Abstract
Cardiomyocyte proliferation is high in early development and decreases progressively with gestation, resulting in the lack of a robust cardiomyocyte proliferative response in the adult heart after injury. Little is understood about how both cell-autonomous and nonautonomous signals are integrated to regulate the balance of cardiomyocyte proliferation during development. In this study, we show that a single transcription factor, Foxp1, can control the balance of cardiomyocyte proliferation during development by targeting different pathways in the endocardium and myocardium. Endocardial loss of Foxp1 results in decreased Fgf3/Fgf16/Fgf17/Fgf20 expression in the heart, leading to reduced cardiomyocyte proliferation. This loss of myocardial proliferation can be rescued by exogenous Fgf20, and is mediated, in part, by Foxp1 repression of Sox17. In contrast, myocardial-specific loss of Foxp1 results in increased cardiomyocyte proliferation and decreased differentiation, leading to increased myocardial mass and neonatal demise. We show that Nkx2.5 is a direct target of Foxp1 repression, and Nkx2.5 expression is increased in Foxp1-deficient myocardium. Moreover, transgenic overexpression of Nkx2.5 leads to increased cardiomyocyte proliferation and increased ventricular mass, similar to the myocardial-specific loss of Foxp1. These data show that Foxp1 coordinates the balance of cardiomyocyte proliferation and differentiation through cell lineage-specific regulation of Fgf ligand and Nkx2.5 expression.
Keywords: Foxp1, Nkx2.5, Fgf16, Fgf20, Sox17, cardiomyocyte, proliferation
Cardiomyocyte proliferation is precisely regulated, resulting in high levels of proliferation early in cardiac development when the heart is growing rapidly. Subsequently, cardiomyocyte proliferation decreases until in the postnatal heart, when these cells have fully exited the cell cycle and, by most measures, are completely quiescent. The precise regulatory network that controls this gradual withdrawal from the cell cycle is poorly understood, but correlates with changes in cell cycle genes, including cyclins and cyclin-dependent kinases (CDKIs). In addition to cell-autonomous mechanisms, cardiomyocyte proliferation is also regulated by signals from the endocardium and epicardium. Secreted factors, including neuregulin and Fgfs9/16/20, are expressed in these nonmyocardial lineages, and promote cardiomyocte proliferation in a cell-nonautonomous fashion (Lavine et al. 2005; Hotta et al. 2008; Lu et al. 2008; Bersell et al. 2009). Genetic loss-of-function studies in mice reveal that both the neuregulin/ErbB2 and Fgf pathways play a critical role in this paracrine regulation (Lee et al. 1995; Lavine et al. 2005). Given the importance of promoting cardiomyocyte proliferation in the adult in settings of injury and repair, a more thorough understanding of the pathways and factors that regulate this process is warranted.
Foxp factors are large modular transcriptional repressors that bind to DNA via their highly conserved forkhead DNA-binding domain (Li and Tucker 1993; Shu et al. 2001; Li et al. 2004a). Efficient DNA binding by Foxp1 factors requires dimerization through a leucine zipper motif (Li et al. 2004a). Foxp1 is expressed in cardiomyocytes, in the endocardium, and in cells underlying the cushion mesenchyme, while Foxp4 expression is observed before embryonic day 10.5 (E10.5) in cardiomyocytes and, later, in the epicardium and endocardium (Lu et al. 2002; Li et al. 2004b; Wang et al. 2004). Previous studies have shown that both Foxp1 and Foxp4 play important roles in regulating various aspects of cardiac development. Loss of Foxp4 leads to a cardia bifida phenotype, showing that fusion of the early cardiac primordia is not required for cardiac chamber development or subsequent morphogenesis (Li et al. 2004b). Loss of Foxp1 results in a complex cardiac phenotype characterized by defects in outflow tract (OFT) septation, increased myocardial proliferation, and thinning of ventricular myocardium (Wang et al. 2004).
The conundrum of increased cell proliferation and loss of ventricular mass in Foxp1 mutants suggested that Foxp1 may regulate cardiomyocyte proliferation in a complex fashion. To address this issue further, we generated a conditional allele of Foxp1. Loss of Foxp1 in endothelial cells using the Tie2-cre line results in embryonic lethality similar to that observed in the global knockout of Foxp1, and this lethality correlates with decreased ventricular mass. However, in contrast to the global loss of function, we observe reduced cardiomyocyte proliferation in the endothelial-specific loss of Foxp1 mutants. This is the result of derepression of Sox17 expression, leading to decreased Fgf3/16/17/20 expression in the endocardium. Loss of Foxp1 specifically in the myocardium leads to increased proliferation, particularly in the trabecular zone, increased ventricular wall thickness, and ventricular septation defects. We found that Foxp1 directly represses Nkx2.5 expression in the myocardium, and that loss of Foxp1 leads to increased Nkx2.5 expression. Moreover, transgenic overexpression of Nkx2.5 results in increased cardiomyocyte proliferation and ventricular wall thickness, similar to that in myocardial-deficient Foxp1 mutants. Together, these data indicate that Foxp1 regulates the balance of myocardial growth in development through both cell-autonomous and nonautonomous mechanisms, resulting in the controlled exit of cardiomyocytes from the cell cycle.
Results
Generation of a Foxp1 conditional allele
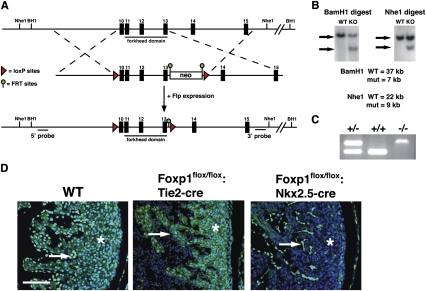
To explore the role of Foxp1 in cardiovascular development in a cell lineage-specific fashion, we generated a conditional Foxp1flox/flox allele by flanking exons 10–13 with loxP sites (Fig. 1A). Southern blot and PCR analysis shows correct targeting of both embryonic stem (ES) cells and gene targeted mice (Fig. 1B,C). Exons 10–13 encode the forkhead DNA-binding domain of Foxp1, and encompass the region deleted in the global knockout animal we reported previously (Wang et al. 2004). Foxp1flox/flox animals are viable and fertile. As reported previously, Foxp1 is expressed in both the myocardium and endocardium (Wang et al. 2004). To assess the efficiency of excision of this Foxp1flox/flox allele in cardiomyocytes and the endothelium, we generated Foxp1flox/flox:Nkx2.5-cre and Foxp1flox/flox:Tie2-cre mutants and performed immunohistochemistry with a Foxp1-specific antibody. These data show that Foxp1 expression is specifically lost in the cardiomyocytes and endocardial cells of Foxp1flox/flox:Nkx2.5-cre and Foxp1flox/flox:Tie2-cre mutants, respectively (Fig. 1D).
Figure 1.
Conditional targeting strategy for Foxp1. (A) Targeting strategy for generating the conditional Foxp1 mouse allele. Exons 10–13 were flanked with loxP sites deleting the entire forkhead DNA-binding domain. Flpe mice were used to delete the FRT-flanked neomycin cassette. (B) Southern blot results from targeted ES clones using the 5′ and 3′ probes indicated in A. (C) PCR genotyping results from germline mice of the indicated genotypes using primers listed in Supplemental Table 1. (D) Immunostaining with Foxp1 antibody on wild type and Foxp1flox/flox:Nkx2.5-cre mutants at E14.5, as well as Foxp1flox/flox:Tie2-cre mutants at E14.5. Asterisk indicates myocardium where expression is missing in Foxp1flox/flox:Nkx2.5-cre mutants, and arrows indicate endocardium where expression is absent in Foxp1flox/flox:Tie2-cre mutants. Bar, 50 μm.
Loss of Foxp1 in the endocardium results in embryonic lethality due to decreased cardiomyocyte proliferation and defects in endocardial cushion development
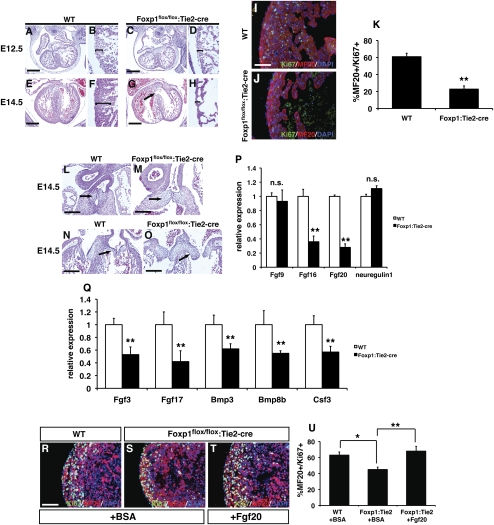
Foxp1flox/flox:Tie2-cre mutants die between E14.5 and E16.5, and none are found after E16.5 (data not shown). Foxp1flox/flox:Tie2-cre mutants were assessed by histology to determine the cause of embryonic death. At E12.5, Foxp1flox/flox:Tie2-cre mutant hearts looked relatively normal, but displayed minor and reproducible ventricular thinning (Fig. 2A–D). By E14.5, most Foxp1flox/flox:Tie2-cre mutants showed extensive ventricular thinning with an associated ventricular septal defect (VSD) (Fig. 2E–H). OFT septation is normal in Foxp1flox/flox:Tie2-cre mutants (data not shown). The Tie2-cre transgene is also expressed in the endothelium of the developing coronary vessels; however, development of these vessels was relatively unperturbed in Foxp1flox/flox:Tie2-cre mutants (Supplemental Fig. 1). Ki67/MF20 coimmunostaining shows a significant loss of proliferation in both the compact zone myocardium as well as the trabecular myocardium (Fig. 2I–K). This loss of ventricular myocardial mass likely leads to reduced cardiac output and subsequent embryonic lethality.
Figure 2.
Foxp1flox/flox:Tie2-cre mutants exhibit endocardial cushion defects and reduced ventricular myocardial development and proliferation, which are due to loss of Fgf16 and Fgf20 expression. H + E staining of wild type (A,B) and Foxp1flox/flox:Tie2-cre mutants (C,D) at E12.5 showing a slight thinning of ventricular myocardium in the mutants. (E–H) By E14.5, Foxp1flox/flox:Tie2-cre mutants exhibit extensive ventricular thinning and pronounced VSDs as compared with wild-type littermates. (I–K) This reduced ventricular myocardial development correlates with a loss in myocardial proliferation, as shown by reduced Ki67/MF20 coimmunostaining at E12.5. Both OFT (L,M) and AV canal (N,O) endocardial cushions are enlarged in Foxp1flox/flox:Tie2-cre mutants, suggesting defective endocardial–mesenchymal transformation. (P) Expression of Fgf16 and Fgf20, but not Fgf9 and neuregulin, are reduced in Foxp1flox/flox:Tie2-cre mutants. (Q) Q-PCR results from growth factor PCR array studies showing decreased Fgf3, Fgf17, Bmp3, Bmp8b, and Csf3 expression in Foxp1flox/flox:Tie2-cre mutants at E12.5. Myocardial proliferation defects in Foxp1flox/flox:Tie2-cre mutants can be rescued with exogenous Fgf20 (T,U) but not BSA (R,S,U). Proliferation assessments were made on three embryos of each indicated genotype/condition, and the data represent the average ± SEM. Q-PCR data represent the average of three different assays performed in triplicate ± SEM. All data are considered significant using Student's t-test unless otherwise noted. (n.s.) Not significant; (*) P < 0.05; (**) P < 0.01. Bars: I,J, 30 μm; R–T, 50 μm.
Cardiac valves form through a process of endocardial–mesenchymal transformation, leading to development of the endocardial cushions, which undergo further remodeling to generate the mature cardiac valves. The global loss of Foxp1 results in defective endocardial cushion development in both the OFT valves and atrio–ventricular (AV) valves (Wang et al. 2004). Foxp1flox/flox:Tie2-cre mutants were examined to determine whether Foxp1 expression in the endocardium was responsible for these defects. Foxp1flox/flox:Tie2-cre mutants have thickened OFT and AV cushions, similar to what is observed in the global Foxp1 mutants (Fig. 2L–O). Together, these data show that Foxp1 expression in the endocardium promotes ventricular myocardial growth and maturation of the endocardial cushions.
Foxp1 is required for Fgf3/Fgf16/Fgf17/Fgf20 expression, and exogenous Fgfs can rescue reduced cardiomyocyte proliferation in Foxp1flox/flox:Tie2-cre mutants
Paracrine signaling from the endocardium by factors such as neuregulin and Fgf9/16/20 have been shown to promote cardiomyocyte proliferation (Lee et al. 1995; Lavine et al. 2005; Bersell et al. 2009). Therefore, we assessed expression of these factors in Foxp1flox/flox:Tie2-cre mutants by quantitative PCR (Q-PCR). While neuregulin and Fgf9 expression were unchanged in Foxp1flox/flox:Tie2-cre mutants, Fgf16 and Fgf20 expression were significantly down-regulated by Q-PCR (Fig. 2P). In situ hybridization shows that Fgf16 and Fgf20 expression is decreased in the endocardium and, to a lesser degree, in the myocardium (Supplemental Fig. 2). To more fully address the effects of loss of Foxp1 in the endocardium, we performed a growth factor-specific microarray. Data from these studies identified decreased expression of two additional Fgf ligands, Fgf3 and Fgf17 (Fig. 2Q; Supplemental Table 1). Moreover, expression of four additional genes—Bmp3, Bmp8b, Spp1, and Csf3—was also decreased in Foxp1flox/flox:Tie2-cre mutants, based on these studies (Fig. 2Q; Supplemental Table 1). These data suggest that, in addition to Fgf signaling, there may also be an important role for Bmp signaling acting downstream from Foxp1.
Given their potent role in promoting cardiomyocyte proliferation, and the finding that expression of multiple ligands are down-regulated, we assessed whether exogenous Fgf could rescue the proliferation defects in Foxp1flox/flox:Tie2-cre mutants. Embryonic hearts from wild type and Foxp1flox/flox:Tie2-cre mutants were cultured in the presence of bovine serum albumin (BSA) or Fgf20, and cardiomyocyte proliferation was assessed after 48 h. These studies showed that exogenous Fgf20 was able to rescue much of the reduction in cardiomyocyte proliferation observed in Foxp1flox/flox:Tie2-cre mutants (Fig. 2R–U). Together, these data suggest that Foxp1 regulates multiple Fgf ligands in the endocardium, and that decreased expression of these Fgf ligands in part leads to ventricular hypoplasia in Foxp1flox/flox:Tie2-cre mutants.
Foxp1 directly regulates Sox17, which represses Fgf16/20 expression
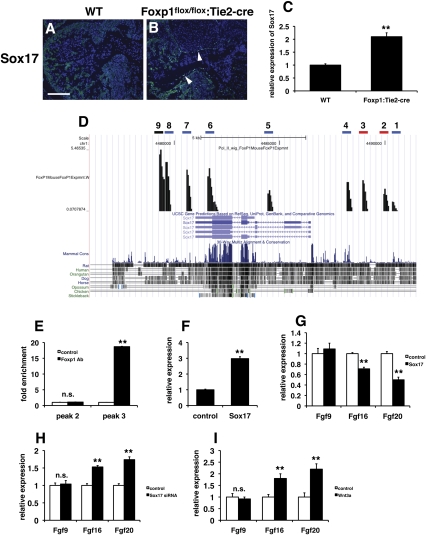
We performed chromatin immunoprecipitation (ChIP) accompanied by high-throughput sequencing (ChIP-seq) to identify potential Foxp1 targets in the embryonic heart. Based on our previous studies, as well as those from other laboratories, Foxp1 is a transcriptional repressor (Shu et al. 2001, 2007; Wang et al. 2003; Li et al. 2004a). Therefore, we searched the ChIP-seq results for possible repressors expressed in the endocardium that Foxp1 would target directly—i.e. act as a repressor of a repressor—leading to decreased Fgf16/20 expression. From these analyses, Sox17 was identified as a potential target of Foxp1. Sox17 is expressed at high levels in the developing endocardium, and has been shown to be important for early heart development (Sakamoto et al. 2007; Francois et al. 2010). Although Sox17 can act as a transcriptional activator, it is also known to repress Wnt/β-catenin signaling by competing for β-catenin interaction with LEF/TCF transcription factors (Zorn et al. 1999). Moreover, Fgf16/20 are known targets of Wnt signaling in cardiac development, providing a potential link between loss of Foxp1 in the endocardium and decreased Fgf16/20 expression (Chamorro et al. 2005; Cohen et al. 2007). Sox17 expression is up-regulated in the endocardium of Foxp1flox/flox:Tie2-cre mutants at E12.5 (Fig. 3A–C). ChIP-seq data revealed two peaks with conserved forkhead-binding sites (Fig. 3D). ChIP analysis showed that Foxp1 can bind readily to peak 3 but not peak 2 (Fig. 3E). To determine if increased expression of Sox17 could repress Fgf16/20 expression, we overexpressed Sox17 in human umbilical vein endothelial cells (HUVECs). Lentiviral-mediated Sox17 overexpression led to an approximately threefold increase in Sox17 expression (Fig. 3F). Sox17 overexpression also led to decreased Fgf16 and Fgf20 expression, but not Fgf9 expression (Fig. 3G). siRNA-mediated inhibition of Sox17 expression resulted in increased Fgf16 and Fgf20 expression in HUVECs (Fig. 3H). A similar increase in Fgf16 and Fgf20 expression is also observed after treatment of HUVECs with Wnt3a (Fig. 3I). Together, these data support the notion that Foxp1 normally represses Sox17 in the endocardium, allowing for the proper expression of the cardiomyocyte mitogens Fgf16 and Fgf20 by acting through Wnt/β-catenin signaling.
Figure 3.
Sox17 is a direct target of Foxp1 in the endocardium, and overexpression of Sox17 represses Fgf16/20 expression. (A,B) Immunostaining shows that Sox17 expression is increased in the endocardium of Foxp1flox/flox:Tie2-cre mutants at E12.5 (arrowheads). (C) Q-PCR shows increased Sox17 expression in Foxp1flox/flox:Tie2-cre mutants at E12.5. (D) ChIP-seq data showing peaks with forkhead DNA-binding sites (red bar), peaks unique to Foxp1 antibody ChIP-seq analysis but lacking forkhead DNA-binding sites (blue bars), and peaks that were also observed in control IgG ChIP-seq analysis (black bar). (E) ChIP analysis shows that Foxp1 binds to the peak 3 region but not the peak 2 region efficiently. (F) Lentiviral overexpression results in a threefold increase of Sox17 expression in HUVECs, as measured by Q-PCR. (G) Overexpression of Sox17 in HUVECs results in decreased expression of Fgf16 and Fgf20 but not Fgf9, as measured by Q-PCR. (H) siRNA inhibition of Sox17 expression leads to increased Fgf16 and Fgf20 expression. (I) Wnt3a treatment of HUVECs results in increased Fgf16 and Fgf20 expression. All data are considered significant using Student's t-test unless otherwise noted. (n.s.) Not significant; (**) P < 0.01. Bar: A,B, 100 μm.
Cardiomyocyte-specific loss of Foxp1 causes increased myocardial mass and defects in OFT septation
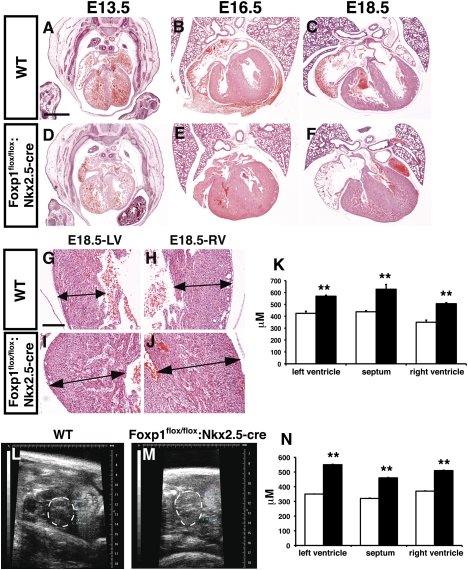
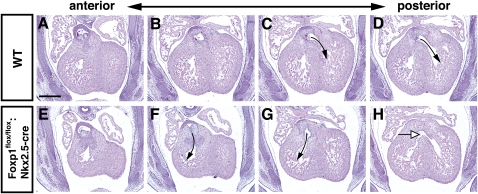
To determine the role of Foxp1 in the developing myocardium, Foxp1flox/flox:Nkx2.5-cre mutants were generated using the previously reported Nkx2.5-cre allele (Moses et al. 2001). All Foxp1flox/flox:Nkx2.5-cre mutants die within 24 h of birth, indicating an important role for Foxp1 in developing cardiomyocytes (data not shown). Histological analysis of Foxp1flox/flox:Nkx2.5-cre mutants at various stages of development show that loss of Foxp1 in cardiomyocytes does not appreciably affect cardiac morphology before E14.5 (Fig. 4A,D). However, starting as early as E16.5, the ventricular walls of Foxp1flox/flox:Nkx2.5-cre mutants appear significantly thicker, with both the left and right ventricular walls as well as the ventricular septum affected (Fig. 4B,C,E–K). Echocardiography was performed at E16.5 to assess whether the difference in ventricular wall thickness was also apparent in utero. These studies show that all three regions—left and right outer ventricular walls and the ventricular septum—were significantly thicker in Foxp1flox/flox:Nkx2.5-cre mutants than in wild-type animals (Fig. 4L–N). This increase in myocardial mass is in contrast to what is observed in the Foxp1flox/flox:Tie2-cre mutants, which exhibit a thinner ventricular wall (Fig. 2). These data show that loss of Foxp1 in the myocardium results in increased ventricular myocardial mass during development.
Figure 4.
Foxp1flox/flox:Nkx2.5-cre mutants exhibit increased ventricular wall thickness. H + E staining of wild type (A–C) and Foxp1flox/flox:Nkx2.5-cre mutants (D–F) from E13.5–E18.5 showing increased ventricular wall thickness at E16.5 and E18.5. (G–K) This increased thickness is observed in the wall of the left and right ventricles, as well as the ventricular septum. (L–N) Increased ventricular wall thickness is also observed in vivo using echocardiography. All data are considered significant using Student's t-test. (**) P < 0.01. Measurements are the average of three embryos of each indicated genotype ± standard deviation. Bars: A–F, 400 μm; G–J, 100 μm.
Our previous work showed that global loss of Foxp1 resulted in OFT defects, including double-outlet right ventricle (DORV) and VSDs (Wang et al. 2004). We assessed whether Foxp1flox/flox:Nkx2.5-cre mutants exhibited OFT defects or VSDs. Foxp1flox/flox:Nkx2.5-cre mutants had DORV as well as VSDs at E14.5 (Fig. 5). However, most Foxp1flox/flox:Nkx2.5-cre mutants did not have VSDs at E16.5, suggesting that ventricular septation was delayed but not permanently inhibited (data not shown). Loss of Foxp1 in cardiac neural crest using the Wnt1-cre line did not result in OFT defects, suggesting that this cell lineage does not play an important role in Foxp1-mediated OFT development (Supplemental Fig. 3). These studies suggest that the loss of Foxp1 in cardiomyocytes causes a delay in myocardial development, leading to a delay in formation of the ventricular septum, and that this may be the cause, at least in part, of the DORV phenotype observed in the global Foxp1-null mutants (Wang et al. 2004).
Figure 5.
Foxp1flox/flox:Nkx2.5-cre mutants exhibit DORV and VSDs. H + E staining of wild type (A–D) and Foxp1flox/flox:Nkx2.5-cre mutants (E–H) at E14.5 showing the aorta exiting from the right ventricle (arrows, F,G) and a VSD (white arrow, H). Bar, 200 μm.
Cardiomyocyte-specific loss of Foxp1 leads to increased cardiomyocyte proliferation and decreased maturation
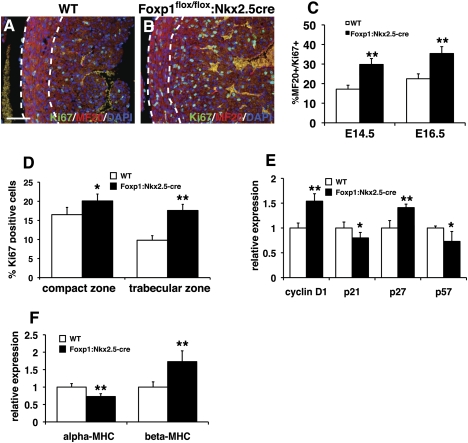
The increase in myocardial mass observed in Foxp1flox/flox:Nkx2.5-cre mutants suggested that myocardial proliferation was affected. We analyzed Ki67 expression as well as expression of the important cell cycle genes cyclin D1, p21, p27, and p57 to assess the proliferation and cell cycle state of Foxp1flox/flox:Nkx2.5-cre mutant cardiomyocytes. Ki67/MF20 coimmunostaining showed increased cardiomyocyte proliferation in Foxp1flox/flox:Nkx2.5-cre mutants (Fig. 6A–C). Moreover, this increased proliferation is most evident in the trabecular layer of the heart, a region that normally exhibits less proliferation than the outer compact zone (Fig. 6D). Expression of the proproliferative cell cycle gene cyclin D1 and the CDKI p27 were up-regulated, while expression of the inhibitory CDKs p21 and p57 was decreased (Fig. 6E).
Figure 6.
Foxp1flox/flox:Nkx2.5-cre mutants have increased myocardial proliferation as well as alterations in myocardial differentiation. (A–C) Ki67/MF20 coimmunostaining in Foxp1flox/flox:Nkx2.5-cre mutants is increased in comparison with wild-type littermates at E14.5 and E16.5. (D) This increased proliferation is observed in both the compact and trabecular zones. (E) Cell cycle factor expression is altered in Foxp1flox/flox:Nkx2.5-cre mutants. (F) Expression of α-MHC and β-MHC is altered in Foxp1flox/flox:Nkx2.5-cre mutants. Proliferation assessments were made on three embryos of each indicated genotype/condition, and the data represent the average ± S.E.M. Q-PCR data represent the average of three different assays performed in triplicate ± SEM. All data are considered significant using Student's t-test. (*) P < 0.05; (**) P < 0.01. Bar, 50 μm.
As cardiomyocytes differentiate, expression of several genes related to maturation of this lineage is altered. In particular, expression of the α and β isoforms of myosin heavy chain (MHC) is well known to correlate with cardiomyocyte maturation (Ng et al. 1991). To assess whether the increase in cardiomyocyte proliferation was associated with a change in cardiomyocyte maturation, we assessed α-MHC and β-MHC expression by Q-PCR. At E16.5, expression of α-MHC was decreased in Foxp1flox/flox:Nkx2.5-cre mutants, while β-MHC expression was increased (Fig. 6F). Moreover, the expression pattern of α-actinin and desmin indicated disrupted sarcomeric structure in Foxp1flox/flox:Nkx2.5-cre mutants, correlating with decreased cardiomyocyte maturity (Supplemental Fig. 4). These data suggest that loss of Foxp1 increases proliferation while inhibiting cardiomyocyte maturation.
Identification of Nkx2.5 as a direct target of Foxp1
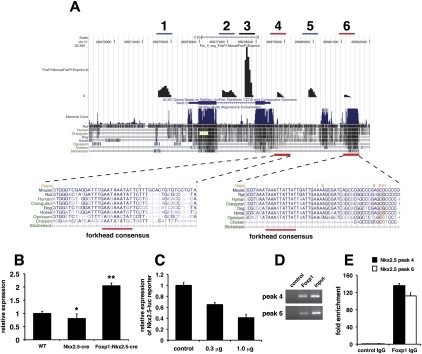
Using our ChIP-seq data, we identified Nkx2.5 as a potential direct target of Foxp1 in cardiomyocytes (Fig. 7A). Nkx2.5 is a critical homeodomain transcription factor required for proper cardiac development and expression of many cardiac-specific genes (Tanaka et al. 1999). Moreover, Nkx2.5 has been shown to be important for regulation of cardiomyocyte proliferation and numbers in the developing heart (Prall et al. 2007; Targoff et al. 2008; Tu et al. 2009). While Nkx2.5 expression is decreased ∼20% in Nkx2.5-cre hearts, Nkx2.5 expression is up-regulated by approximately twofold in Foxp1flox/flox:Nkx2.5-cre mutants, which correlates with Foxp1 acting as a transcriptional repressor (Fig. 7B). This would suggest that the increase in Nkx2.5 expression due to loss of Foxp1 expression would be even greater on a Nkx2.5 wild-type background. ChIP-seq data identified two regions within ∼7 kb of the transcriptional start site of Nkx2.5 that were specific for the Foxp1 antibody versus a control IgG (Fig. 7A). One of these peaks, peak 4, is located in a previously identified enhancer region of Nkx2.5 that was shown to direct Nkx2.5 expression to the heart (Searcy et al. 1998). Consensus forkhead DNA-binding sites were located in both peaks 4 and 6 (Fig. 7A). Foxp1 can repress a luciferase reporter containing the previously identified −3-kb mouse Nkx2.5 promoter and enhancer that contains site 4 (Fig. 7C). ChIP analysis using both agarose gel electrophoresis and real-time Q-PCR showed that Foxp1 binding to peaks 4 and 6 was enriched by >100-fold over control antibody (Fig. 7D,E). Since Foxp1 is a known transcriptional repressor, these data suggested that Foxp1 normally restricts Nkx2.5 expression in the heart, leading to increased myocardial proliferation in the loss-of-function Foxp1flox/flox:Nkx2.5-cre mutants.
Figure 7.
ChIP-seq analysis identifies Nkx2.5 as a target of Foxp1 transcriptional repression. ChIP-seq data showing peaks with forkhead DNA-binding sites (red bar), peaks unique to Foxp1 antibody ChIP-seq analysis but lacking forkhead DNA-binding sites (blue bars), and peaks that were also observed in control IgG ChIP-seq analysis (black bar). (A) ChIP-seq analysis identifies two peaks in the Nkx2.5 locus that contain conserved forkhead DNA-binding sites (peaks 4 and 6). (B) Expression of Nkx2.5 is decreased by ∼20% in Nkx2.5-cre hearts but is elevated in Foxp1flox/flox:Nkx2.5-cre hearts at E14.5. Of note, the Nkx2.5-cre mice were used as controls in these experiments to control for the loss of one Nkx2.5 allele. (C) Foxp1 expression represses the −3-kb mouse Nkx2.5 promoter/enhancer in a dose-dependent manner. Directed ChIP using primers in Supplemental Table 1 show high-level association of Foxp1 with peak regions 4 and 6 using agarose gel analysis (D) and real-time Q-PCR (E). Q-PCR data represent the average of three different assays performed in triplicate ± SEM. All data are considered significant using Student's t-test. (*) P < 0.05; (**) P < 0.01.
Increased expression of Nkx2.5 in cardiomyocytes in vivo results in increased myocardial mass and proliferation
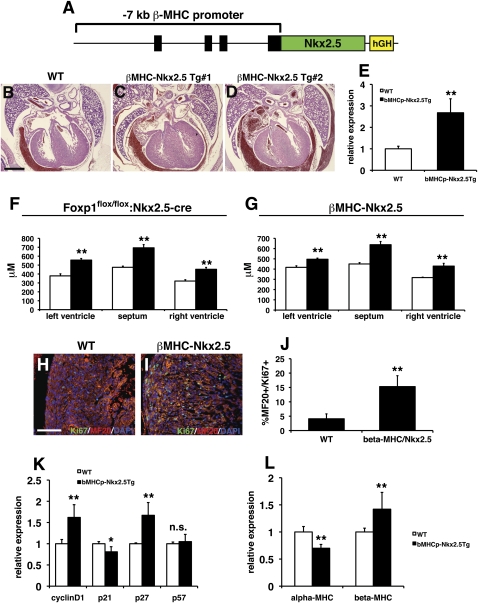
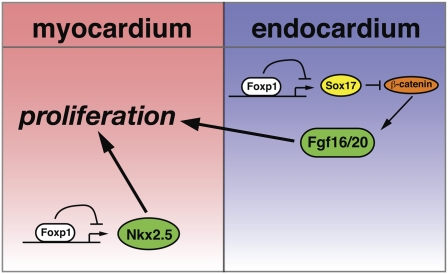
Nkx2.5 has been implicated previously in regulating cardiomyocyte proliferation in development (Prall et al. 2007; Targoff et al. 2008; Tu et al. 2009). Our data suggest that Foxp1 normally restricts Nkx2.5 gene expression, and that loss of Foxp1 in cardiomyocytes results in increased Nkx2.5 expression and increased myocardial proliferation. To directly assess whether increased expression of Nkx2.5 in the myocardium would result in increased proliferation, as observed in Foxp1flox/flox:Nkx2.5-cre mutants, we generated a transgenic overexpression model using the β-MHC promoter to drive expression of Nkx2.5 in developing myocardium (Fig. 8A; Rindt et al. 1993). Seven βMHC:Nkx2.5 transgenics were generated and collected at E18.5, and used for histological characterization. Compared with their nontransgenic littermates, five of the seven βMHC:Nkx2.5 animals exhibited significantly increased ventricular mass (Fig. 8B–D). Ventricular wall thickness was also increased in βMHC:Nkx2.5 transgenics, and was similar to that of Foxp1flox/flox:Nkx2.5-cre mutants at E16.5 (Fig. 8F,G). Three additional βMHC:Nkx2.5 transgenics were generated and used for Q-PCR assessment of gene expression, which shows an ∼2.5-fold increase in Nkx2.5 gene expression in transgenic hearts (Fig. 8E). Ki67/MF20 coimmunostaining shows that proliferation is significantly increased in cardiomyocytes of βMHC:Nkx2.5 transgenics (Fig. 8H–J). This increase in expression is coupled to changes in cyclin D1 and p27 expression, similar to that observed in Foxp1flox/flox:Nkx2.5-cre mutants (Fig. 8K). Changes in α-MHC and β-MHC expression, similar to the Foxp1flox/flox:Nkx2.5-cre mutants, were observed in βMHC:Nkx2.5 transgenics (Fig. 8L). βMHC:Nkx2.5 transgenics did not exhibit DORV or VSDs, suggesting that some but not all aspects of the Foxp1flox/flox:Nkx2.5-cre phenotype are recapitulated upon overexpression of Nkx2.5 (data not shown). Together, these data indicate that Foxp1 restricts cardiomyocyte proliferation in a cell-autonomous manner, in part through repression of Nkx2.5, and that this balances the function of Foxp1 in the endocardium that activates a subset of Fgf ligands, including Fgf16 and Fgf20, through inhibition of Sox17 expression, leading to the promotion of cardiomyocyte proliferation in early development (Fig. 9).
Figure 8.
Increased Nkx2.5 expression leads to increased ventricular wall thickness and myocardial proliferation. (A) The β-MHC promoter was use to overexpress Nkx2.5 in transgenic mice. (B–D) Genotype-positive embryos exhibited increased ventricular wall thickness compared with wild-type littermates. (E) Expression of Nkx2.5 is increased in βMHC-Nkx2.5 transgenic hearts. (F,G) The increase in ventricular wall thickness in βMHC-Nkx2.5 transgenic hearts at E16.5 is similar to that observed in Foxp1flox/flox:Nkx2.5-cre mutants at the same age. (H–J) βMHC-Nkx2.5 transgenic hearts exhibit a significant increase in proliferation, as measured by Ki67/MF20 coimmunostaining. (K) Expression of cyclin D1 and p27 is elevated in βMHC-Nkx2.5 transgenic hearts. (L) Expression of α-MHC is decreased and expression of β-MHC is increased in βMHC-Nkx2.5 transgenic hearts. Proliferation assessments were made on two embryos of each indicated genotype/condition, and the data represent the average ± SEM. Q-PCR data represent the average of three different assays performed in triplicate ± SEM. All data are considered significant using Student's t-test unless otherwise noted. (n.s.) Not significant; (*) P < 0.05; (**) P < 0.01. Bars: B–D, 300 μm; H,I, 100 μm.
Figure 9.
Model of how Foxp1 regulates the balance of cardiomyocyte proliferation by targeting different pathways in the endocardium and myocardium. Foxp1 expression in the myocardium targets Nkx2.5 expression, leading to increased Nkx2.5 expression, increased proliferation, and decreased maturation. Conversely, Foxp1 expression in the endocardium represses Sox17 expression, which, through regulation of β-catenin activity, controls Fgf16/20 expression.
Discussion
Proliferation and maturation of cardiomyocytes is orchestrated in a temporal-specific fashion that results in high rates of proliferation in early development, but an almost completely quiescent cell in the adult heart. We show that Foxp1 controls this process, in part, by regulating expression of Fgf16 and Fgf20 in the endocardium, promoting cardiomyocyte proliferation in a non-cell-autonomous fashion in early cardiac development, while restricting expression of Nkx2.5 and inhibiting proliferation in a cell-autonomous fashion later in development. These studies highlight the importance of maintaining the correct balance of signals that allow for appropriate cardiomyocyte proliferation and differentiation, and demonstrate that Foxp1 plays a critical role in regulating this process.
Although cardiomyocytes are thought to permanently exit the cell cycle postnatally, recent evidence shows that there is a very small level of cell turnover in adult humans. Studies using radiocarbon dating have revealed cardiomyocyte turnover in humans at a rate of 1% per year in young adults, which decreases to <0.5% a year in older adults (Bergmann et al. 2009). Even at this rate, <50% of cardiomyocytes would be renewed over an average life span. This turnover appears to involve simple replacement of cardiomyocytes through proliferation and not through activation of resident cardiac stem/progenitor populations. Our finding that Foxp1 represses Nkx2.5 gene expression, and that increased Nkx2.5 expression leads to increased cardiomyocyte proliferation and myocardial mass, implies a new role for these factors in regulating myocardial proliferation and maturation. Previous work has implicated Nkx2.5 in the regulation of early cardiomyocyte proliferation, and shows that loss of Nkx2.5 results in decreased proliferation in early cardiomyocytes (Prall et al. 2007; Tu et al. 2009). However, whether increased Nkx2.5 expression would increase cardiomyocyte proliferation was, until now, unknown. Our data show that loss of Foxp1 leads to increased Nkx2.5 expression due to direct repression of Nkx2.5 by Foxp1. This in turn leads to increased myocardial proliferation, which can be mimicked by transgenic overexpression of Nkx2.5 in cardiomyocytes during development.
The present study suggests that there is a subset of Fgf ligands whose expression is positively regulated by Wnt/β-catenin signaling. We show that expression of Fgf3, Fgf16, Fgf17, and Fgf20 is reduced upon loss of Foxp1 in the endocardium. Previous reports have demonstrated that several Fgf ligands—including Fgf10, Ffg16, and Fgf20—are targets of Wnt/β-catenin signaling (Chamorro et al. 2005; Cohen et al. 2007). The ability of Foxp1 to positively regulate Fgf3/Fgf16/Fgf17/Fgf20 expression resides in its repression of Sox17, a known inhibitor of Wnt/β-catenin signaling (Zorn et al. 1999). Examination of other potential mediators of Foxp1 function in the endocardium and endothelium, including Bmp signaling, will be important to further elucidate the mechanism by which this factor regulates cardiovascular development.
Foxp1/2/4 are highly related forkhead transcription factors expressed in a diverse array of tissues, including the heart, lung, brain, and hematopoietic lineages. Foxp factors have been shown to regulate cell proliferation in a cell context-dependent fashion (Wang et al. 2004; Shu et al. 2007). Previous studies have implicated Foxp1 as a tumor suppressor gene, and correlate its expression with decreased tumorigenicity (Fox et al. 2004). Combined loss of Foxp1 and Foxp2 in the lung leads to decreased proliferation of both epithelial and mesenchymal cell lineages (Shu et al. 2007). Since these studies were performed with the global loss-of-function mice, cell type-specific increases in proliferation could be counterbalanced by non-cell-autonomous effects of loss of Foxp1 in neighboring cells. Such a counterbalance effect could be important in regulating cardiomyocyte maturation and proliferation in late gestation and early postnatal life.
Other Foxp factors also regulate important cell proliferation processes. Loss of the related factor Foxp3 leads to the mouse mutant scurfy and the human lymphoproliferative disease IPEX, which is characterized by excessive proliferation and decreased maturation of CD4+, CD25+ regulatory T cells (Brunkow et al. 2001). This phenotype can be rescued by re-expression of Foxp3 in the CD4+, CD25+ T-cell compartment via transgenesis. How Foxp factors regulate cell proliferation is unclear, and may be through multiple mechanisms. The present study suggests that Foxp1 regulates expression of Nkx2.5 in cardiomyocytes to suppress cardiomyocyte proliferation, while, in the endocardium, Foxp1 regulates Fgf16 and Fgf20 through direct repression of Sox17, resulting in increased Sox17 expression, which in turn inhibits β-catenin function, causing decreased Fgf16 and Fgf20 expression. While this pathway is one mechanism for Foxp1 function in cardiac development, additional mechanisms may also be involved, and the complex structure of the Foxp proteins likely plays an important role. All Foxp proteins contain a similar repression domain, which is characterized by the presence of leucine zipper and zinc finger motifs. Previous data have shown that C-terminal-binding protein-1 (CtBP-1), a corepressor, binds to Foxp1 and Foxp2 in this region (Li et al. 2004a). CtBP-1/2 have been shown to regulate cell cycle progression, resulting in decreased proliferation (Bergman et al. 2009). Interestingly, Foxp1 has been shown to interact with the corepressor SMRT to coregulate cardiac development (Jepsen et al. 2008). Whether or not SMRT plays a critical role with Foxp1 to repress Nkx2.5 gene expression in cardiomyocytes will be important to explore in future studies. Identification and characterization of additional factors that bind to the repression domain of Foxp factors and regulate their function will likely provide important insights into the mechanisms of cell proliferation responses orchestrated by the Foxp family.
Given the need for a better understanding of how cardiomyocyte cell proliferation is regulated, our finding that loss of Foxp1 in differing cell lineages within the heart results in different proliferative responses shows how expression of a single factor in different lineages is critical to balance the autonomous and non-cell-autonomous regulation of cardiomyocyte proliferation. Future studies of how Foxp1 regulates cardiomyocyte proliferation—as well as how expression levels of Fgf16, Fgf20, and Nkx2.5 are carefully regulated—during development and in the adult will increase our knowledge of this molecular pathway in cardiac development and regeneration.
Materials and methods
Generation of Foxp1flox/flox mice
The vector for generating the floxed allele of murine Foxp1 was produced using recombineering techniques (Liu et al. 2003). The 5′ loxP site was introduced in the intron between exons 9 and 10, and the 3′ loxP site was introduced in the intron between exons 13 and 14. The targeting vector was linearized and electroporated into R1 ES cells. ES cell clones were isolated and DNA was extracted using standard techniques. Southern blot analysis using both 5′ and 3′ probes shown in Figure 1 were used to verify correct integration of both loxP sites. Two correctly targeted ES clones were injected into blastocysts to generate chimeric mice, which were further bred to C57BL/6 mice to generate germline transmission of the mutant Foxp1 allele. The neomycin cassette was removed using Flpe mice (JAX ACTFLPe-9205Dym/J). The Tie2-cre, Nkx2.5-cre, and Wnt1-cre mouse lines have been described previously (Danielian et al. 1998; Kisanuki et al. 2001; Moses et al. 2001).
Histology
Embryos and tissues were fixed in 4% paraformaldehyde for 24 h, dehydrated in increasing concentrations of ethanol, and embedded in paraffin for tissue sectioning. In situ hybridization was performed as described (Morrisey et al. 1997a,b). Immunohistochemistry was performed using the following antibodies: rabbit anti-Foxp1 (Lu et al. 2002), rat anti-Ki67 (clone TEC-3, Dako), MF20 (University of Iowa Hybridoma Bank).
Echocardiography
Pregnant mice were lightly anesthetized with 1%–2% isoflurane, and embryonic echocardiography was performed using a high-resolution Vevo 770 microultrasound system (VisualSonics, Inc.) equipped with an RMV-704 transducer with a center frequency of 40 MHz. Cardiac four-chamber view of the embryo was acquired by rotating the transducer ∼45° counterclockwise from the longitudinal axis of the embryo body. End-diastolic and end-systolic left ventricular internal diameters (LVIDd and LVIDs, respectively) were measured from the left ventricular short axis view with two-dimensional-orientated M-mode imaging. Measurements are the average of three embryos of each indicated genotype ± standard deviation.
Growth factor microarray analysis
Hearts from Foxp1flox/flox:Tie2-cre mutants and control littermates were collected at E12.5, and RNA was extracted and converted to cDNA and used for PCR array analysis using a Mouse Growth Factor PCR Array (SABiosciences, Inc.). The array was performed in duplicate. Of note, Fgf16 and Fgf20 were not represented on this array. Duplicates of each sample were analyzed and the results averaged. Expression of genes that changed significantly (>20%) were confirmed by standard Q-PCR, and data shown are the average of three PCR assays performed in triplicate ± standard error of the mean (SEM). Primer sequences are available on request.
Fgf20 rescue studies and HUVEC studies
Hearts from Foxp1flox/flox:Tie2-cre mutants and control littermates were collected at E11.5 and cultured in the presence of Fgf20 (100 ng/mL) or 0.1% BSA for 48 h on Transwell filters as described previously (Lavine et al. 2005). Explanted hearts were then processed for immunohistochemistry as described above for other tissues. HUVECs were transduced with a Sox17-expressing lentivirus or transfected with siRNA against human Sox17 using Lipofectamine 2000 (Invitrogen, Inc.). For Wnt3a treatment, HUVECs were treated with 100 ng/mL recombinant Wnt3a for 48 h (R&D Systems, Inc.).
ChIP-seq analysis
For ChIP-seq analysis, each biological sample was generated from 20–30 E12.5 embryonic hearts. The chromatin DNA was prepared as per Upstate Biotechnologies ChIP Assay Kit instructions (Millipore, catalog no. 17-295), except protein-G agarose beads were used to preclear chromatin. In brief, the manually minced tissues were cross-linked in 1% formaldehyde, and the previously characterized Foxp1 polyclonal antibody as well as control rabbit IgG were used to immunoprecipitate protein–DNA complexes from chromatin. The cross-links were reversed using 5 M NaCl, and the chromatin DNA was recovered using phenol/chloroform extraction. Following the manufacturer's protocol (Illumina), the immunoprecipitated DNA was end-repaired, ligated to sequencing adapters, and enriched by PCR. Gel-purified amplified ChIP DNA was sequenced on the Illumina Genome Analyzer II. Genome Analyzer sequencing output was analyzed using Illumina Genome Analyzer. Sequence alignment to the mouse genome (mm9) following pipeline processing was performed. Peaks within a region covering 5 kb upstream of and downstream from the transcriptional start site unique to the Foxp1 IgG and not found in the normal rabbit IgG analysis were used to assess the regulatory region of Sox17 and Nkx2.5 for Foxp1-binding sites. ChIP-seq-binding data was confirmed by directed ChIP in independent DNA samples using primers shown in Supplemental Table 2, and the data presented are the average of three ChIP assays performed in triplicate ± SEM.
βMHC:Nkx2.5 transgenic model
The mouse Nkx2.5 cDNA was cloned upstream of the βMHC promoter, which consists of 7 kb of upstream regulatory sequences, including exons 1–4 (Rindt et al. 1993). Transgenic DNA was purified away from plasmid backbone DNA, and was injected into CD-1 oocytes as described previously (Shu et al. 2005). Seven independent F0 founders were isolated and analyzed by histology, and three F0 founders were collected and used for Q-PCR assessment of mRNA transcript levels.
Acknowledgments
We thank Yixin Shen for his help in generation of the βMHC-Nkx2.5 transgenic mice. The Morrisey laboratory is supported by funding from the NIH (HL071589 and HL100405) and the American Heart Association Jon DeHaan Myogenesis Center. Y.T. is supported by a Post-doctoral Fellowship from the American Heart Association.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1929210.
Supplemental material is available at http://www.genesdev.org.
References
- Bergman LM, Birts CN, Darley M, Gabrielli B, Blaydes JP 2009. CtBPs promote cell survival through the maintenance of mitotic fidelity. Mol Cell Biol 29: 4539–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. 2009. Evidence for cardiomyocyte renewal in humans. Science 324: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B 2009. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138: 257–270 [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27: 68–73 [DOI] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE 2005. FGF-20 and DKK1 are transcriptional targets of β-catenin and FGF-20 is implicated in cancer and development. EMBO J 24: 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE 2007. Wnt/β-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest 117: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, Banham AH 2004. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor α and improved survival in primary human breast carcinomas. Clin Cancer Res 10: 3521–3527 [DOI] [PubMed] [Google Scholar]
- Francois M, Koopman P, Beltrame M 2010. SoxF genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol 42: 445–448 [DOI] [PubMed] [Google Scholar]
- Hotta Y, Sasaki S, Konishi M, Kinoshita H, Kuwahara K, Nakao K, Itoh N 2008. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev Dyn 237: 2947–2954 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Gleiberman AS, Shi C, Simon DI, Rosenfeld MG 2008. Cooperative regulation in development by SMRT and FOXP1. Genes Dev 22: 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M 2001. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol 230: 230–242 [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM 2005. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell 8: 85–95 [DOI] [PubMed] [Google Scholar]
- Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C 1995. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378: 394–398 [DOI] [PubMed] [Google Scholar]
- Li C, Tucker PW 1993. DNA-binding properties and secondary structural model of the hepatocyte nuclear factor 3/fork head domain. Proc Natl Acad Sci 90: 11583–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE 2004a. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24: 809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhou D, Lu MM, Morrisey EE 2004b. Advanced cardiac morphogenesis does not require heart tube fusion. Science 305: 1619–1622 [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MM, Li S, Yang H, Morrisey EE 2002. Foxp4: A novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech Dev 119: S197–S202 doi: 10.1016/S0925-4773(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Lu SY, Sheikh F, Sheppard PC, Fresnoza A, Duckworth ML, Detillieux KA, Cattini PA 2008. FGF-16 is required for embryonic heart development. Biochem Biophys Res Commun 373: 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS 1997a. GATA-5: A transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol 183: 21–36 [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Parmacek MS 1997b. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J Biol Chem 272: 8515–8524 [DOI] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ 2001. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31: 176–180 [DOI] [PubMed] [Google Scholar]
- Ng WA, Grupp IL, Subramaniam A, Robbins J 1991. Cardiac myosin heavy chain mRNA expression and myocardial function in the mouse heart. Circ Res 68: 1742–1750 [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. 2007. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128: 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindt H, Gulick J, Knotts S, Neumann J, Robbins J 1993. In vivo analysis of the murine β-myosin heavy chain gene promoter. J Biol Chem 268: 5332–5338 [PubMed] [Google Scholar]
- Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P, Kanai Y 2007. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun 360: 539–544 [DOI] [PubMed] [Google Scholar]
- Searcy RD, Vincent EB, Liberatore CM, Yutzey KE 1998. A GATA-dependent nkx-2.5 regulatory element activates early cardiac gene expression in transgenic mice. Development 125: 4461–4470 [DOI] [PubMed] [Google Scholar]
- Shu W, Yang H, Zhang L, Lu MM, Morrisey EE 2001. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem 276: 27488–27497 [DOI] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. 2005. Wnt/β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol 283: 226–239 [DOI] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE 2007. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134: 1991–2000 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126: 1269–1280 [DOI] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D 2008. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol 322: 314–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CT, Yang TC, Tsai HJ 2009. Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebrafish embryos. PLoS One 4: e4249 doi: 10.1371/journal.pone.0004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lin D, Li C, Tucker P 2003. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem 278: 24259–24268 [DOI] [PubMed] [Google Scholar]
- Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, Tucker PW 2004. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131: 4477–4487 [DOI] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE 1999. Regulation of Wnt signaling by Sox proteins: XSox17α/β and XSox3 physically interact with β-catenin. Mol Cell 4: 487–498 [DOI] [PubMed] [Google Scholar]