Abstract
Human solid tumors frequently have pronounced heterogeneity of both neoplastic and normal cells on the histological, genetic, and gene expression levels. While current efforts are focused on understanding heterotypic interactions between tumor cells and surrounding normal cells, much less is known about the interactions between and among heterogeneous tumor cells within a neoplasm. In glioblastoma multiforme (GBM), epidermal growth factor receptor gene (EGFR) amplification and mutation (EGFRvIII/ΔEGFR) are signature pathogenetic events that are invariably expressed in a heterogeneous manner. Strikingly, despite its greater biological activity than wild-type EGFR (wtEGFR), individual GBM tumors expressing both amplified receptors typically express wtEGFR in far greater abundance than the ΔEGFR lesion. We hypothesized that the minor ΔEGFR-expressing subpopulation enhances tumorigenicity of the entire tumor cell population, and thereby maintains heterogeneity of expression of the two receptor forms in different cells. Using mixtures of glioma cells as well as immortalized murine astrocytes, we demonstrate that a paracrine mechanism driven by ΔEGFR is the primary means for recruiting wtEGFR-expressing cells into accelerated proliferation in vivo. We determined that human glioma tissues, glioma cell lines, glioma stem cells, and immortalized mouse Ink4a/Arf−/− astrocytes that express ΔEGFR each also express IL-6 and/or leukemia inhibitory factor (LIF) cytokines. These cytokines activate gp130, which in turn activates wtEGFR in neighboring cells, leading to enhanced rates of tumor growth. Ablating IL-6, LIF, or gp130 uncouples this cellular cross-talk, and potently attenuates tumor growth enhancement. These findings support the view that a minor tumor cell population can potently drive accelerated growth of the entire tumor mass, and thereby actively maintain tumor cell heterogeneity within a tumor mass. Such interactions between genetically dissimilar cancer cells could provide novel points of therapeutic intervention.
Keywords: Glioblastoma, EGFR, ΔEGFR, IL-6, LIF, gp130, tumor heterogeneity
Tumor cells are similar to members of other societies in that the ways in which they interact depend on their potential and the circumstances in which they find themselves. Emigrating immune cells, like the indigenous stromal and vascular components, influence and are influenced by their tumor neighbors; thus, a complete picture of neoplastic growth for most solid cancers is most aptly described as a heterogeneous composite of tumor and host cell constituents. While this tumor–host cell interaction has become a major focus of interest in such cancers as breast and pancreatic (Allinen et al. 2004; Tian et al. 2009), the roles of intratumoral interactions between tumor cells harboring different genetic alterations are poorly understood.
Glioblastoma multiforme (GBM)—a highly aggressive primary brain cancer typified by uncontrolled cellular proliferation, intense resistance to cell death, diffuse infiltration, robust angiogenesis, and associated vascular edema—is well recognized for such intratumoral heterogeneity. The heterogeneous nature of GBM cancer cells manifests as mixed cytological subtypes, regional differences in gene expression, and nonuniform representation of key gene mutations and genomic alterations (Jung et al. 1999; Maher et al. 2001; Kleihues et al. 2002; Furnari et al. 2007; Network 2008). Whether such marked intratumoral heterogeneity is the result of an inherent interactivity between tumor cells, genomic instability, or stochastic noise at the level of transcription, translation, or post-translational modifications remains unclear.
Chromosomal amplifications of epidermal growth factor receptor gene (EGFR) are seen in some or most cells of ∼50% of primary GBMs, and are associated with poor prognosis (Hurtt et al. 1992; Jaros et al. 1992; Schlegel et al. 1994). Approximately half of the tumors that have cells with amplified EGFR also contain cells that have sustained intragenic EGFR gene rearrangements that generate truncated, constitutively active mutant variants analogous to the viral v-erbB oncogene. The most common of these involves deletion of exons 2–7 (referred to as ΔEGFR [but also known as de2-7], EGFRvIII, and EGFR*), which causes an in-frame deletion and loss of a portion of the extracellular domain. The expression of ΔEGFR has been correlated with wild-type EGFR (wtEGFR) expression, and expression of both receptors within a tumor has been determined to confer a worse prognosis than wtEGFR expression alone (Shinojima et al. 2003; Heimberger et al. 2005). Interestingly, limitations of the specificity of available reagents have not allowed conclusive proof that the same cells within a GBM tumor express both receptors. The identification of uncommon tumors in which only one or the other receptor is expressed indicates that coexpression within the same tumor cells, although a possibility, is not required (Shinojima et al. 2003; Nishikawa et al. 2004). Experimentally, transfer of ΔEGFR into established glioma cell lines causes several cell-intrinsic effects, such as constitutive autophosphorylation, constitutive association with and activation of the Shc-Grb2-Ras and class I phosphoinositide-3-kinase (PI3K) pathways (Huang et al. 1997; Narita et al. 2002), enhanced tumorigenicity (Huang et al. 1997), increased cellular proliferation (Narita et al. 2002), and resistance to apoptosis induced by DNA-damaging chemotherapeutic drugs through modulation of Bcl-XL expression (Nagane et al. 1998). Importantly, none of these promalignant biological properties are conferred by overexpression of wtEGFR. For instance, wtEGFR cannot substitute for ΔEGFR in driving infiltrative glioma formation in genetically engineered mice (Hesselager and Holland 2003; Zhu et al. 2009) or in Ink4a/Arf−/− mouse neural stem cells or astrocytes (Holland et al. 1998; Bachoo et al. 2002), except when EGF ligand is infused at a high concentration into the injection site of wtEGFR-transduced cells (Bachoo et al. 2002).
The potent tumor-promoting cell-intrinsic function of ΔEGFR demonstrated using human glioma cell lines (Nishikawa et al. 1994; Nagane et al. 1996) and mouse models (Bachoo et al. 2002; Zhu et al. 2009) predicts that this receptor should be the predominant amplified EGFR variant in clinical samples. However, ΔEGFR expression is actually rare in the absence of wtEGFR amplification (Shinojima et al. 2003; Biernat et al. 2004; Nishikawa et al. 2004), raising the possibility that ΔEGFR is derived as a byproduct from amplified EGFR. However, inconsistent with this notion are the rare occurrences of ΔEGFR-positive/wtEGFR-negative GBMs (Shinojima et al. 2003; Nishikawa et al. 2004), suggesting that cells expressing ΔEGFR do not necessarily express amplified wtEGFR.
Given this infrequent occurrence of ΔEGFR-only GBMs, one may postulate that this mutant receptor occurs late in tumor progression, and that there is insufficient time for ΔEGFR-expressing cells to emerge as the dominant population in these rapidly fatal tumors. Here, we explored an alternative possibility: The minority ΔEGFR-expressing cancer cell subpopulation might enhance their own intrinsic tumorigenic abilities as well as extrinsically potentiate the proliferation of neighboring majority cells expressing amplified wtEGFR. We provide evidence that the intratumoral heterogeneity of GBM is actively maintained primarily by a cooperative tumor cell–tumor cell interaction involving ΔEGFR-expressing cells releasing IL-6 or leukemia inhibitory factor (LIF), which activate wtEGFR-expressing cells by a paracrine cytokine signaling circuit mediated by a novel gp130–wtEGFR interaction.
Results
ΔEGFR and wtEGFR cooperate to promote tumor growth
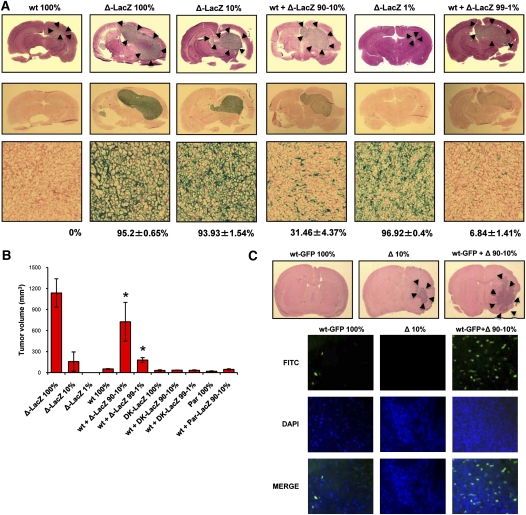
In GBM clinical samples, expression of ΔEGFR in some cells is frequently associated with expression of amplified levels of wtEGFR, and the coexpression of both receptors within the tumor mass confers a worse prognosis (Shinojima et al. 2003; Heimberger et al. 2005). We attempted to model this heterogeneous receptor coexpression (Pandita et al. 2004) and determine the effect on tumorigenic behavior. In a first series of experiments, human U87MG glioma cells, which express low levels of wtEGFR (∼1 × 105 receptors per cell) (Nishikawa et al. 1994; Wikstrand et al. 1997), were engineered to overexpress wtEGFR (U87wt) or ΔEGFR (U87Δ-LacZ) at levels (0.5 × 106 to 1 × 106 receptors per cell) (Nishikawa et al. 1994) that are consistent with the amplified levels found in GBM tumors and glioma xenografts prepared directly from patient material (0.2 × 106 to 0.7 × 106 receptors per cell) (Wikstrand et al. 1997), and we engrafted these intracranially into animals as single homogenous populations or in mixed ratios of U87wt:U87Δ-LacZ cells (90%:10% or 99%:1%). Twelve days after orthotopic injection, the mice were sacrificed and their brains were processed for H&E staining (Fig. 1A). The coinjection of U87wt and U87Δ cells produced a noticeable tumor growth enhancement (Fig. 1A), which resulted in a shorter survival over U87wt (Supplemental Fig. 1a–d). In contrast, very little, if any, tumor enhancement was observed when U87wt cells were mixed with U87MG parental (U87Par) or U87 cells expressing a dead kinase version of ΔEGFR (U87DK) (Supplemental Fig. 2; data not shown), or when U87DK was mixed with U87Δ-LacZ (Supplemental Fig. 1e). Tumor compositions were analyzed by X-Gal staining, and demonstrated that U87wt cells (LacZ-negative) remained the predominant population in the mixed tumor (68.54% ± 4.37% for 90:10 ratio; 93.16% ± 1.41% for 99:1 ratio; mean ± SEM) (Fig. 1A) and were not overgrown by the U87Δ-LacZ cells. Similarly, subcutaneous injection into nude mice of U87wt cells alone or mixed with U87Par-LacZ, U87Δ-LacZ, or U87DK-LacZ in ratios of 90%:10% or 99%:1% demonstrated that U87wt cells formed tumors poorly when mixed with U87Par-LacZ or U87DK-LacZ or when injected alone; however, a significant increase in their tumor growth was achieved when wtEGFR-overexpressing cells were mixed with a minor proportion of ΔEGFR-expressing cells (P = 0.002). Tumors obtained from these mixtures were significantly larger than the expected tumor volume obtained by the sum of the tumor volumes of the different cell populations injected alone (U87wt 100% and U87Δ 10% tumors) (Fig. 1B; Supplemental Fig. 3a). Additionally, as seen in the intracranial injection, the lack of growth enhancement imparted by U87DK-LacZ confirms that the catalytic kinase activity of the ΔEGFR is required for this effect.
Figure 1.
Tumor growth enhancement induced by mixing of wtEGFR and ΔEGFR-expressing cells. (A, top) H&E at day 12 after intracranial injection of U87wt (wt), U87Δ-LacZ (Δ-LacZ), or U87wt mixed with U87Δ-LacZ at 90:10 or 99:1 ratios (wt + Δ-LacZ 90%–10% or wt + Δ-LacZ 99%–1%; 100% = 5 × 105 cells). (Bottom) Whole-brain sections and X-Gal staining of tumor samples; Fast red counterstain. LacZ-positive percentage mean of each tumor sample is indicated below X-Gal staining pictures. (B) Tumor volume after subcutaneous injection of U87wt cells alone or mixed with U87Δ-LacZ, U87Par-LacZ, or U87DK-LacZ at ratios of 90:10 or 99:1 (100% = 1 × 106 cells). Error bars represent mean ± SEM; n = 6. (*) P < 0.05. (C, top) H&E of mouse brains 22 d after intracranial injection of mAstr-Ink4/Arf−/−-wtEGFR-GFP astrocytes (wt-GFP) alone, mAstr-Ink4/Arf−/−-ΔEGFR (Δ) alone, or mAstr-Ink4/Arf−/−-wtEGFR-GFP astrocytes and mAstr-Ink4/Arf−/−-ΔEGFR mixed at a 90:10 ratio (100% = 5 × 105 cells). (Bottom) mAstr-Ink4/Arf−/−-wtEGFR-GFP cells as GFP immunofluorescence. DAPI (blue) labels nuclei.
In a second and complementary series of experiments, we took advantage of the observation that primary astrocytes derived from Ink4a/Arf−/− mice that overexpress wtEGFR (mAstr-Ink4a/Arf−/−-wtEGFR) are not tumorigenic upon intracranial injection unless superphysiological levels of EGF are infused into the injection site, upon which they form infiltrative glioblastomas (Bachoo et al. 2002). We used this cellular system to test whether cells overexpressing ΔEGFR were competent to promote mAstr-Ink4/Arf−/−-wtEGFR participation in tumor formation. GFP-tagged mAstr-Ink4a/Arf−/−-wtEGFR (mAstr-Ink4a/Arf−/−-wtEGFR-GFP) cells were injected into nude mice intracranially alone or mixed with astrocytes overexpressing ΔEGFR (mAstr-Ink4a/Arf−/−-ΔEGFR) in a 90%:10% ratio, respectively. Twenty-two days after injection, mice were sacrificed and brains were processed for H&E and GFP immunofluorescence (Fig. 1C). As expected, mAstr-Ink4a/Arf−/−-wtEGFR-GFP did not form tumors upon injection. In contrast, mAstr-Ink4a/Arf−/−-ΔEGFR cells did form tumors, but tumor volumes were strikingly larger when the two cell populations were mixed together (Fig. 1C; Supplemental Fig. 4). Importantly, GFP staining revealed the presence of mAstr-Ink4a/Arf−/−-wtEGFR-GFP cells within these tumors, despite being unable to engraft by themselves (Fig. 1A). Thus, murine astrocytes and human glioma cells each showed that the mixture of a small proportion of ΔEGFR-expressing cells with those overexpressing wtEGFR resulted in a more aggressive, heterogeneous tumor phenotype when compared with the injection of either separately.
ΔEGFR-expressing glioma cells enhance the proliferation of cells overexpressing wtEGFR in vivo and not vice versa
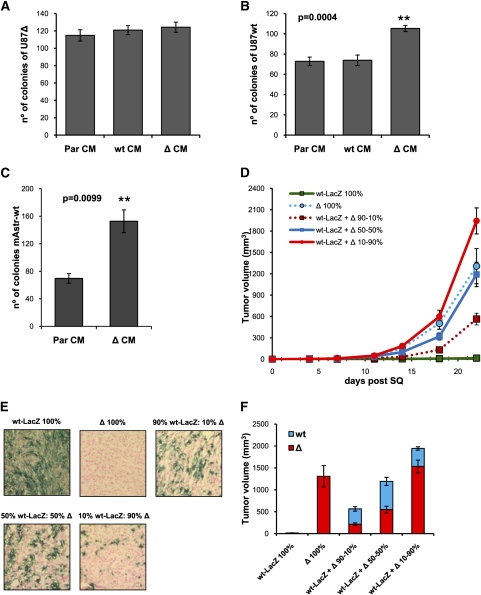
The potent tumor growth enhancement observed when wtEGFR and ΔEGFR cells are mixed together suggests that either or both cell populations secrete factors that promote cell growth and survival in vivo. To verify whether this cross-talk is unidirectional or bidirectional, we performed soft agar colony formation assays of wtEGFR cells treated with conditioned media (CM) derived from ΔEGFR cells and vice versa. Treatment with parental cell CM or normal media (Neg) was also included (Fig. 2A–C). ΔEGFR CM significantly increased wtEGFR colony formation in soft agar, indicating that ΔEGFR secrete one or more factors able to induce wtEGFR growth (Fig. 2B,C). However, no difference in soft agar colony formation was observed when ΔEGFR cells were treated with wtEGFR CM, forming colonies with equal efficiency irrespective of the source of the CM (Fig.2A).
Figure 2.
ΔEGFR cells enhance wtEGFR cell growth in vivo and in vitro. (A) Soft agar colony formation assay quantification of U87Δ cells treated with normal media (Neg), U87Par CM (ParCM), U87wt CM (wtCM), or U87Δ CM (ΔCM). (B) Soft agar colony formation assay quantification of U87wt cells treated with normal media (Neg), U87Par CM (ParCM), U87wt CM (wtCM), or U87Δ CM (ΔCM). (C) Soft agar colony formation assay quantification of mAstr-Ink4/Arf−/−-wtEGFR treated with mAstr-Ink4/Arf−/− CM (ParCM) or mAstr-Ink4/Arf−/−-ΔEGFR CM (ΔCM). (D) Tumor growth kinetics after subcutaneous injection of U87wt-LacZ (wt-LacZ), U87Δ (Δ), or U87wt-LacZ mixed with U87Δ at ratios of 90:10, 50:50, or 10:90 (100% = 2 × 105 cells). (E) Representative X-Gal staining images of subcutaneous tumors obtained at day 22 after subcutaneous injection of U87wt-LacZ (wt-LacZ), U87Δ (Δ), or U87wt-LacZ mixed with U87Δ at ratios of 90:10, 50:50, or 10:90. Fast red counterstain. (F) Relative tumor volume after analysis of tumor composition by X-Gal staining and Image Pro-Analyzer 6.2 software of tumors obtained at day 22 after subcutaneous injection of indicated mixtures of U87wt and U87Δ. Error bars in all experiments represent mean ± SEM. One-way ANOVA and two-tail t-test were used to compare samples. (**) P < 0.001. n = 6 for subcutaneous injection; n = 3 for soft agar assay.
These results suggest that ΔEGFR unidirectionally promotes wtEGFR cell growth. To verify this hypothesis in vivo, U87wt tagged with LacZ (U87wt-LacZ) and U87Δ cells were injected subcutaneously into nude mice alone or mixed at different ratios, and then tumor volumes and compositions were determined. As expected, U87wt-LacZ formed tumors poorly compared with U87Δ, but when U87wt-LacZ and U87Δ were mixed together, a potent tumor growth enhancement was observed (Fig. 2D,F). Tumors derived from engraftment of U87wt-LacZ:U87Δ at ratios of 50:50 or 10:90 were not significantly different (P ≥ 0.05) in size when compared with tumors derived from 100% U87Δ (Fig. 2F). Analysis of tumor composition by X-Gal staining revealed that U87wt-LacZ cells were rendered able to grow at the same rate as U87Δ in the 50:50 ratio engraftment, or even faster in 10:90 ratio, where the final proportion of U87wt-LacZ was 21.14% ± 3.39% (Fig. 2E,F; Supplemental Fig. 5). These results demonstrate that, by increasing the amount of ΔEGFR cells in mixed tumor cell engraftments, there is a corresponding increase in wtEGFR cell growth (Supplemental Fig. 5b). In contrast, the tumor volume attributable to ΔEGFR cells was proportional to the ratio injected. Similar results were obtained using a flow cytometry procedure designed to discriminate between wtEGFR- and ΔEGFR-expressing cells that also demonstrated a substantial unidirectional growth enhancement effect of U87Δ tumor cells on U87wt tumor cells (Supplemental Fig. 5c).
ΔEGFR activates proliferation and survival pathways in wtEGFR cells in vivo and in vitro
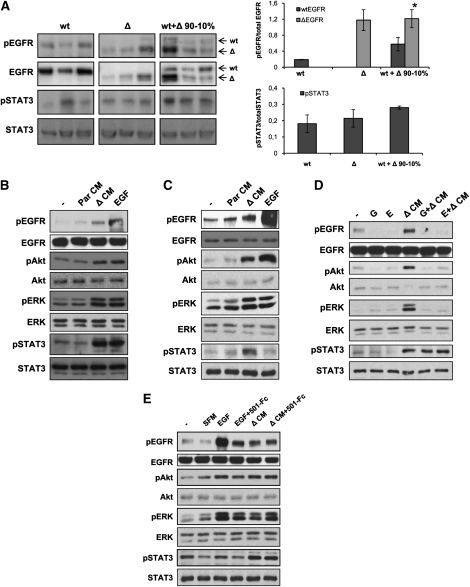
We observed a modest enhancement of tumorigenicity when U87Par cells were mixed with U87Δ cells (data not shown), illustrating that levels of wtEGFR expression, and probably activation state, might be important parameters in heterogeneous tumor growth potentiation. Analysis of intracranial and subcutaneous tumor lysates by Western blot and densitometric quantification revealed a stronger activation of wtEGFR in tumors originating from the coinjection of wtEGFR and ΔEGFR cells than in tumors that formed from any of the unmixed cell populations (U-Mann Whitney test, P = 0.0406) (Fig. 3A; Supplemental Fig. 6). Furthermore, in support of the unidirectionality of the cross-talk between these receptors, no increase in ΔEGFR activation was detected in engrafted tumors composed of wtEGFR- and ΔEGFR-expressing cells.
Figure 3.
EGFR and STAT3 are activated in mixed tumors and after in vitro treatment of wtEGFR cell with ΔEGFR cell CM. (A) Analysis of EGFR and STAT3 phosphorylation by Western blot (left) and densitometry (right) of tumor lysates obtained after intracranial injection of indicated U87 cell line derivative mixtures. Every lane represents a different tumor sample. Arrows indicate wtEGFR or ΔEGFR bands. Error bars represent mean ± SEM (n = 3). Mann-Whitney U-test was used to compare results. (*) P < 0.05. (B) Western blot analysis of U87wt lysates untreated (−) or treated with 50 ng/mL EGF, or U87Par CM (ParCM) or U87Δ CM (ΔCM) showing activation of EGFR, ERK, Akt, and STAT3 pathways when treated with ΔCM. (C) Western blot analysis of mAstr-Ink4/Arf−/−-wtEGFR lysates untreated (−) or treated with mAstr-Ink4/Arf−/−-Par CM (ParCM), mAstr-Ink4/Arf−/−-ΔEGFR CM (ΔCM), or 2.5 ng/mL EGF. (D) Western blot analysis of lysates of U87wt treated with U87Δ CM (ΔCM), with or without pretreatment with 2 μM EGFR inhibitors gefitinib (G) or erlotinib (E). (E) Western blot analysis of lysates of U87wt untreated (−) or treated with serum-free medium (SFM), EGF, or U87Δ CM (ΔCM), with or without preincubation with EGFR ligand trap (501-Fc). Actin and/or total protein were used as loading controls.
We attempted to model this intercellular communication in vitro to determine which pathways might be responsible for tumor enhancement. To recapitulate the function of ΔEGFR cells in vitro, wtEGFR cells were treated with CM from serum-starved ΔEGFR cells. Western blot analysis of the lysates revealed activation of EGFR and three known major signal transduction proteins involved in GBM tumorigenesis—Akt, ERK1/2, and STAT3 (Tyr705)—in both U87wt (Fig. 3B) and mAstr-Ink4a/Arf−/−-wtEGFR (Fig. 3C) cells, suggesting the possibility of equivalent mechanisms across species to promote tumor growth enhancement in the two distinctly different cellular systems. No activation of STAT3 at Ser727 was detected in any case (data not shown). Similar results were also obtained when analogous experiments were performed with U373 and U178 human GBM cell lines (Supplemental Fig. 7). In accord with the in vitro results, enhanced STAT3 activation was also observed in intracranial and subcutaneous mixed tumors (Fig. 3A; Supplemental Fig. 6). Notably, no differences in activation of STAT3 or wtEGFR were observed when U87wt cells were mixed with U87Par or U87DK (Supplemental Fig. 6) or treated with parental CM (Fig. 3B,C).
wtEGFR activation is independent of its natural ligands
To determine whether the activation of these intracellular pathways is dependent on the kinase activity of EGFR, we analyzed the effect of ΔCM in the presence of the EGFR inhibitors gefitinib and erlotinib (Fig. 3D). Pretreatment of U87wt cells with either EGFR inhibitor at 2 μM completely abolished the activation of EGFR, as well as Akt and ERK activation, while no effect was observed for STAT3 activation, suggesting that ΔEGFR cells produce soluble factors that activate Akt and ERK directly through EGFR, while STAT3 activation is presumably transduced by another receptor.
In order to test whether the EGFR activation is caused by any of its known ligands, we attempted to neutralize them in ΔCM using a recombinant protein consisting of the extracellular portion of EGFR that binds the known EGFR ligands with high affinity (Adams et al. 2009). As shown in Figure 3E, preincubation of recombinant EGF with the ligand trap 501-Fc reduced the activation of EGFR as well as Akt and ERK. However, no effect on these pathways was observed when ΔCM was pretreated with the ligand trap, suggesting that EGFR activation in treated wtEGFR cells is not caused by any of the EGFR ligands. As expected, STAT3 activation was not affected by the presence of the ligand trap. Furthermore, analysis of CM by ELISA revealed no significant increase in secretion by U87Δ of any of the EGFR ligands analyzed (Supplemental Fig. 8). Confirmation of these results was achieved by incubation of ΔCM with neutralizing antibodies against each EGFR ligand; these also failed to block the ability of the CM to activate EGFR (data not shown).
IL-6 and/or LIF are up-regulated in ΔEGFR cells
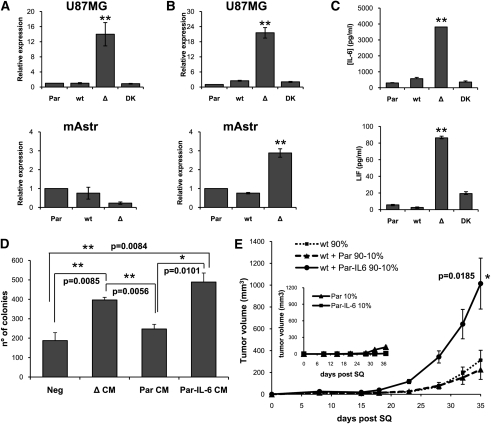
To identify candidate soluble factors expressed by ΔEGFR cells that could potentially mediate intercellular communication with, and promote the proliferation of, wtEGFR cells, a cytokine array was used to qualitatively detect 79 human cytokines and growth factors in supernatants of cultured cells. IL-6 was found to be significantly up-regulated in U87Δ cells compared with the other U87 cell lines (data not shown). Real-time PCR and ELISA analysis of supernatants demonstrated that IL-6 expression was significantly higher in U87 and U178 cells overexpressing ΔEGFR (P = 0.0014) (Fig. 4A,C; Supplemental Fig. 9). Since the IL-6 family of cytokines are known activators of STAT3, and we observed a strong STAT3-stimulating activity in ΔCM, we surveyed by PCR the expression of a panel of IL-6-related cytokines in the U87 series of cells, and found that the IL-6 family member LIF was also significantly up-regulated in ΔEGFR cells (P = 0.0008) (Supplemental Fig 10). Real-time PCR and multiplex analysis not only confirmed this result in U87 cells, but also revealed that LIF specifically is significantly up-regulated in both the mAstr-Ink4a/Arf−/−-ΔEGFR cells (P = 0.0007) and the U373 glioma cell line (Fig. 4B,C; Supplemental Fig. 9).
Figure 4.
IL-6 and LIF are up-regulated in ΔEGFR-expressing cells, and IL-6 overexpression enhances wtEGFR tumor growth and soft agar colony formation. Real-time PCR for IL-6 (A) and LIF (B) expression in U87MG and mAstr-Ink4/Arf−/− (Par) cells, or cells engineered to overexpress wtEGFR (wt), ΔEGFR (Δ), or ΔEGFR with a dead kinase domain (DK). (C) ELISA quantification of IL-6 (top) and multiplex quantification of LIF (bottom) in supernatants of U87Par (Par), U87wt (wt), U87Δ (Δ), and U87DK (DK) cultures after 48 h of starvation. (D) Soft agar colony formation assay quantification of U87wt cells treated with normal media (Neg) U87Δ CM (ΔCM), U87Par CM (ParCM), or U87Par-IL6 CM (Par-IL-6 CM). (E) Tumor growth kinetics after subcutaneous injection of U87wt 100%, or U87wt mixed with U87Par or U87Par-IL6 at a ratio of 90%:10% (100% = 1 × 106 cells). Inserted in the graph is the tumor growth kinetics for U87Par 10% and U87Par-IL6 10%. Error bars in all experiments represent mean ± SEM. One-way ANOVA and two-tail t-test were used to compare samples. (*) P < 0.05; (**) P < 0.001. n = 3 for soft agar assay; n = 6 for subcutaneous injections.
IL-6 enhances heterogeneous tumorigenic growth
To determine whether IL-6 was a candidate mediator for the enhanced proliferation of wtEGFR-expressing cells, we engineered U87Par cells, which lack the ability to enhance U87wt tumor growth (Fig. 1B), to overexpress IL-6 (U87Par-IL6), and their CM was applied to U87wt in soft agar colony formation assays (Fig. 4D,E). CM generated from U87Par-IL6 was able to potently enhance U87wt colony formation with the same efficiency as U87Δ CM (Fig. 4D). In contrast, significant colony enhancement failed to occur when U87wt cells were treated with control U87Par CM. To verify the potential role of IL-6 in tumor growth enhancement, U87wt cells were injected subcutaneously into nude mice after mixing with U87Par or U87Par-IL6 (in lieu of U87Δ) at 90:10 ratios (Fig. 4E). U87Par-IL6 did not itself grow faster than U87Par; however, when they were mixed with U87wt cells, a potent tumor growth enhancement was observed, demonstrating a paracrine tumor enhancement effect mediated by IL-6 secretion in these composite tumors.
ΔEGFR-induced cytokines operate through gp130 to promote wtEGFR-mediated growth enhancement
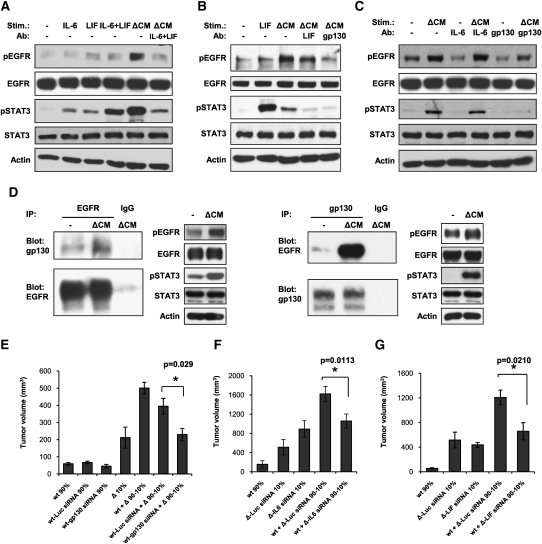
To determine if IL-6 and LIF were the factors present in ΔEGFR cell CM responsible for the signaling signature activated in wtEGFR cells, we sought to neutralize their activity by preincubating ΔEGFR CM with blocking antibodies targeting these cytokines prior to treatment of wtEGFR cells. Both antibodies were able to reduce, but not completely block, ΔEGFR CM-induced STAT3 activation in U87wt cells, suggesting an additive effect of the two cytokines (Supplemental Fig. 11). In contrast, U87Δ CM treated with either antibody retained the ability to activate EGFR and its downstream Akt and MAPK pathways (Supplemental Fig. 11; data not shown); however, the combination of the two neutralizing antibodies not only further inhibited the STAT3 activation, but also strongly prevented EGFR activation (Fig. 5A), showing that the two cytokines have a redundant stimulatory effect on the wtEGFR. To further validate these data, we similarly used a blocking antibody targeting gp130, the receptor subunit common to all IL-6 family cytokines. U87wt and mAstr-Ink4a/Arf−/−-wtEGFR cells preincubated with the anti-gp130 antibody and then treated with the respective ΔCM did not show any activation of STAT3, and EGFR activation was reduced dramatically (Fig. 5B,C). Similar results were achieved with U178 and U373 glioma cell lines (Supplemental Fig. 7).
Figure 5.
IL-6 and LIF are responsible for wtEGFR stimulation and growth enhancement via gp130–EGFR interaction. (A) Western blot analysis of EGFR and STAT3 phosphorylation in U87wt lysates untreated (−) or treated with recombinant IL-6, LIF, or both, or with U87Δ CM (ΔCM) pretreated or not with anti-IL-6-neutralizing and anti-LIF-neutralizing antibodies (IL-6n + LIFn). (B) Western blot analysis of EGFR and STAT3 phosphorylation in mAstr-Ink4/Arf−/−-wtEGFR lysates untreated (−) or treated with recombinant LIF or with mAstr-Ink4/Arf−/−-ΔEGFR CM (ΔCM) pretreated or not with anti-LIF-neutralizing (LIFn) or anti-gp130-neutralizing (gp130n) antibodies. (C) Western blot analysis of U87wt lysates after stimulation with U87Δ CM (ΔCM) pretreated or not with anti-IL-6-neutralizing (IL-6n) or anti-gp130-neutralizing (gp130n) antibodies. (D) Coimmunoprecipitation of EGFR and gp130 in U87wt lysates untreated (−) or treated with U87Δ CM (ΔCM). Total lysates were loaded as controls. IgG immunoprecipitation was used as negative control. (E) Tumor volumes 17 d after subcutaneous injection of U87wt (wt) nontransfected or transfected with 25 nM luciferase siRNA (wt-Luc siRNA) or gp130 siRNA (wt-gp130 siRNA) alone or mixed with U87Δ at a ratio 90:10, respectively. (F,G) Tumor volumes 24 d (F) or 21 d (G) after subcutaneous injection of U87wt alone or mixed with U87Δ transfected with 25 nM luciferase siRNA (Δ-Luc siRNA) and IL-6 siRNA (Δ-IL6 siRNA) (F), or LIF siRNA (Δ-LIF siRNA) (G) at a ratio of 90:10, respectively (100% = 1 × 106 cells). Error bars represent mean ± SEM. One-way ANOVA and two-tail t-test were used for statistical analysis; n = 6. (*) P < 0.05; (**) P < 0.001.
Although a direct cross-talk between gp130 and EGFR has not been described previously, it has been shown that gp130 can interact with ErbB2, another member of the EGFR family (Qiu et al. 1998). We hypothesized that gp130 might also interact directly with overexpressed EGFR and induce its activation. Coimmunoprecipitation experiments demonstrated that the two receptors do interact, and that their association is stimulated by ΔCM treatment (Fig. 5D). Since all of the ΔCM-activated signaling pathways that we observed in vitro depend on the stimulation of gp130, we speculated that gp130 and its ligands play a central role in the in vivo tumor growth enhancement observed in mixed tumors. To address this, we knocked down the expression of gp130 in U87wt cells by siRNA transfection (Supplemental Fig. 12a), and injected these cells subcutaneously alone or mixed with U87Δ cells at a ratio of 90:10%. U87wt cells that were not transfected or those transfected with an irrelevant luciferase siRNA were used as negative controls. There was only a small and insignificant (P = 0.1487) difference in tumor volumes between U87wt cells treated with gp130 siRNA compared with the controls when injected alone (Fig. 5E; Supplemental Fig. 13). However, tumor growth enhancement driven by the presence of ΔEGFR cells was abolished completely when gp130 was knocked down in U87wt cells (P = 0.029) (Fig. 5E; Supplemental Fig. 13), indicating that, in the absence of gp130, wtEGFR cells are unresponsive to the growth-promoting stimuli produced by ΔEGFR cells.
To further demonstrate that IL-6-type cytokines are the soluble factors responsible for the paracrine proliferation of U87wt cells, U87Δ cells were transfected with a combination of two siRNAs targeting IL-6 (Supplemental Fig. 12b), or luciferase siRNA as a negative control, and then injected subcutaneously into nude mice alone (10%) or mixed with U87wt cells at a ratio of 90%:10% (Fig. 5F; Supplemental Fig. 13). U87Δ cells transfected with IL-6 siRNAs formed tumors as efficiently as luciferase siRNA controls when injected alone (P > 0.05); however, they were unable to stimulate the growth of U87wt cells when mixed together (P = 0.0113) (Fig. 5F; Supplemental Fig. 13). Similar results were obtained by knocking down LIF expression in U87Δ cells and subcutaneous engraftment (P = 0.0210) (Fig. 5G; Supplemental Figs. 12c, 13). Taken together, these results show that the amplified levels of wtEGFR expressed in glioma can be activated through a paracrine mechanism initiated by ΔEGFR activity. This activity results in the production of IL-6/LIF family member cytokines that engage their cognate common receptor, gp130, and promote cross-talk interaction with EGFR, the net outcome of which is an active maintenance of tumor heterogeneity.
IL-6 and LIF expression in GBM clinical samples and glioma stem cells
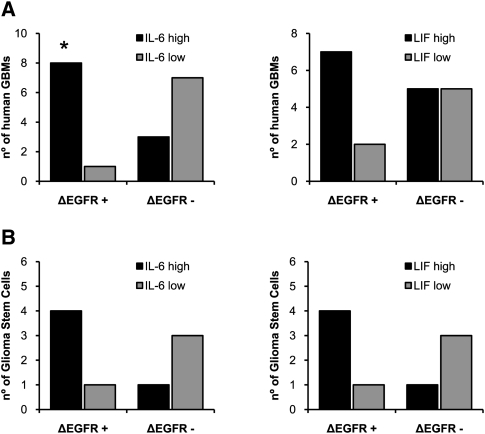
To determine whether these findings had clinical correlates, 19 human GBM tumor samples were analyzed for wtEGFR, ΔEGFR, IL-6, and LIF RNA expression by real-time PCR, and were compared with normal brain tissue (Fig. 6A, left). Notably, we observed a very significant correlation between ΔEGFR and IL-6 expression in the GBM tumor samples (P = 0.0198). High LIF expression was also observed frequently in ΔEGFR-positive clinical samples, but did not reach statistical analysis significance (P = 0.2101) (Fig. 6A, right). Each of the tumor samples analyzed that was positive for ΔEGFR, except one, expressed high levels of IL-6 and/or LIF (Fig. 6A). In contrast, no correlation was found between the two cytokines and wtEGFR expression (Supplemental Fig. 14). Nine different glioma stem cell lines were similarly analyzed by real-time PCR, and all of the lines expressing ΔEGFR also had higher expression of IL-6 (four out of five) and/or LIF (four out of five) (Fig. 6B). In contrast, lines not expressing ΔEGFR did not express high levels of IL-6 or LIF (three out of four), and only one line with amplification of wtEGFR also showed somewhat increased levels of IL-6 and LIF.
Figure 6.
IL-6 and/or LIF expression correlates with ΔEGFR-positive human GBM clinical samples and glioma stem cells. (A) Analysis of ΔEGFR and IL-6 (left) or LIF (right) expression by real-time PCR in 19 human GBM samples. (B) Analysis of ΔEGFR and IL-6 (left) or LIF (right) expression by real-time PCR in nine human glioma stem cell lines. Error bars in all experiments represent mean ± SEM. Expression levels three times or more the expression in normal brain were considered as high expression. (*) P < 0.05., Fisher's exact test.
Discussion
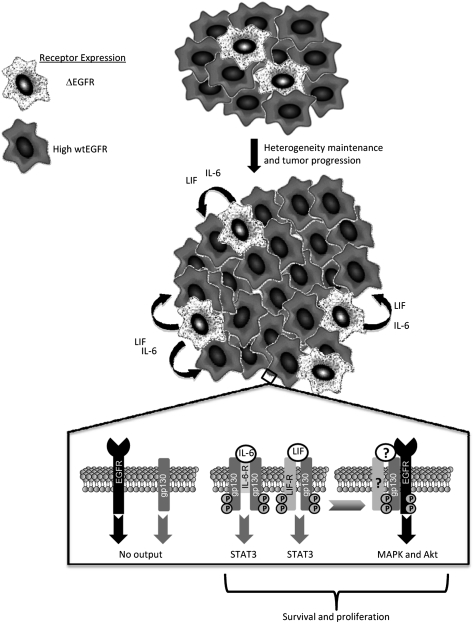
While much attention has been placed on tumors as architecturally heterogeneous systems that differ regionally in vasculature, host infiltrates, and connective tissue components (Allinen et al. 2004), far less is known about how genetically different cancer cells of an individual tumor interact in the malignant process. A cardinal feature of GBM is its profound intratumoral genetic heterogeneity, a property that challenges our understanding of its pathobiology and our ability to effect meaningful therapeutic responses to targeted agents. Here, we show that a signature genetic event of GBM, ΔEGFR, provides a mechanism to actively maintain this complex tumor biology by both its cell-intrinsic growth-promoting activity and a cell-extrinsic function that serves to promote the proliferation of wtEGFR-expressing glioma cells via ΔEGFR-mediated production of IL-6 family cytokines (Fig. 7). While our results support a paracrine mechanism, whereby cytokines are produced from ΔEGFR-expressing cells to act upon neighboring cells expressing amplified wtEGFR, it is formerly possible that an autocrine mechanism may also exist in these tumors where ΔEGFR and wtEGFR are coexpressed on the same cell. This would not invalidate our finding of a major role for a paracrine mechanism induced by ΔEGFR-positive cells, as it has been demonstrated that, within GBMs, there are regions of predominant wtEGFR-only expression (Nishikawa et al. 2004; Heimberger et al. 2005; Jeuken et al. 2009), and so these cells would also benefit from the cytokines secreted by ΔEGFR-positive cells. Therefore, it remains possible that both paracrine and autocrine circuitries coexist to maintain receptor heterogeneity.
Figure 7.
Model of paracrine-mediated tumor enhancement and maintenance of glioma heterogeneity. (Top panel) Cells expressing ΔEGFR (stippled) secrete elevated levels of IL-6 family cytokines IL-6 and/or LIF, which promote the in vivo growth of cells with high levels of wtEGFR (dark grey), thus maintaining the heterogenic composition of the tumor. At the molecular level, IL-6 and LIF bind to their respective receptors, IL-6R and LIFR, which form oligomeric complexes with two or one subunits, respectively, of the common signal transducer gp130. Upon ligand binding, gp130 complexes recruit and activate JAK kinases (not shown), which phosphorylate gp130, LIFR, and STAT3 at Tyr705, inducing its transcriptional activation function. Activated gp130 interacts with and transactivates EGFR, leading to receptor-mediated signaling pathway activation (Akt and MAPK activation). (?) The participation of IL-6R and LIFR, as well as their ligands, in gp130:EGFR complexes has yet to be determined. Coordinate activation of STAT3, Akt, and MAPK enhance the survival and proliferation of cells expressing amplified wtEGFR.
Our findings mirror well the cytokine expression profiles in ΔEGFR-expressing GBM clinical samples and derivative glioma stem cells (Fig. 6). This novel function of ΔEGFR provides a rational explanation for the receptor heterogeneity often detected in human brain tumors. We propose that tumor heterogeneity can be maintained by cooperative interactions among tumor cells of differing genotypes and proliferative capacities, and that the elucidation of such interactions provides a new avenue for therapeutic intervention. Together, our findings refine the clonal evolution model of cancer positing a stepwise natural selection for the genetically fittest, most aggressive cancer cells, which leave in their wake less aggressive ancestral cells, and instead support the existence of nonautonomous mechanisms supporting growth across the cancer cell population. Our results do not address the origins of heterogeneity, and leave open the issue of how amplified EGFR and ΔEGFR arise during the establishment of a glioma. Possible explanations for this might be that, at an early stage during glioma development, amplified wtEGFR is better tolerated then ΔEGFR, and that other lesions need to occur before cells expressing ΔEGFR can survive. Alternatively, ΔEGFR might more easily arise from an amplified EGFR locus.
ΔEGFR expression is rare in tumors that do not also possess cells with wtEGFR overexpression, and the presence of this mutant receptor in a subpopulation of cancer cells of a GBM confers poor clinical outcome (Shinojima et al. 2003; Heimberger et al. 2005; Jeuken et al. 2009). In concordance, we observed that coinjection of wtEGFR cells with a smaller number of ΔEGFR cells potently enhances tumorigenicity and aggressiveness in several different cellular systems. To determine whether the cellular cross-talk demonstrated in this model was due to a ΔEGFR-prompted secreted factor, we assessed the ability of CM generated from ΔEGFR cells to activate overexpressed wtEGFR on recipient cells. While the CM activated wtEGFR, using various methods, we were unable to detect the presence or altered expression of any known EGF ligand. These results are in contrast to Ramnarain et al. (2006), who detected autocrine activation of wtEGFR by HB-EGF secreted by U251 glioma cells expressing ΔEGFR, possibly due to the different genetic backgrounds of U251 (mutated for p53) and U87MG and mouse astrocytes (wild type for p53) used in our study that more accurately represent the genetic profile of most primary GBMs expressing ΔEGFR (Furnari et al. 2007). Additionally, analysis of U87Δ-generated CM and FACS analysis for receptor composition in cocultures of U87wt with U87Δ cells or tumors derived from the mix of these cells failed to demonstrate the transfer of ΔEGFR protein shed as vesicles (Al-Nedawi et al. 2008; data not shown).
Overexpressed wtEGFR conveys little intrinsic tumorigenic enhancement to glioma cells (Huang et al. 1997), and overexpressed levels of this receptor are not sufficient to drive glioma formation with Ink4a/Arf−/− mouse astrocytes (Bachoo et al. 2002) unless EGF is infused into the intracranial engraftment site. This result suggests that the abundant expression of this wild-type receptor in GBMs, unlike the mutant ΔEGFR, requires additional inputs, such as ligand or cross-talk activation. By analyzing lysates from xenografts composed of wtEGFR cells alone or mixed with ΔEGFR cells, we found that wtEGFR was significantly more phosphorylated in the latter. To ascertain if ΔEGFR cells could provoke sufficient wtEGFR signaling, thus promoting tumorigenic growth, we tested CM produced by ΔEGFR cells for the ability to activate the wtEGFR. Regardless of the cell type used, CM derived from ΔEGFR-engineered cells was capable of activating wtEGFR as well as signature growth-promoting pathways (Akt, MAPK, and STAT3) typically associated with EGFR activation and gliomagenesis (Guha and Mukherjee 2004; Brantley and Benveniste 2008; Huang et al. 2009). Inhibition of EGFR activation by erlotinib or gefitinib blocked Akt and MAPK activation generated by U87Δ CM, while STAT3 activation was still achieved. As we found the CM to be devoid of EGF ligands, we conclude that receptor cross-talk is responsible for ΔEGFR CM-prompted EGFR activation (Gschwind et al. 2001). Furthermore, given the inability of CM produced from wtEGFR-expressing cells to promote enhanced colony formation of ΔEGFR-expressing cells, we conclude that cellular cross-talk is unidirectional.
We analyzed supernatants of human glioma cell lines, immortalized mouse astrocytes, and glioma stem cells engineered to overexpress ΔEGFR, and, among several soluble factors that we found to be up-regulated, the IL-6-type cytokines were the most frequent and constant across the different cell types. Notably, we observed up-regulation of only two of the cytokines of this family: IL-6 and LIF. Several previous studies (Van Meir et al. 1990; Rolhion et al. 2001; Trikha et al. 2003) have demonstrated that IL-6 expression increases with glioma malignancy grade, is associated with shorter survival of GBM patients (Sasaki et al. 2001; Tchirkov et al. 2001, 2007; Chang et al. 2005), promotes proliferation of glioma cells (Goswami et al. 1998) and neuroectodermal tumors (Candi et al. 1997), and is required for glioma development in the mouse (Weissenberger et al. 2004). However, few reports have indicated a possible involvement of LIF in CNS tumor growth (Liu et al. 1999, 2000). Only recently has LIF been identified as a crucial mediator of the self-renewal capacity of glioma stem cells (Penuelas et al. 2009), in agreement with its previously known effect on normal neural stem cells (Bauer and Patterson 2006). In the same study, it was demonstrated that LIF can actively increase the oncogenic potential of glioma stem cells (Penuelas et al. 2009). Importantly, gp130 (Wang et al. 2009) and receptors for IL-6 (IL6-R) (Wang et al. 2009) and LIF (LIFR) (Krona et al. 2005) are expressed in glioma tissues and derived stem cell cultures, thus underscoring that all the components of the cytokine circuitry we identified are associated with the tumor cell itself. Recently, it has been shown that IL-6 also increases neurosphere self-renewal (Wang et al. 2009). These results raise the intriguing question of whether ΔEGFR-prompted expression of IL-6 and/or LIF might enhance tumorigenicity by assisting in the maintenance of tumor stem cell populations. Moreover, we found that IL-6 and LIF expression correlate with ΔEGFR in glioma stem cells, suggesting a potential autocrine self-renewal mechanism.
Overexpression of IL-6 in our model confirmed the ability of this cytokine to increase tumor growth of wtEGFR cells. Similarly, knocking down this cytokine by siRNA in ΔEGFR cells ablated their capacity to enhance wtEGFR tumor growth, raising the possibility of a novel therapeutic approach to uncouple cross-talk between ΔEGFR and wtEGFR-expressing cells by inhibiting IL-6 activity using antibodies that bind IL-6R (Kudo et al. 2009). IL-6-type cytokines are well known potent activators of the JAK/STAT3 pathway (Schaefer et al. 2000; Heinrich et al. 2003), the inhibition of which has been shown to promote apoptosis in GBM cells (Rahaman et al. 2002). Indeed, these cytokines proved to be responsible for STAT3 activation induced by ΔCM. However, the blockade of these cytokines or their common receptor subunit, gp130, by neutralizing antibodies drastically reduced the ability of ΔCM to activate wtEGFR, demonstrating the existence of a previously unknown mechanism of cross-talk between these two receptors. It is noteworthy that the treatment of wtEGFR cells with recombinant IL-6 or LIF induced poor activation, if any, of EGFR, while they potently activated STAT3 phosphorylation. This disconnect might be explained by the presence of a third factor necessary to produce the stimulation, perhaps simply an extracellular matrix component (Rathjen et al. 1990). In this view, IL-6 and LIF would be necessary but not sufficient to activate EGFR. Another possibility could be that these cytokines must be sufficiently glycosylated to convey full biological activity (May et al. 1989). Although the most common mechanism of transactivation of EGFR implies the cleavage and release in active form of membrane-bound EGFR ligand precursors present on the target cells, a process known as ectodomain shedding (Gschwind et al. 2001), we found that ligand traps failed to even slightly reduce the stimulatory activity of ΔCM, and by ELISA we did not detect any increase in EGFR ligands in ΔCM pre- and post-application to wtEGFR-expressing cells (data not shown). We thus hypothesized and demonstrated that the transactivation could be the result of the interaction between EGFR and the only subunit common to IL-6 and LIF receptor complexes: gp130. This is, to our knowledge, the first reported evidence of a physical interaction between these two receptors. It had been shown previously by Qiu et al. (1998) that gp130 does dimerize with another member of the ErbB receptor family, ErbB2, and that an inhibitor of ErbB2 tyrosine kinase activity prevented its phosphorylation, suggesting that gp130 can induce autophosphorylation of ErbB receptors. We observed that ΔCM can also induce this previously described gp130–ErbB2 complex formation in U87 cells; however, we failed to prevent EGFR activation by using antibodies that block the EGFR-ErbB2 dimerization (data not shown). These results indicate that ErbB2 is not required to activate EGFR, and that the gp130–EGFR interaction, which promotes enhanced tumorigenicity, is most likely direct and potently driven by glioma-relevant levels of wtEGFR. Indeed, similarly for ΔEGFR, the levels of this mutant receptor expressed per cell also seem to be a critical determinant for its signaling and tumor-enhancing potential, as we showed recently for threshold-level activation of c-Met and pathway switching from MAPK and STAT3 to PI3K at a high level of ΔEGFR expression (Huang et al. 2007; Johns et al. 2007). Lastly, cross-talk between the JAK/STAT pathway and EGFR has also been described, in which JAK2 has been shown to directly phosphorylate Tyr1068 on EGFR when activated by growth hormone (Yamauchi et al. 1997). However, JAK kinase inhibitors had no effect on the ability of ΔCM to activate EGFR, while they were potent in blocking STAT3 activation (data not shown).
In summary, we identified a network actively driven by ΔEGFR that promotes and maintains the receptor heterogeneity commonly found in GBMs. Our analyses of different cellular systems provided evidence that the more tumorigenic cells overexpressing ΔEGFR generate a communication link through IL-6 family cytokines to neighboring cells that converges on gp130, thus revealing a new cell-extrinsic role for this mutant receptor as a mediator of tumor growth-promoting stimuli. Our analysis of 19 GBM tumor samples and nine glioma stem cell lines further supports the hypothesized role for these cytokines and their clinical relevance. Knockdown of gp130 expression in wtEGFR completely prevented the tumor growth enhancement usually achieved in mixed composition tumors. These observations suggest gp130 as a potential new target for the treatment of gliomas. Further clinical assessment of targeting these secreted molecules should illuminate the role of cytokines or other factors in driving this complex glioma phenotype, and provide new avenues for therapeutic intervention in which the interacting receptors and the cytokine signals they use are targeted conjointly.
Materials and methods
Cell lines and plasmids
U87Par, U87Δ, U87DK, U87wt, mAstr-Ink4a/Arf−/−-wtEGFR, and mAstr-Ink4a/Arf−/−-ΔEGFR were obtained as described previously (Nishikawa et al. 1994). Where indicated, cells were infected with retroviruses expressing the LacZ gene. The pCEP4/H2B-EGFP plasmid (gift from Dr Shi-Yuan Cheng, University of Pittsburgh) was used to tag astrocytes expressing wtEGFR with nuclear GFP. Glioma stem cell cultures were generated from fresh tumor tissue using standard techniques from patients who consented prior to surgery under IRB-approved protocol. Minced tumor tissue was placed in Accumax for 45 min at room temperature, and gently dissociated every 15 min with a 5-mL serological pipette. Supernatant was removed by centrifugation, and cells were resuspended in Complete NeuroCult NS-A Proliferation Medium that contains NeuroCult NS-A Basal Medium, NeuroCult NS-A Proliferation Supplements, 20 ng/mL rh-EGF, 10 ng/mL rh-bFGF, and 2 μg/mL heparin (StemCell Tehcnologies). Resuspended cells were passed through a 70-μm strainer and plated in six-well plates, and were maintained and expanded in Complete NeuroCult Medium.
Intracranial and subcutaneous injection
A total of 5 × 105 cells (100%) in a 5-μL volume were injected intracranially into 4- to 5-wk-old athymic nude mice using a guide screw system according to the protocol described by Lal et al. (2000). For subcutaneous injection, 1 × 106 cells (100%) suspended in 100 μL of PBS were injected into the right flank of 4- to 5-wk-old athymic nude mice. Tumors were measured with a vernier caliper, and volumes were calculated using width (a) and length (b) measurements (V = a2 × b/2, where a ≤ b). Mice were euthanized in accordance with our institutional guidelines for animal welfare and experimental conduct. Survival until the onset of neurologic sequelae was used for intracranial injections. For subcutaneous injections, mice were euthanized when tumor volume reached 1500 mm3 or tumors became ulcerated.
GFP immunofluorescence and X-Gal staining
Tumor samples were fixed in 4% PFA and embedded in OCT, and 6-μm cryostat sections were obtained. GFP immunofluorescence and X-Gal staining were performed by conventional methods. Images were analyzed using Image Pro-Analyzer 6.2 software.
Antibodies
Antibodies used were as follows: anti-phospho-tyrosine (pEGFR) (4G10) and anti-gp130 (immunoprecipitation and Western blot) were obtained from Upstate Biotechnologies; anti-phospho-Akt (S473), total Akt, phospho-STAT3 (Y705), total STAT3, phospho-ERK (T202/Y204), and total ERK were obtained from Cell Signaling; antiβ-actin was obtained from Sigma; anti-IL-6, anti-gp130 (neutralization), and anti-LIF were obtained from R&D Systems; rabbit-anti-GFP was obtained from Molecular Probes; and anti-EGFR was obtained from BD Biosciences (Western blot) and Calbiochem (immunoprecipitation).
Flow cytometry analysis
Tumors harvested from euthanized mice were processed for flow cytometry using a Medimachine system and 50-μm Medicon filters (BD Biosciences). Resulting single-cell suspensions were stained for wtEGFR and ΔEGFR, and for wtEGFR with Ab-1 (clone 528) and Ab-5 (clone EGFR.1) (Calbiochem), respectively. After 1 h of incubation at 4°C, cells were washed in 5% BSA and incubated for 1 h at 4°C with IgG isoform-specific secondary antibodies conjugated with either FITC (anti-IgG 2a) or APC (anti-IgG 2b) (Jackson ImmunoResearch). Stained cells were analyzed with a FACS LSR II using FACS DIVA software (BD Biosciences).
Western blot and immunoprecipitation
Cells were seeded at equal densities, serum-starved for 48 h, treated with CM or recombinant proteins for 15 min at 37°C, and harvested in RIPA buffer, and cell lysates were analyzed by Western blot. CM was prepared fresh from identically prepared cells, and was collected and cleared of debris by centrifugation. Where indicated, CM was preincubated with IL-6- and/or LIF-neutralizing antibodies for 30 min at 4°C. For gp130 neutralization, cells were incubated with antibody for 30 min prior to treatment. Gefitinib and erlotinib were added to the cells 15 min before the treatment at 2 μM concentration. For immunoprecipitation, cells were lysated in 1% NP-40 or 1% Brij97. Two milligrams of proteins was precleared and incubated overnight with 2 μg of antibody plus 25 μL of protein G-sepharose, washed, and resolved by SDS-PAGE.
ELISA and Multiplex
Cells were seeded in 24-well plates at 4 × 105 cells per well and serum-starved. Serum-free media was changed daily. On the second day, media was collected and centrifuged to remove cell debris. ELISA was performed in 96-well plates (Nunc), coated overnight with capture antibodies. Plates were blocked, incubated with standards and CM samples, and incubated with the biotinylated detection antibodies followed by streptavidin-HRP (Biosource). The conversion of the substrate tetramethylbenzidine (Sigma), terminated with 1N sulfuric acid, was measured by a Tecan Genios-Pro microplate reader at 450 nm with wavelength correction set to 540 nm. The absorbance readings from standards were converted using a four-parameter logistic curve. Bio-Rad human LIF multiplex kit was used according to the manufacturer's instructions.
siRNA transfection and ex vivo delivery
Cells were transfected with 25 nM siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and were injected subcutaneously into nude mice. Luciferase siRNA was used as negative control. Luciferase, LIF, and gp130 siRNAs were purchased from Dharmacon. IL-6 siRNAs were synthesized by Alnylam Pharmaceuticals on an ABI3900 DNA synthesizer using standard procedures, purified by AEX HPLC, and annealed in PBS (Ambion). IL-6 siRNA sequences were siRNA#1: sense, 5′-AuGAGcGuuAGGAcAcuAudTsdT-3′, and antisense, 5′-AuAGUGUCCuAACGCUcAUdTsdT-3′; siRNA#2: sense, 5′- GAGcGuuAGGAcAcuAuuudTsdT-3′, and antisense, 5′-AAAuAGUGUCCuAACGCUCdTsdT-3′ (lowercase letters indicate 2′-O-methyl modification, S indicates phosphorothioate linkage, and dT indicates deoxythymidine). Effectiveness of siRNAs was tested by Western blot, ELISA, and/or real-time PCR (Supplemental Fig. 12).
Soft agar colony formation assay
Soft agar colony formation assays were performed in six-well plates, with a bottom layer containing 0.6% agar in 1× DMEM/10% FBS. The upper layer, containing 2.5 × 103 wtEGFR cells, was prepared by mixing equal volumes of 1.2% agar, 2× DMEM/20% FBS, and CM or normal media. CM treatment media was added on top of the agar and replaced weekly. Every treatment was performed in triplicate. After 3–6 wk, plates were stained with 0.005% crystal violet solution, and colonies were counted using Image Pro-Analyzer 6.2 Software.
RNA extraction and retrotranscription
Total RNA from 19 frozen human GBM tumor samples and one normal brain tissue was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA from cell lines was extracted using RNeasy Plus Mini Kit (Qiagen). Two micrograms to 5 μg of RNA was retrotranscribed using SuperScript II transcriptase (Invitrogen).
Real time PCR
Real-time PCR was performed with 2 μL of diluted cDNA using iQTM SYBR Green Supermix (Bio-Rad) on an iCycler (Bio-Rad) following the manufacturer's instructions. All reactions were performed in duplicate and repeated at least three times. Primers used for real-time PCR are described in the Supplemental Material (Supplemental Table 1). Relative quantification was performed for each sample and normalized with GAPDH expression for comparison.
Statistical analysis
All data were analyzed for significance using GraphPad Prism 5.0 software, where P < 0.05 was considered statistically significant. One-way ANOVA and two-tail t-test were used to compare groups. Real-time PCR results were analyzed by Fisher's exact test. Densitometry results were analyzed by Mann-Whitney U-test.
Acknowledgments
We thank Dr. Keith Ligon for helpful discussions, Dr. Tim Fenton for technical assistance and advice, and Dr. Jayne Stommel and Alicia Pedraza for providing neurosphere cultures. This work was supported by an award from the Goldhirsh Foundation (to F.B.F.) and NIH Grant P01-CA95616 (to W.K.C., R.A.D., F.B.F., C.B.). W.K.C. is a fellow of the National Foundation for Cancer Research. M.M.I. thanks the Gobierno de Navarra, Spain, and the American Brain Tumor Association in Honor of Walter Terlik for the fellowships received. R.B. was supported by a fellowship from Federazione Italiana Ricerca Cancro. R.A.D. is supported by the Robert A. and Renee E. Belfer Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1890510.
Supplemental material is available at http://www.genesdev.org.
References
- Adams TE, Koziolek EJ, Hoyne PH, Bentley JD, Lu L, Lovrecz G, Ward CW, Lee FT, Scott AM, Nash AD, et al. 2009. A truncated soluble epidermal growth factor receptor-Fc fusion ligand trap displays anti-tumour activity in vivo. Growth Factors 27: 141–154 [DOI] [PubMed] [Google Scholar]
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. 2004. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6: 17–32 [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J 2008. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10: 619–624 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269–277 [DOI] [PubMed] [Google Scholar]
- Bauer S, Patterson PH 2006. Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J Neurosci 26: 12089–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H 2004. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol 14: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantley EC, Benveniste EN 2008. Signal transducer and activator of transcription-3: A molecular hub for signaling pathways in gliomas. Mol Cancer Res 6: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Knight RA, Spinedi A, Guerrieri P, Melino G 1997. A possible growth factor role of IL-6 in neuroectodermal tumours. J Neurooncol 31: 115–122 [DOI] [PubMed] [Google Scholar]
- Chang CY, Li MC, Liao SL, Huang YL, Shen CC, Pan HC 2005. Prognostic and clinical implication of IL-6 expression in glioblastoma multiforme. J Clin Neurosci 12: 930–933 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Goswami S, Gupta A, Sharma SK 1998. Interleukin-6-mediated autocrine growth promotion in human glioblastoma multiforme cell line U87MG. J Neurochem 71: 1837–1845 [DOI] [PubMed] [Google Scholar]
- Gschwind A, Zwick E, Prenzel N, Leserer M, Ullrich A 2001. Cell communication networks: Epidermal growth factor receptor transactivation as the paradigm for interreceptor signal transmission. Oncogene 20: 1594–1600 [DOI] [PubMed] [Google Scholar]
- Guha A, Mukherjee J 2004. Advances in the biology of astrocytomas. Curr Opin Neurol 17: 655–662 [DOI] [PubMed] [Google Scholar]
- Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K 2005. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 11: 1462–1466 [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 374: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselager G, Holland EC 2003. Using mice to decipher the molecular genetics of brain tumors. Neurosurgery 53: 685–694 [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12: 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK 1997. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem 272: 2927–2935 [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci 104: 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Xu AM, White FM 2009. Oncogenic EGFR signaling networks in glioma. Sci Signal 2: re6 doi: 10.1126/scisignal.287re6 [DOI] [PubMed] [Google Scholar]
- Hurtt MR, Moossy J, Donovan-Peluso M, Locker J 1992. Amplification of epidermal growth factor receptor gene in gliomas: Histopathology and prognosis. J Neuropathol Exp Neurol 51: 84–90 [DOI] [PubMed] [Google Scholar]
- Jaros E, Perry RH, Adam L, Kelly PJ, Crawford PJ, Kalbag RM, Mendelow AD, Sengupta RP, Pearson AD 1992. Prognostic implications of p53 protein, epidermal growth factor receptor, and Ki-67 labelling in brain tumours. Br J Cancer 66: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken J, Sijben A, Alenda C, Rijntjes J, Dekkers M, Boots-Sprenger S, McLendon R, Wesseling P 2009. Robust detection of EGFR copy number changes and EGFR variant III: Technical aspects and relevance for glioma diagnostics. Brain Pathol 19: 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns TG, Perera RM, Vernes SC, Vitali AA, Cao DX, Cavenee WK, Scott AM, Furnari FB 2007. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res 13: 1911–1925 [DOI] [PubMed] [Google Scholar]
- Jung V, Romeike BF, Henn W, Feiden W, Moringlane JR, Zang KD, Urbschat S 1999. Evidence of focal genetic microheterogeneity in glioblastoma multiforme by area-specific CGH on microdissected tumor cells. J Neuropathol Exp Neurol 58: 993–999 [DOI] [PubMed] [Google Scholar]
- Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK 2002. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol 61: 215–225 [DOI] [PubMed] [Google Scholar]
- Krona A, Jarnum S, Salford LG, Widegren B, Aman P 2005. Oncostatin M signaling in human glioma cell lines. Oncol Rep 13: 807–811 [PubMed] [Google Scholar]
- Kudo M, Jono H, Shinriki S, Yano S, Nakamura H, Makino K, Hide T, Muta D, Ueda M, Ota K, et al. 2009. Antitumor effect of humanized anti-interleukin-6 receptor antibody (tocilizumab) on glioma cell proliferation. Laboratory investigation. J Neurosurg 111: 219–225 [DOI] [PubMed] [Google Scholar]
- Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF 2000. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg 92: 326–333 [DOI] [PubMed] [Google Scholar]
- Liu J, Li JW, Gang Y, Guo L, Li H 1999. Expression of leukemia-inhibitory factor as an autocrinal growth factor in human medulloblastomas. J Cancer Res Clin Oncol 125: 475–480 [DOI] [PubMed] [Google Scholar]
- Liu J, Guo L, Luo Y, Li JW, Li H 2000. All trans-retinoic acid suppresses in vitro growth and down-regulates LIF gene expression as well as telomerase activity of human medulloblastoma cells. Anticancer Res 20: 2659–2664 [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA 2001. Malignant glioma: Genetics and biology of a grave matter. Genes Dev 15: 1311–1333 [DOI] [PubMed] [Google Scholar]
- May LT, Santhanam U, Tatter SB, Ghrayeb J, Sehgal PB 1989. Multiple forms of human interleukin-6. Phosphoglycoproteins secreted by many different tissues. Ann N Y Acad Sci 557: 114–119 [PubMed] [Google Scholar]
- Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ 1996. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res 56: 5079–5086 [PubMed] [Google Scholar]
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ 1998. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci 95: 5724–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK 2002. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res 62: 6764–6769 [PubMed] [Google Scholar]
- Network CGAR 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ 1994. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci 91: 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M 2004. Immunohistochemical analysis of the mutant epidermal growth factor, ΔEGFR, in glioblastoma. Brain Tumor Pathol 21: 53–56 [DOI] [PubMed] [Google Scholar]
- Pandita A, Aldape KD, Zadeh G, Guha A, James CD 2004. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer 39: 29–36 [DOI] [PubMed] [Google Scholar]
- Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J, et al. 2009. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15: 315–327 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Ravi L, Kung HJ 1998. Requirement of ErbB2 for signalling by interleukin-6 in prostate carcinoma cells. Nature 393: 83–85 [DOI] [PubMed] [Google Scholar]
- Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ 2002. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 21: 8404–8413 [DOI] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. 2006. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res 66: 867–874 [DOI] [PubMed] [Google Scholar]
- Rathjen PD, Toth S, Willis A, Heath JK, Smith AG 1990. Differentiation inhibiting activity is produced in matrix-associated and diffusible forms that are generated by alternate promoter usage. Cell 62: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Rolhion C, Penault-Llorca F, Kemeny JL, Lemaire JJ, Jullien C, Labit-Bouvier C, Finat-Duclos F, Verrelle P 2001. Interleukin-6 overexpression as a marker of malignancy in human gliomas. J Neurosurg 94: 97–101 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ishiuchi S, Kanda T, Hasegawa M, Nakazato Y 2001. Analysis of interleukin-6 gene expression in primary human gliomas, glioblastoma xenografts, and glioblastoma cell lines. Brain Tumor Pathol 18: 13–21 [DOI] [PubMed] [Google Scholar]
- Schaefer LK, Menter DG, Schaefer TS 2000. Activation of stat3 and stat1 DNA binding and transcriptional activity in human brain tumour cell lines by gp130 cytokines. Cell Signal 12: 143–151 [DOI] [PubMed] [Google Scholar]
- Schlegel J, Merdes A, Stumm G, Albert FK, Forsting M, Hynes N, Kiessling M 1994. Amplification of the epidermal-growth-factor-receptor gene correlates with different growth behaviour in human glioblastoma. Int J Cancer 56: 72–77 [DOI] [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. 2003. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res 63: 6962–6970 [PubMed] [Google Scholar]
- Tchirkov A, Rolhion C, Bertrand S, Dore JF, Dubost JJ, Verrelle P 2001. IL-6 gene amplification and expression in human glioblastomas. Br J Cancer 85: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchirkov A, Khalil T, Chautard E, Mokhtari K, Veronese L, Irthum B, Vago P, Kemeny JL, Verrelle P 2007. Interleukin-6 gene amplification and shortened survival in glioblastoma patients. Br J Cancer 96: 474–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, de Sauvage FJ 2009. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci 106: 4254–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trikha M, Corringham R, Klein B, Rossi JF 2003. Targeted anti-interleukin-6 monoclonal antibody therapy for cancer: A review of the rationale and clinical evidence. Clin Cancer Res 9: 4653–4665 [PMC free article] [PubMed] [Google Scholar]
- Van Meir E, Sawamura Y, Diserens AC, Hamou MF, de Tribolet N 1990. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res 50: 6683–6688 [PubMed] [Google Scholar]
- Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, et al. 2009. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 27: 2393–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenberger J, Loeffler S, Kappeler A, Kopf M, Lukes A, Afanasieva TA, Aguzzi A, Weis J 2004. IL-6 is required for glioma development in a mouse model. Oncogene 23: 3308–3316 [DOI] [PubMed] [Google Scholar]
- Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD 1997. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res 57: 4130–4140 [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, et al. 1997. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature 390: 91–96 [DOI] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, et al. 2009. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci 106: 2712–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]