to uncover the significance of this protein class in ABA signaling (Wasilewska et al. 2008). GTG1 and GTG2 are membrane proteins with homology with noncanonical G protein-coupled receptors (GPCRs) with nine transmembrane domains that hydrolyze GTP (Pandey et al. 2009). Double gtg1gtg2 mutants retain an ABA response, but have a partially reduced sensitivity to ABA at the level of seed germination and stomatal responses, consistent with the existence of alternative ABA perception pathways (i.e., by PYR/RCARs). A proposed GPCR (GCR2) was also proposed to act as an ABA receptor, but this has been disputed (e.g., Guo et al. 2008), and so will not be discussed further here.
Capturing the message—ABA binding and interactions with PP2Cs
The strategies described above showed that PYR/RCARs acted with PP2Cs to confer ABA-induced inhibition of PP2C activity in vitro. Next, it was critical to determine if PYR/RCAR proteins bind ABA directly, and thus act as receptors. Initial evidence for ABA binding of PYR1 was obtained through heteronuclear single quantum coherence nuclear magnetic resonance studies (Park et al. 2009) and isothermal titration calorimetry analyses (Ma et al. 2009), but whether PYR/RCARs and PP2Cs functioned together as ABA coreceptors remained unknown. Direct ABA binding to PYR/PYLs was subsequently established through the elucidation of PYR1, PYL1, and PYL2 crystal structures in the presence of ABA (Melcher et al. 2009; Miyazono et al. 2009; Nishimura et al. 2009; Santiago et al. 2009a; Yin et al. 2009). The ligand-binding site of PYR/RCAR proteins lies within a large internal cavity. The majority of protein interactions with the ABA molecule are through nonpolar contacts; however, the ring carbonyl, central hydroxyl, and carboxylic acid groups of ABA are held in place through water-mediated hydrogen bonds (Melcher et al. 2009; Miyazono et al. 2009; Nishimura et al. 2009; Santiago et al. 2009a; Yin et al. 2009). The carboxylate forms a buried salt bridge with an inward-facing lysine side chain amine.
The nonnaturally occurring ABA enantiomer R-(−)-ABA has biological activity (Milborrow 1974), in contrast to many other signaling molecules that are highly stereospecific. ABA is a near-symmetrical molecule, with rotation around the chiral center resulting in a swap of the relative positions of the mono- and dimethyl groups on the ring (Milborrow 1974; Cutler et al. 2010). The crystal structure of PYR1 bound to both the S-(+)-ABA and R-(−)-ABA stereoisomers revealed that the chirality difference is accommodated within the ABA receptor-binding pocket by the flipping of the ABA ring by ∼180° (Nishimura et al. 2009). The ability to bind alternative stereoisomers may not translate to equal biological activity in all PYR/RCAR family members; PYR1 interactions with the HAB1 PP2C are promoted by S-(+)-ABA but not R-(−)-ABA in yeast two-hybrid interaction tests (Park et al. 2009). It is worth noting that neither ChlH/GUN5 nor GTG1/GTG2 are able to bind the nonnatural but biologically active R-(−)-ABA isomer (Shen et al. 2006; Pandey et al. 2009), and therefore cannot account for R-(−)-ABA-induced responses. The relative stereo specificity of different PYR/RCAR proteins and their ability to bind alternative ligands could be used to explore the link between biochemical activities and physiological responses, and may also provide useful tools to manipulate ABA signaling in both an experimental and agronomical context.
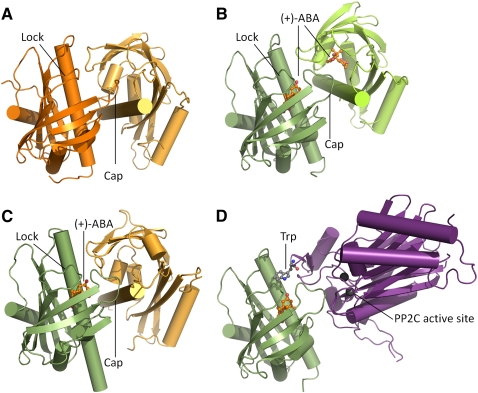
Crystal structures of PYR1/PYL1/PYL2 in the absence of PP2Cs led to the resolution of both ABA-unbound and ABA-bound structures, thus revealing the protein conformational changes upon ABA binding (Fig. 2; Nishimura et al. 2009; Santiago et al. 2009a; Yin et al. 2009). Two loops flanking the entrance to the ligand-binding pocket, termed the “proline cap” or “gate” (consensus sequence SGLPA) and the “leucine lock” or “latch” (consensus sequence HRL), will be referred to here as the “cap” and “lock,” respectively. The lock is completely conserved among all PYR/RCAR members, and the cap differs from the consensus sequence in only two of 14 isoforms (PYL12 and PYL13). When ABA binds to the protein, the cap closes over ABA, sealing the hormone within the protein cavity and sequestering it from solvent. The lock rearranges to secure the position of the cap, and an α-helix extends (recoils) in PYR1 to complete lid closure over the ABA molecule. These conformational changes both seal ABA inside the protein and reshape the protein surface, therefore impacting subsequent protein–protein interactions.
Figure 2.
Structural mechanism of ABA–PYR/RCAR–PP2C interactions. The structures of PYR/RCAR proteins in both ABA-unbound conformations (gold) and ABA-bound conformations (green), and in complex with PP2C. The following structures are shown: a PYL2 homodimer in the absence of ABA (A), and a symmetrical PYL2 dimer with both ABA molecules shown in orange (B). Note that the cap and lock have changed position to come in closer contact with the ABA molecule, while reducing dimer interaction. (C) An asymmetrical PYR1 dimer exhibiting “closed” hormone-bound (green) and “open” hormone-free (gold) subunit conformations. (D) A PYL2–HAB1 (PP2C) complex: A tryptophan residue from the PP2C (purple) inserts into the gap between the cap and lock to interact with the ABA molecule. The cap makes contact with the Mg2+-containing active site of the PP2C, therefore preventing phosphatase activity in the presence of ABA. The structures of PYR1 (Nishimura et al. 2009), PYL2, and the HAB1–PYL2 complex (Melcher et al. 2009) are oriented to align PYR/RCAR (shown at left).
One of the interesting findings of the structural studies was that PYR/RCAR proteins are able to dimerize; PYR1, PYL1, and PYL2 all form homodimers within the crystal lattice (Nishimura et al. 2009; Santiago et al. 2009a; Yin et al. 2009). As crystallization may introduce artifacts, PYR1 homodimerization was independently assayed and confirmed in solution (Nishimura et al. 2009; Santiago et al. 2009a). Furthermore, PYR1 homodimerization was found in planta through coimmunoprecipitation experiments (Nishimura et al. 2009). ABA binding also alters the dimer assembly; the relative orientation between the two PYR/RCAR subunits changes (Yin et al. 2009), and the dimer becomes flatter and more compact in solution, as shown by small-angle X-ray scattering (Nishimura et al. 2009). The number of van der Waals contacts and hydrogen bonds between the two proteins decreases in response to ABA binding, which has been suggested to weaken the homodimeric interface (Nishimura et al. 2009; Yin et al. 2009).
The conformational changes in the PYR/RCAR protein upon ABA binding create a new interface on the protein surface, providing a novel site for interaction with PP2C proteins. The PYR/RCAR–PP2C complexes that have been crystallized—PYL1–ABI1 (Miyazono et al. 2009; Yin et al. 2009) and PYL2–HAB1 (Melcher et al. 2009)—identify a conserved tryptophan residue from the PP2C that inserts between the cap and lock of the PYR/RCAR protein, forming a novel hydrogen bond between the indole ring of the tryptophan and the ketone group of ABA. ABA binding is thus stabilized further by the presence of the PP2C, which is reflected in the ∼10-fold increase in PYR/RCAR–ABA binding affinity in the presence of PP2Cs (Ma et al. 2009; Santiago et al. 2009b). For example, the binding affinity of S-(+)-ABA for RCAR1/PYL9 shifts from ∼600 nM to 60 nM in the presence of ABI2 (Ma et al. 2009). This may be a function of the ABA off-rate, with PP2C binding favoring PYR/RCAR lid closure, and thus decreasing the rate of ABA dissociation from the protein (Nishimura et al. 2009). The PYR/RCAR proteins occlude access to the catalytic site of the PP2C (Melcher et al. 2009; Miyazono et al. 2009; Yin et al. 2009), therefore providing a direct structural explanation for PYR/RCAR inhibition of PP2C activity (Ma et al. 2009; Park et al. 2009).
The structures provide a framework for mapping known ABA signaling mutations to biological function. The abi1-1 (G180D) and abi2-1 (G168D) mutations result in the loss of interaction with PYR/RCARs (Ma et al. 2009; Park et al. 2009) and dominant ABA insensitivity. In the PYR/RCAR–PP2C complex, the G180/G168 residues are in close proximity to the cap; thus, in the mutant protein, the bulkier aspartic acid residue would disrupt hydrogen bonding and introduce a steric constraint to PYR/RCAR and PP2C binding (Yin et al. 2009). Mutations in the PYR/RCAR cap or lock (e.g., PYR1 P88, R116) also reduce ABA sensitivity (Melcher et al. 2009; Miyazono et al. 2009; Nishimura et al. 2009; Park et al. 2009; Yin et al. 2009), underlining the importance of these loops for ABA binding and receptor function. Mutations of residues that make direct contact with the ABA molecule (e.g., PYR1 K59, R116; PYL2 L91) reduce ABA binding and disrupt ABA-induced PYR1 interactions with PP2Cs (Melcher et al. 2009; Nishimura et al. 2009), underlining the importance of ABA-induced conformational changes. ABA binding can, however, be functionally separated from PP2C interaction and inhibition; the PYR1P88S (corresponding to the cap) mutant protein retains ABA binding, but loses ABI1 binding (Park et al. 2009)—a pattern repeated in the PYR1H115A (lock) mutant protein (Melcher et al. 2009). The ABA-dependent root growth phenotype of Arabidopsis pyr1pyl1pyl2pyl4 seedlings is complemented by overexpression of the wild-type PYR1 but not PYR1H115A (Melcher et al. 2009).
Interestingly, there are biochemical differences in the PYR/RCAR–PP2C interaction, depending on the PYR/RCAR isoform, indicating possible functional specialization among the family members. Some PYR/RCARs interact with ABI1 in the absence of ABA treatment, whereas, for others, the interaction is induced by exogenous ABA (Ma et al. 2009; Park et al. 2009; Santiago et al. 2009b); for PYR1 and PYL1–4, the PYR–PP2C interaction is ABA-dependent, while, for other members of the family (PYL5–12), the interaction is constitutive in yeast two-hybrid analysis (Ma et al. 2009; Park et al. 2009). This pattern was also observed in Arabidopsis plants; without adding exogenous ABA, the most abundant ABI1 interactors were PYL5–12, whereas exogenous ABA treatment increased PYR1/PYL1–4 interactions with ABI1 (Nishimura et al. 2010). Some evidence indicates the possibility that formation of the protein–protein complex between PYR/RCAR and PP2C can be functionally separated from PP2C inhibition: For RCAR1 (PYL9), the interaction with ABI1 and ABI2 is constitutive, but ABA is still required for inhibition of phosphatase activity (Ma et al. 2009).
Handing on the signal—SnRK2s as positive regulators
As PYR/RCARs function through ABA-dependent inhibition of PP2C activity, targets of PP2Cs represent the next stage of the signaling pathway. The SnRK2 Ser/Thr kinase OST1 (OST1/SnRK2.6/SnRK2E) is a known positive regulator of ABA-dependent stomatal movements (Mustilli et al. 2002; Yoshida et al. 2002), and is closely related to the ABA-activated protein kinase (AAPK) of V. faba (Li et al. 2000). The Arabidopsis genome encodes 10 SnRK2s, of which SnRK2.6/OST1, SnRK2.2, and SnRK2.3 have been associated with ABA signaling. Triple snrk2.2snrk2.3snrk2.6 mutants are almost completely unresponsive to ABA (Fujii and Zhu 2009; Fujita et al. 2009; Nakashima et al. 2009), indicating that these SnRK2s form a major hub in the ABA signaling network. The C termini of SnRK2.2, SnRK2.3, and SnRK2.6 contain an Asp-enriched domain (Domain II) (Table 1) required for both ABA-specific activation of the kinase (Belin et al. 2006) and interaction with ABI1 (R Yoshida et al. 2006). HAB1 dephosphorylates the kinase within this activation domain to repress kinase activity (Belin et al. 2006; Boudsocq et al. 2007; Vlad et al. 2009). Consistent with this, an unbiased phosphopeptide array screening approach to identify targets of the PP2Cs HAB1, ABI1, and ABI2 isolated OST1 as a PP2C target (Vlad et al. 2009). The PYR/RCAR-mediated inhibition of PP2C activity therefore results in SnRK2 kinase activation, allowing the phosphorylation of downstream targets. Some PP2C–SnRK2 interactions may be constitutive, as ABI1 and SnRK2.3 interact in both the presence and absence of ABA (Umezawa et al. 2009; Nishimura et al. 2010). In contrast, yeast three-hybrid analyses indicate the ABA dependency of ABI2–SnRK2.6 and HAB1–SnRK2.6 interactions with PYL8 and PYL5, respectively (Fujii et al. 2009). More thorough testing in planta may reveal interesting isoform-specific differences in complex assembly.
Emerging questions relating to the central signaling module
The central PYR/RCAR–PP2C–SnRK2 signaling module provides an elegant model of early events in ABA signal transduction at a molecular and structural level. However, to understand the early signaling pathway more fully, the following important questions should be addressed.
Does PP2C binding dissociate the PYR1/PYL1,2 dimer?
Structural and mutational analyses indicate significant overlap between PYR1/PYL1,2 residues located in the dimer interface and those participating in the PYR1/PYL1,2–PP2C complex, suggesting that PYR1/PYL1,2 homodimers and PYR1/PYL1,2–PP2C complexes are mutually exclusive (Fig. 2). The model therefore implies that formation of PYR1/PYL1,2–PP2C complexes is correlated with dissociation of PYR1/PYL1,2 homodimers, but does not directly distinguish whether dimer dissociation precedes PP2C complex formation or vice versa. As described above, binding of ABA energetically weakens the homodimeric interface between the PYR1/PYL1,2s; however, ABA-bound PYR1/PYL1,2 dimers are stable enough to form both in concentrated solutions of purified proteins and when overexpressed in plant tissues (Nishimura et al. 2009; Santiago et al. 2009a; Yin et al. 2009). The protein and hormone concentrations could tune the association/dissociation equilibria for homodimers and PP2C inhibition complexes, to appropriately respond to the plant's environmental conditions.
Do PYR/RCAR heterodimers occur and regulate ABA sensitivity?
The PYR/RCAR family proteins in Arabidopsis are very closely related in sequence and structure, suggesting the possibility of heterodimer formation. Different PYR/RCAR isoforms have different ligand and protein affinities; for example, PYR1 interacts with HAB1 only in the presence of (+)-ABA, while PYL2 and HAB1 interact in the presence of both the natural and unnatural ABA isomers (Park et al. 2009). PYR/RCAR heterodimers may further modulate ABA sensitivity, and provide increased flexibility in the signaling cascade.
How is ABA dependency of PYR/RCAR–PP2C complexes achieved?
As described earlier, PYL5–12 (excluding PYL8) form constitutive complexes with PP2Cs (Ma et al. 2009; Park et al. 2009), yet RCAR1 (PYL9)-mediated inhibition of ABI1 and ABI2 is ABA-dependent (Ma et al. 2009). Constitutive PYL5–PYL12 (excluding PYL8) interaction with PP2Cs suggests that these PYR/RCARs may not energetically favor receptor–receptor dimer formation. The crystal structures indicate that PYR/RCAR–PP2C interactions occur at the surface created by the closing of the ligand-binding pocket, which raises the question of how ABA can access the receptor protein if binding of the PP2C has already closed the cap and lock loops. All structural studies thus far have considered PYR/RCAR proteins of the same subfamily (PYR1–PYL2); structure studies of PYL5–12 isoforms in the presence and absence of ABA and in complex with PP2Cs should help to resolve this uncertainty in mechanism.
Which PYR/RCAR and PP2C isoforms form functional receptor complexes in planta?
The vast majority of research on PYR/RCAR–PP2C interactions has been done in vitro or through heterologous expression in tobacco. Under these conditions, multiple combinations of PYR/RCAR–PP2C can form complexes; for example, ABI1 interacts with PYR1, PYL1, PYL8 (RCAR3), and RCAR1 (PYL9) (Ma et al. 2009; Miyazono et al. 2009; Park et al. 2009; Yin et al. 2009; Szostkiewicz et al. 2010). However, the diversity of interactions indicated in vitro does not necessarily occur in intact plants. In vitro analysis indicates that the specific PYR/RCAR–PP2C isoforms in a complex determine its biochemical parameters; for example, half maximal inhibition of PP2C activity occurs at a lower [ABA] in a PYL9–ABI1 complex than a PYL9–ABI2 complex (Szostkiewicz et al. 2010). Determination of the major interactions in vivo is therefore required to relate biochemical and physiological data. Identification of PYR/RCAR–ABI1 interactions in Arabidopsis leaves indicates that several combinations of interactions may occur at any one time (Nishimura et al. 2010). This question needs to be considered in a cell- or tissue-specific manner for full characterization of the physiologically relevant pathways. Similarly, identification of the most relevant in planta PP2C–SnRK2 interactions is required.
How are SnRK2s activated?
Given that PP2C-mediated inhibition of OST1 occurs through dephosphorylation, the origin of the activating phosphorylation should be considered. Two models of SnRK2 activation can be envisaged: autophosphorylation or phosphorylation by upstream kinases. In-gel analysis indicates that SnRK2s autophosphorylate (Belin et al. 2006; Boudsocq et al. 2007; Fujii et al. 2009; Vlad et al. 2009), yet OST1 can be activated by osmotic stress in the abi1-1 and abi2-1 backgrounds, indicating a role for an unknown protein kinase (R Yoshida et al. 2006). The current model predicts that SnRK2s are constitutively active and lose their ABA dependency in the absence of PP2Cs. Consistent with this, in an abi1hab1pp2ca mutant, SnRK2.2 and SnRK2.3 (and SnRK2.6, to a lesser extent) activity is up-regulated (Fujii et al. 2009). SnRK2.6/OST1 is reported to mediate ABA-independent responses, indicating additional possible regulation mechanisms (Zheng et al. 2010). Testing the model that inhibition of PP2C activity alone is sufficient for SnRK2 activation in planta (Fujii et al. 2009) is important; additional components might reduce the energetic cost incurred by continual removal of activating phosphates by PP2Cs.
Do PP2Cs target proteins other than SnRK2s?
The identification of SnRK2s as direct targets of PP2Cs reveals one downstream pathway for ABA signaling through PP2Cs, but does not rule out others. Genetic analyses indicate both independent and overlapping functions for the closely related PP2Cs ABI1 and ABI2 (e.g., Gilmour and Thomashow 1991; Gosti et al. 1995; Pei et al. 1997; Murata et al. 2001; Rubio et al. 2009). Phosphopeptide array studies (Vlad et al. 2009) identified additional HAB1 targets, including enzymes involved in primary metabolism and proton pumps. Other PP2C-interacting proteins identified include the chromatin remodeling factor SWI3 (Saez et al. 2008), several calcineurin-B-like protein (CBL)-interacting protein kinases (CIPK) (Guo et al. 2002; Ohta et al. 2003), and the calcium-dependent protein kinase 21 (CPK21) and CPK23 (Geiger et al. 2010). PP2Cs may therefore have a broad range of substrates, and, as such, may represent the first major branchpoint in the ABA signaling network; these alternate targets should therefore be considered in more detail.
Location of ABA perception
The identification of the PYR/RCAR proteins as ABA receptors provides clear evidence that ABA perception occurs primarily intracellularly through small soluble proteins. The site of ABA perception has been debated for several years, with evidence for both intracellular and extracellular receptors. Early studies suggested the presence of a transmembrane ABA receptor; ABA binding to proteins in V. faba is abolished in the presence of trypsin, suggesting an ABA-binding site on the external face of the membrane (Hornberg and Weiler 1984). An extracellular model of ABA perception was also suggested through experiments using ABA–protein conjugates that were unable to enter the cell, but were able to induce gene expression in rice suspension cells (Schultz and Quatrano 1997) and barley aleurone cells (Gilroy and Jones 1994), and ion channel activity in cultured Arabidopsis suspension cells (Jeannette et al. 1999); in some studies, microinjection of ABA failed to stimulate responses (Anderson et al. 1994; Gilroy and Jones 1994). However, the extracellular pH dependence of ABA responses also indicated an intracellular site of ABA action in one of these studies (Anderson et al. 1994). More directly, several independent studies in guard cells have suggested that ABA action can be intracellular. Patch-clamp analysis of guard cells showed that cytosolic ABA application is sufficient for rapid regulation of ion channels (Schwartz et al. 1994; Schwarz and Schroeder 1998; Levchenko et al. 2005). Microinjection of either ABA or photolyzable ABA stimulated stomatal closure (Allan et al. 1994; Schwartz et al. 1994), again suggesting an intracellular site of ABA perception. Some of the conflicting data may be resolved with the recent identification of two ABC transporters that mediate transmembrane ABA flux. AtABCG40 and AtABCG25 are both members of the ABCG subfamily, with AtABCG40/PDR12 being a full-length transporter and AtABCG25 being a half-size ABC transporter. AtABCG40 increases the rate of ABA uptake into cells when expressed heterologously in either yeast or cultured tobacco BY2 cells (Kang et al. 2010), whereas AtABCG25 mediates ABA efflux (Kuromori et al. 2010). AtABCG40 is expressed primarily in guard cells and AtABCG25 is expressed primarily in the leaf vasculature, suggesting that the two transporters act coordinately to control ABA flux in mature plants, with AtABCG25 mediating ABA efflux from the site of ABA synthesis, and AtABCG40 allowing ABA uptake at the guard cell plasma membrane. Lack of effects of microinjected ABA may therefore be explained through the cellular export of ABA.
Potential alternative ABA receptors have different cellular locations than the PYR/RCARs, which may contribute to the diversity in predicted ABA response sites. GTG1 and GTG2 are plasma membrane proteins (Pandey et al. 2009), and therefore might be responsible for ABA perception at the plasma membrane. ChlH/GUN5, being part of the chlorophyll biosynthesis pathway, is located within the chloroplast. As ABA is synthesized from intermediates in the carotenoid biosynthesis pathway, which is also chloroplast-located, there is the potential for a plastid-specific ABA perception mechanism that may therefore link light perception to ABA responses.
How does the central signaling module interact with other ABA-dependent processes?
As described above, ABA activates downstream signaling through the PYR/RCAR-mediated inactivation of PP2Cs, resulting in the activation of SnRK2s. Several targets of SnRK2.6/OST1 have been identified—including the ion channels SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1/RCD3) (Geiger et al. 2009; Lee et al. 2009; Vahisalu et al. 2010) and the inward-rectifying K+ CHANNEL IN ARABIDOPSIS THALIANA (KAT1) (Sato et al. 2009), the ROS-generating enzyme RESPIRATORY BURST OXIDASE HOMOLOG F (RbohF) (Sirichandra et al. 2009), and the bZIP transcription factors ABI5 (Nakashima et al. 2009) and ABA-RESPONSIVE ELEMENT-BINDING FACTOR 2 (ABF2) (Fujii et al. 2009). While many of the targets identified contain the SnRK2 consensus target sequence R-X-X-[S/T] (Furihata et al. 2006; Vlad et al. 2008), SLAC1 and KAT1 are phosphorylated at nonconsensus sites (Geiger et al. 2009; Sato et al. 2009), suggesting greater diversity of SnRK2 targets than indicated previously. The identification of both ion channels and transcription factors as targets of OST1 connects ABA to terminal responses through a PYR–PP2C–SnRK2 pathway, reflected in the rapid activation of stomatal closure after ABA application (Siegel et al. 2009).
Regulation of ion channels
Stomatal aperture is regulated by the coordinated actions of ion channels at the plasma membrane and tonoplast (for review, see Schroeder et al. 2001). ABA induces a large depolarization at the plasma membrane that is controlled through (1) the inhibition of the plasma membrane H+-ATPase (Goh et al. 1996), (2) inhibition of the K+ influx channel (Schroeder and Hagiwara 1989; Lemtiri-Chlieh and MacRobbie 1994), and (3) activation of the slow (S-type) anion channel (Schroeder and Hagiwara 1989). This depolarization activates K+ efflux from the cell, resulting in a net loss of solutes and a decrease in turgor. SnRK2 phosphorylation is now associated with two of these major regulatory events at the plasma membrane; namely, the inhibition of K+ influx channels and the activation of S-type anion channels. Thus, the central signaling module interfaces with membrane proteins required for stomatal closure.
Electrophysiological analyses established that S-type anion channels are dependent on ATP and sensitive to the Ser/Thr kinase inhibitor K252a, providing early evidence that phosphorylation is required for ABA responses and channel activation in guard cells (Schmidt et al. 1995). Consistent with this, activation of S-type anion channels is abolished in abi1-1 and abi1-2 (Pei et al. 1997). SLAC1, identified recently as the S-type anion channel, is a 10-transmembrane domain protein with an extended cytosolic N-terminal region (Negi et al. 2008; Vahisalu et al. 2008) that includes an OST1 phosphorylation site required for channel activity (Geiger et al. 2009; Lee et al. 2009; Vahisalu et al. 2010). Expression of SLAC1 and OST1, but not SLAC1 alone, resulted in channel activity (Geiger et al. 2009; Lee et al. 2009). OST1 needed to be constitutively bound to SLAC1 via split YFP (bimolecular fluorescence complementation [BiFC]) constructs for strong anion channel activation (Geiger et al. 2009). The addition of ABI1, ABI2, or PP2CA to the system abolished channel activity (Geiger et al. 2009; Lee et al. 2009), demonstrating that the PP2C–SnRK2 regulatory module can be reconstituted. Expression of RCAR1/PYL9 conferred ABA-dependent phosphorylation of the SLAC1 N terminus when coexpressed with ABI1 and OST1 (Geiger et al. 2010). ABA-dependent activation of the channel in this heterologous system remains to be demonstrated.
Guard cell K+ uptake is also SnRK2-dependent. Kin+ currents are inhibited by protein phosphatase inhibitors (Li et al. 1994; Thiel and Blatt 1994), indicating that inward-rectifying K+ uptake channels KAT1 (Schachtman et al. 1992) and KAT2 are targets of a phosphorylation-dependent pathway. Both KAT1 and KAT2 facilitate K+ influx in guard cells, with dominant-negative mutations in both channel proteins inhibiting K+ uptake (Kwak et al. 2001; Lebaudy et al. 2008). Following previous conflicting reports arguing for (Mori et al. 2000) and against (Li et al. 1998) phosphorylation of KAT1 by the OST1 homolog AAPK in V. faba, OST1 has been shown recently to phosphorylate the C terminus of KAT1 in Arabidopsis; mutation of the phosphorylated residue inhibits both K+ conductance and complementation of a K+-deficient yeast strain, indicating that this site is critical for channel activity (Sato et al. 2009).
ROS and Ca2+ signaling
ABA-dependent stomatal closure is associated with small signaling molecules (also known as secondary messengers), including nitric oxide (Garcia-Mata et al. 2003), ROS (Pei et al. 2000), and cytosolic free calcium (McAinsh et al. 1990). ABA-induced stomatal closure is dependent on two plasma membrane NADPH oxidases (RbohD and RbohF) (Kwak et al. 2003) that generate ROS in the cell wall space. ABA-induced increases in ROS (Pei et al. 2000; Mustilli et al. 2002; Suhita et al. 2004) are abolished in the ost1-2 mutant (Mustilli et al. 2002; Suhita et al. 2004), indicating that SnRK2s function upstream of ROS production (Fig. 1). A recent study showed that RbohF is a direct target of OST1; RbohF and OST1 interact physically in BiFC assays, and OST1 phosphorylates two sites in the extended N terminus (Sirichandra et al. 2009). One of the target motifs (R-X-X-S) is conserved between all Arabidopsis Rboh isoforms, suggesting that SnRK2 activation may be a widespread mechanism of Rboh regulation.
Analysis of ABA-induced stomatal closure in Arabidopsis under conditions in which increases in [Ca2+]cyt are inhibited indicates that [Ca2+]cyt-dependent mechanisms are responsible for ∼70% of stomatal closure (Siegel et al. 2009), and several Ca2+-associated proteins are required for normal stomatal closure, including CPKs, CBLs, and CIPKs) (Table 1; for review, see Dodd et al. 2010; Kim et al. 2010; Kudla et al. 2010). However, the central PYR/RCAR–PP2C–SnRK2 signaling module is not thought to be Ca2+-dependent; ABI1 contains a Ca2+-binding EF hand domain, suggesting Ca2+ sensitivity of the phosphatase (Leung et al. 1994; Meyer et al. 1994), but expression of truncated ABI1 provides tentative evidence that the EF hand may not have physiological significance (Sheen 1998). This raises the question: How is Ca2+ sensitivity of stomatal closure achieved if the seemingly Ca2+-independent central signaling module directly targets channels responsible for ion efflux? The identification of RbohF as an OST1 phosphorylation target provides a possible mechanistic link between the central signaling module and [Ca2+]cyt-dependent processes. H2O2 stimulates Ca2+-permeable channels (ICa) at the plasma membrane, and H2O2-induced stomatal closure requires external Ca2+, showing that the ROS–ICa–[Ca2+]cyt pathway is involved in ABA signaling (Pei et al. 2000; Murata et al. 2001; Kwak et al. 2003; Suhita et al. 2004). Ca2+-dependent and Ca2+-independent pathways can function in parallel to regulate cellular components; for example, the N terminus of SLAC1 is phosphorylated by both SnRK2.6/OST1 (Geiger et al. 2009) and the Ca2+-dependent kinases CPK21 and CPK23 (Geiger et al. 2010).
Transient increases in [Ca2+]cyt are observed in guard cells under resting or nonstimulated conditions (for example, see Young et al. 2006), which might appear to contradict a model of [Ca2+]cyt being required for ABA-induced stomatal closure. However, recent studies suggest that physiological stimuli such as ABA and CO2 increase or “prime” the sensitivity of Ca2+-dependent processes (Young et al. 2006). Notably, in the presence of the stimulus, ABA or CO2, downstream target signaling proteins are more Ca2+-responsive (Young et al. 2006). For example, [Ca2+]cyt-dependent activation of S-type anion channels and Kin+ channel down-regulation are induced by pre-exposure to ABA (Siegel et al. 2009; Chen et al. 2010), and also by pre-exposure to high extracellular [Ca2+] (Allen et al. 2002). This novel Ca2+ sensitivity priming model could produce specificity in plant [Ca2+]cyt signaling, in light of the diversity and large number (>200) of Ca2+ sensors encoded in the Arabidopsis genome (Day et al. 2002). How priming functions at a mechanistic level is an important unresolved question, but several nonmutually exclusive mechanisms can be envisaged, including subcellular relocalization of Ca2+ sensors, post-translational modification, protein–protein interactions, coincidence detection of parallel signaling pathways, and transcriptional reprogramming (Fig. 3; for review, see Kim et al. 2010). Further analysis of potential priming mechanisms is required to fully understand the mechanisms of [Ca2+]cyt-dependent ABA signaling.
Figure 3.
Possible mechanisms of Ca2+ sensitivity priming by ABA. Exposure to ABA enhances the Ca2+ sensitivity of the signaling pathway through several potential mechanisms; a calcium sensor is shown as the point of priming for clarity, but the priming mechanism may potentially involve downstream components. (i) ABA allows the appropriate subcellular localization of the sensor protein. (ii) ABA treatment causes the appropriate post-translational protein modifications for activity; removal of a negatively regulating modification is shown, but addition of a positively regulating modification is equally possible. (iii) ABA treatment provides the appropriate additional components of a protein complex to associate with the sensor; removal of a negative regulator is indicated, but ABA may also recruit positive interactors. (iv) ABA-stimulated transcriptional changes include expression of the sensor protein.
Transcriptional responses
ABA treatment induces changes in gene expression in more than ∼10% of the Arabidopsis genome (Sánchez et al. 2004; Seki et al. 2007; Yang et al. 2008; Zeller et al. 2009), resulting in the increased expression of stress-associated and signaling component transcripts. The major ABA-dependent cis-regulatory element associated with expression of abiotic stress-responsive genes is the ABA response element (ABRE; ACGTGT) (for review, see Yamaguchi-Shinozaki and Shinozaki 2005). Analysis of synthetic promoters suggests that, although a single ABRE may be insufficient, two copies of the ABRE or a single ABRE combined with the related ABRE-coupling element (ABRE-CE; ACGCGT/G/C) renders a promoter ABA-sensitive (Hobo et al. 1999; Zhang et al. 2005). Several bZIP transcription factors—including SnRK2 phosphorylation targets ABI5, AREB1/ABF2, and AREB2/ABF4 (Johnson et al. 2002; Furihata et al. 2006; Fujii et al. 2009; Nakashima et al. 2009)—bind the ABRE and induce ABA-dependent gene expression. Correspondingly, ABA-induced gene expression is compromised in snrk2.2snrk2.3snrk2.6 mutants (Fujii and Zhu 2009; Nakashima et al. 2009). However, transcriptome analysis indicates that, despite significant overlap between genes misregulated in snrk2.2snrk2.3snrk2.6 mutants and abi5 mutants, some transcripts are misregulated in the snrk2 triple mutant and not abi5, and vice versa (Nakashima et al. 2009). Thus, a full description of the complexity of ABA-induced transcriptional regulation may also include the action of the central signaling module on alternative transcriptional activators binding to other ABA-related cis-elements. One possible class of alternate transcriptional regulators is the Ca2+-regulated CALMODULIN-BINDING TRANSCRIPTIONAL ACTIVATORS (CAMTAs) (Bouché et al. 2002; Finkler et al. 2007), which bind the ABRE-CE (Doherty et al. 2009), and therefore may contribute to ABA-dependent transcriptional regulation or function in a parallel stress signaling pathway. An additional layer of complexity in ABA-induced control of gene expression is provided by links to chromatin remodeling. HAB1 interacts with the A. thaliana homolog of the yeast SWI3 subunit of SWI/SNF chromatin remodeling complexes, and swi3b knockout plants have reduced sensitivity to ABA in seed germination and growth assays (Saez et al. 2008).
Conclusions
Recently, a revolution has occurred in our understanding of ABA perception and signaling, allowing signaling cascades from ABA binding to target responses to be mapped out. As with all major discoveries, the identification of the PYR/RCAR–PP2C signaling mechanisms raises interesting new questions. ABA signaling issues still needing to be resolved include the detailed biochemical and structural mechanisms of dimer dissociation and PP2C association, and ABA-receptor responses in those PYR/RCARs that interact constitutively with PP2Cs. The considerable evidence reviewed here suggests that ABA signaling is mediated by a complex protein network, so other known and potentially unknown components remain to be integrated with PYR/RCAR receptor signaling. To complement the biochemical and structural details, the central signaling module also needs to be considered in a physiological context, as it is likely that cell and tissue specificity exists within the network. In addition to enabling new approaches to understanding the fundamental biology of ABA signaling, the identification of PYR/RCARs may enable new strategies for the development of drought-resistant crops.
Acknowledgments
We thank Mark Estelle, TaeHoun Kim, Sean Cutler, and Maik Böhmer for reading and comments on the manuscript. We apologize to those whose work was not discussed here due to space constraints. Research in our laboratories was supported by NIH (GM060396), NSF (MCB0918220), and, in part, DOE (DEFG02-03ER15449) grants to J.I.S., and by NIH (GM37684) grant to E.D.G.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1953910.
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J 2001. The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Weinl S, Blazevic D, D'Angelo C, Batistic O, Kolukisaoglu Ü, Bock R, Schulz B, Harter K, Kudla J 2003. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36: 457–470 [DOI] [PubMed] [Google Scholar]
- Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ 1994. Two transduction pathways mediate rapid effects of abscisic acid in commelina guard cells. Plant Cell 6: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI 1999. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Murata Y, Chu SP, Nafisi M, Schroeder JI 2002. Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1-2. Plant Cell 14: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BE, Ward JM, Schroeder JI 1994. Evidence for an extracellular reception site for abscisic acid in commelina guard cells. Plant Physiol 104: 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti DS, Naylor RL 2009. Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323: 240–244 [DOI] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S 2006. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141: 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Scharlat A, Snedden W, Bouchez D, Fromm H 2002. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C 2007. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Chen Z-H, Hills A, Lim CK, Blatt MR 2010. Dynamic regulation of guard cell anion channels by cytosolic free Ca2+ concentration and protein phosphorylation. Plant J 61: 816–825 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim K-N, Pandey GK, Gupta R, Grant JJ, Luan S 2003. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim B-G, Lee S-C, Kudla J, Luan S 2007. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Choi H-I, Park H-J, Park JH, Kim S, Im M-Y, Seo H-H, Kim Y-W, Hwang I, Kim SY 2005. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol 139: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR 2010. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Day I, Reddy V, Shad Ali G, Reddy A 2002. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 3: research0056 doi: 10.1186/gb-2002-3-10-research0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delk NA, Johnson KA, Chowdhury NI, Braam J 2005. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 139: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla Jr, Sanders D 2010. The language of calcium signalling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF 2009. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C 2008. Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Finkler A, Kaplan B, Fromm H 2007. Ca2+-responsive cis elements in plants. Plant Signal Behav 2: 17–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK 2007. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. 2009. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K 2006. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR 2003. Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci 100: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. 2009. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KAS, Grill E, et al. 2010. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Thomashow MF 1991. Cold-acclimation and cold-regulated gene-expression in ABA mutants of Arabidopsis thaliana. Plant Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones RL 1994. Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L) aleurone protoplasts. Plant Physiol 104: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh CH, Kinoshita T, Oku T, Shimazaki K 1996. Inhibition of blue light-dependent H pumping by abscisic acid in Vicia guard-cell protoplasts. Plant Physiol 111: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Bertauche N, Vartanian N, Giraudat J 1995. Abscisic-acid-dependent and abscisic-acid-independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol Gen Genet 246: 10–18 [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song C-P, Gong D, Halfter U, Zhu J-K 2002. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Guo J, Zeng Q, Emami M, Ellis BE, Chen J-G 2008. The GCR2 gene family is not required for ABA control of seed germination and early seedling development in Arabidopsis. PLoS ONE 3: e2982 doi: 10.1371/journal.pone.0002982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T 1999. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Hornberg C, Weiler EW 1984. High-affinity binding sites for abscisic acid on the plasmalemma of Vicia faba guard cells. Nature 310: 321–324 [Google Scholar]
- Iyer LM, Koonin EV, Aravind L 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Protein Struct Funct Genet 43: 134–144 [DOI] [PubMed] [Google Scholar]
- Jeannette E, Rona J-P, Bardat F, Cornel D, Sotta B, Miginiac E 1999. Induction of RAB18 gene expression and activation of K+ outward rectifying channels depend on an extracellular perception of ABA in Arabidopsis thaliana suspension cells. Plant J 18: 13–22 [DOI] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK 2002. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y 2010. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI 2010. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu J-K 2010. ABA receptors: The START of a new paradigm in phytohormone signalling. J Exp Bot 61: 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan FN 1997. Global drought watch from space. Bull Am Meteorol Soc 78: 621–636 [Google Scholar]
- Koornneef M, Reuling G, Karssen CM 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kudla J, Batistic O, Hashimoto K 2010. Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI 2006. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K 2010. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI 2001. Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol 127: 473–485 [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Moon J-H, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI 2002. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei Z-M, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaudy A, Vavasseur A, Hosy E, Dreyer I, Leonhardt N, Thibaud J-B, Véry A-A, Simonneau T, Sentenac H 2008. Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci 105: 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S 2009. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA 1994. Role of calcium in the modulation of Vicia guard cell potassium channels by abscisic acid: A patch-clamp study. J Membr Biol 137: 99–107 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI 2004. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris P, Guerrier D, Chefdor F, Giraudat J 1994. Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J 1997. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 Genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V, Konrad KR, Dietrich P, Roelfsema MRG, Hedrich R 2005. Cytosolic abscisic acid activates guard cell anion channels without preceding Ca2+ signals. Proc Natl Acad Sci 102: 4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Luan S, Schreiber SL, Assmann SM 1994. Evidence for protein phosphatase 1 and 2A regulation of K+ channels in two types of leaf cells. Plant Physiol 106: 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Julie Lee Y-R, Assmann SM 1998. Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol 116: 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X-Q, Watson MB, Assmann SM 2000. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Ma SY, Wu WH 2007. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol 65: 511–518 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud J-P, Aldon D 2008. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Marris E 2008. Water: More crop per drop. Nature 453: 273–277 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM 1990. Abscisic acid-induced elevation of guard-cell cytosolic Ca2+ precedes stomatal closure. Nature 343: 186–188 [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. 2009. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J 2002. Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube M, Grill E 1994. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Milborrow BV 1974. The chemistry and biochemistry of abscisic acid. Annu Rev Plant Physiol 25: 259–307 [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. 2009. Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Mori IC, Uozumi N, Muto S 2000. Phosphorylation of the inward-rectifying potassium channel KAT1 by ABR kinase in Vicia guard cells. Plant Cell Physiol 41: 850–856 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang Y-F, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. 2006. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327 doi: 10.1371/journal.pbio.0040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AH, Hansson M 2009. The barley magnesium chelatase 150-kD subunit is not an abscisic acid receptor. Plant Physiol 150: 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Pei Z-M, Mori IC, Schroeder J 2001. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J 2002. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. 2009. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K 2008. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T 2007. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. 2010. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu J-K 2003. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim B-G, Li LG, Luan S 2008. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol Plant 1: 238–248 [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM 2009. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI 1997. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI 2000. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E 1998. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T-H, Santiago J, Flexas J, Schroeder JI, Rodriguez PL 2009. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL 2004. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL 2008. HAB1–SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez J-P, Duque P, Chua N-H 2004. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 38: 381–395 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA 2009a. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park S-Y, Márquez JA, Cutler SR, Rodriguez PL 2009b. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. 2009. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF 1992. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258: 1654–1658 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Schelle I, Liao YJ, Schroeder JI 1995. Strong regulation of slow anion channels and abscisic acid signaling in guard cells by phosphorylation and dephosphorylation events. Proc Natl Acad Sci 92: 9535–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Hagiwara S 1989. Cytosolic calcium regulates ion channels in the plasma-membrane of Vicia faba guard-cells. Nature 338: 427–430 [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ 2001. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Quatrano RS 1997. Evidence for surface perception of abscisic acid by rice suspension cells as assayed by Em gene expression. Plant Sci 130: 63–71 [Google Scholar]
- Schwartz A, Wu WH, Tucker EB, Assmann SM 1994. Inhibition of inward K+ channels and stomatal response by abscisic acid: An intracellular locus of phytohormone action. Proc Natl Acad Sci 91: 4019–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Schroeder JI 1998. Abscisic acid maintains S-type anion channel activity in ATP-depleted Vicia faba guard cells, indicating phosphatase inhibition. FEBS Lett 428: 177–182 [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K 2007. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Sheen J 1998. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y-Y, Wang X-F, Wu F-Q, Du S-Y, Cao Z, Shang Y, Wang X-L, Peng C-C, Yu X-C, Zhu S-Y, et al. 2006. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Siegel RS, Xue S, Murata Y, Yang Y, Nishimura N, Wang A, Schroeder JI 2009. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K channels in Arabidopsis guard cells. Plant J 59: 207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. 2009. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A 2004. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E 2010. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Thiel G, Blatt MR 1994. Phosphatase antagonist okadaic acid inhibits steady-state K+ currents in guard cells of Vicia faba. Plant J 5: 727–733 [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmaki A, Brosche M, Moldau H, Desikan R, et al. 2008. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Puzõrjova I, Brosché M, Valk E, Lepiku M, Moldau H, Pechter P, Wang Y-S, Lindgren O, Salojärvi J, et al. 2010. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J 62: 442–453 [DOI] [PubMed] [Google Scholar]
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S 2008. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J 55: 104–117 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frey NFd, Leung J 2008. An update on abscisic acid signaling in plants and more. Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Wu F-Q, Xin Q, Cao Z, Liu Z-Q, Du S-Y, Mei C, Zhao C-X, Wang X-F, Shang Y, Jiang T, et al. 2009. The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: New evidence in Arabidopsis. Plant Physiol 150: 1940–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K 2005. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel R, Schroeder J 2008. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6 doi: 10.1186/1746-4811-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N 2009. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K 2002. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K 2006. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T 2006. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Mehta S, Israelsson M, Godoski J, Grill E, Schroeder JI 2006. CO2 signaling in guard cells: Calcium sensitivity response modulation, a Ca2+-independent phase, and CO2 insensitivity of the gca2 mutant. Proc Natl Acad Sci 103: 7506–7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S 2009. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Zhang D-P, Wu Z-Y, Li X-Y, Zhao Z-X 2002. Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol 128: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho T-D, You Y, Yu T, Quatrano RS 2005. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21: 3074–3081 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xu X, Crosley RA, Greenwalt SA, Sun Y, Blakeslee B, Wang L, Ni W, Sopko MS, Yao C, et al. 2010. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol 153: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S-Y, Yu X-C, Wang X-J, Zhao R, Li Y, Fan R-C, Shang Y, Du S-Y, Wang X-F, Wu F-Q, et al. 2007. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]