Abstract
Profound epigenetic differences exist between genomes derived from male and female gametes; however, the nature of these changes remains largely unknown. We undertook a systematic investigation of chromatin reorganization during gametogenesis, using the model eukaryote Saccharomyces cerevisiae to examine sporulation, which has strong similarities with higher eukaryotic spermatogenesis. We established a mutational screen of histones H3 and H4 to uncover substitutions that reduce sporulation efficiency. We discovered two patches of residues—one on H3 and a second on H4—that are crucial for sporulation but not critical for mitotic growth, and likely comprise interactive nucleosomal surfaces. Furthermore, we identified novel histone post-translational modifications that mark the chromatin reorganization process during sporulation. First, phosphorylation of H3T11 appears to be a key modification during meiosis, and requires the meiotic-specific kinase Mek1. Second, H4 undergoes amino tail acetylation at Lys 5, Lys 8, and Lys 12, and these are synergistically important for post-meiotic chromatin compaction, occurring subsequent to the post-meiotic transient peak in phosphorylation at H4S1, and crucial for recruitment of Bdf1, a bromodomain protein, to chromatin in mature spores. Strikingly, the presence and temporal succession of the new H3 and H4 modifications are detected during mouse spermatogenesis, indicating that they are conserved through evolution. Thus, our results show that investigation of gametogenesis in yeast provides novel insights into chromatin dynamics, which are potentially relevant to epigenetic modulation of the mammalian process.
Keywords: Histone, acetylation, phosphorylation, sporulation, gametogenesis
More than two decades ago, now classic experiments demonstrated the nonequivalence of DNA derived from male and female gametes (McGrath and Solter 1984; Surani et al. 1984). Indeed, microinjection of nuclei derived from two gametes produced by the same sex, either both male or both female, leads to early embryonic death (McGrath and Solter 1984; Surani et al. 1984). Thus, fertilized eggs must contain two haploid genomes originating from a paternal and a maternal gamete in order to proceed through embryonic development. Because all females are born from union of a male and a female gamete, which share the same chromosome composition, it must be the nongenetic content of the male and female gametes that is required for normal development.
Epigenetic features, and specifically chromatin, are likely to be the crucial inherited nongenetic property of the male and female gametes. Twenty-five years after these pivotal observations, the epigenetic organization of gametes is only starting to be unraveled. The chromatin structure of oocytes remains obscure, beyond initial observations of hypoacetylation of histones H3 and H4 during meiosis (Kim et al. 2003). However, epigenetic changes in the male gamete have been more amenable to analysis. Following meiotic divisions and DNA recombination, dramatic chromatin remodeling occurs during maturation of spermatids, and is associated with extensive chromatin restructuring and compaction. In many species, the majority of histones are removed and replaced by a new group of chromatin-associated proteins, called protamines (Govin et al. 2004; Kimmins and Sassone-Corsi 2005; Gaucher et al. 2009). Protamines are specialized chromatin proteins only 50 amino acids long, highly basic, and exclusively detected in male gametes, where they compact >90% of the DNA (Balhorn 1982). They are critical, as mice bearing even a heterozygous protamine gene deletion are sterile, preventing transmission of the wild-type allele (Cho et al. 2001).
A recent study has provided new insights into the chromatin organization of male gametes (Hammoud et al. 2009), showing that the remaining histones in human sperm are not distributed randomly, but rather are associated with master regulators of early embryonic development, such as HOX genes, in a manner that potentially influences the developmental process. Moreover, epigenetic marks, such as the DNA and histone methylation patterns, have a specific distribution, which may also be linked to early embryonic development. Thus, the male gamete has a unique chromatin structure, likely related to its function after fertilization.
There are a number of additional alterations to chromatin in the mammalian male gamete. For example, several testis-specific histones are incorporated into chromatin at different stages of maturation (Martianov et al. 2005; Tanaka et al. 2005; Govin et al. 2007). Histone modifications are also involved in chromatin reorganization, such as histone hyperacetylation as spermatids start to elongate (Hazzouri et al. 2000). A testis-specific bromodomain protein, Brdt, binds to hyperacetylated histones and compacts the chromatin (Pivot-Pajot et al. 2003; Moriniere et al. 2009). However, it remains unknown how most histones are removed from the genome, and how certain specific regions, such as the HOX genes, are protected against histone loss and protamine deposition. Moreover, the role of post-translational modifications during the steps of this dramatic process of chromatin restructuring have not yet been addressed in a systematic analysis.
Lower eukaryotes undergo a differentiation pathway that is similar to higher eukaryotic spermatogenesis. Upon nutrient starvation, diploid yeast sporulate to generate gametes. The first step consists of meiosis, which ensures mixing of inherited genetic information. Haploid cells then differentiate into mature spores, which develop a thick spore wall providing protection from harsh environmental conditions, such as exposure to chemicals and dehydration. Similar to higher eukaryotic sperm, spore nuclei are reorganized and compacted. This spore differentiation pathway is dependent on coordination of an intricate network of events, including a transcriptional program involving >1000 genes, and other genome-wide chromatin events. Histone modifications have a role at several steps: H3 phosphorylation on Ser 10 is detected during meiosis and then disappears, followed by H4 phosphorylation on Ser 1. This latter mark helps in chromatin compaction, and is conserved through evolution, as it is also found during post-meiotic spermatogenesis (Krishnamoorthy et al. 2006). Beyond these observations, however, little is known about the chromatin transactions that occur during this process.
Our interest is to broadly assess the role of chromatin changes during gametogenesis. We took an unbiased approach, using systematic substitution mutagenesis of histones H3 and H4 in the yeast Saccharomyces cerevisiae, since technical limitations associated with gamete differentiation in higher eukaryotic organisms restrict such screening. Our analysis uncovered several novel and apparently distinct chromatin pathways involved in sporulation, and, strikingly, we find that aspects of these pathways are evolutionarily conserved during mammalian spermatogenesis.
Results
Creation of a new histone library in a yeast strain background able to synchronously and efficiently sporulate
Systematic substitution mutagenesis of histones H3 and H4 in S. cerevisiae has been used recently to examine effects on vegetative growth (Dai et al. 2008; Nakanishi et al. 2008). However, the strain backgrounds used for these mutant collections are not optimal for a sporulation screen. For example, the yeast genetic background strains s288c and W303 sporulate with poor efficiencies: Less than 50% of the initial vegetative cells form tetrads of spores, the cells require up to 5–6 d to reach sporulation, and individual cells proceed nonsynchronously through the process. In contrast, the genetic background SK1 is typically used for sporulation studies because the cells proceed synchronously and nearly completely, and sporulate within 24–48 h.
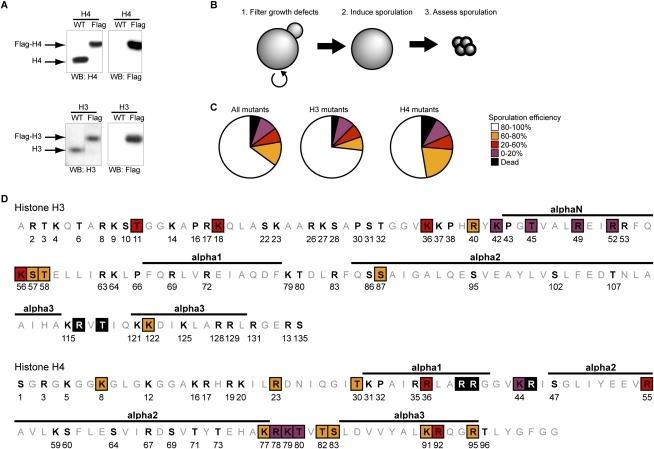
Therefore, we created a “histone shuffle strain” in the SK1 background by deleting both genomic genes encoding the H3 and H4 genes (HHT1, HHT2, HHF1, and HHF2), and providing the histone genes on a plasmid containing a URA3 selectable marker (Supplemental Fig. S1A). The H3 and H4 substitution mutations on a second marked plasmid were transformed, and the original wild-type plasmid was evicted (by negative selection of URA3 on 5-FOA), which ensures that the entire H3 and H4 population is expressed from the new plasmid. Examination of the new strain shows that endogenous H3 and H4 genes are absent, and can be replaced by Flag-tagged versions of either of the histones (Fig. 1A). Importantly, the doubling time (Supplemental Fig. S1B) and sporulation efficiency (Supplemental Fig. S1C) of the shuffle strain are comparable with a wild-type strain.
Figure 1.
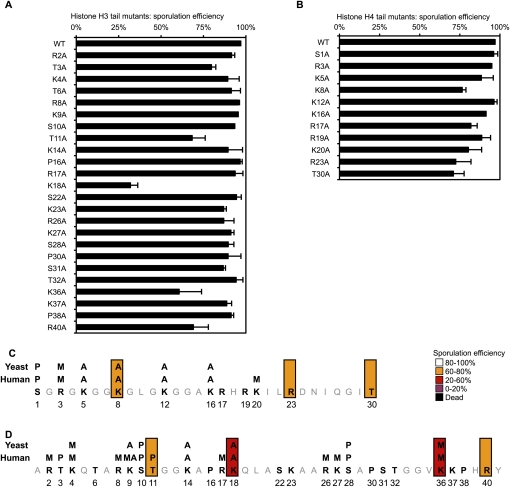
A systematic screen identifies H3 and H4 residues essential for sporulation completion. (A) Creation and validation of a histone shuffle strain in the SK1 background. The strain was validated by replacing a wild-type (WT) plasmid with a plasmid expressing Flag-tagged H3 or H4. Detection of H3 and H4 by Western blot confirms that neither genomic copy of the H3 and H4 genes remains, and that the entire population of H3 and H4 is expressed from the plasmid. (B) Schematic of the rationale used for the screen. More than 100 substitution mutants have been created and validated by sequencing. Several substitution mutations were lethal, and were not analyzed further. Sporulation was induced and spore formation was quantified in all other mutants. (C) Statistical analysis of the screen results. The severity of the sporulation defects is described using the following color code: a sporulation efficiency included within 0%–20%, 20%–60%, 60%–80%, and 80%–100% is represented in purple, red, orange, and white, respectively. Lethal mutations are represented in black. This same color-coding is used throughout the figures. (D) Graphic representation of the substitution mutants affected in sporulation. The histone fold is represented at the top of each histone, and is based on data from White et al. (2001).
Next, in the new SK1 histone shuffle background, we created a collection of strains containing alanine substitution mutations at modifiable Ser, Thr, Lys, and Arg residues, using plasmids that were screened previously during vegetative growth (Nakanishi et al. 2008). Plasmids within the strain collection were recovered and sequenced to confirm the mutations. Prior to induction of sporulation, the mutants were grown in “rich” media—YPD (glucose) and then YPA (acetate) (Fig. 1B)—to eliminate any mutants showing growth defects, since these would suggest a general loss of histone function, such as histone loading or folding into the nucleosome. Indeed, in agreement with previous analyses (Dai et al. 2008; Nakanishi et al. 2008), ∼5% of the substitutions resulted in lethality (Fig. 1C). Since histones H3 and H4 are highly conserved through evolution (Supplemental Fig. S2A–C), this low lethality suggests that the integrity of each amino acid is, in general, not crucial for nucleosome function in optimal growth conditions.
Identification of two patches of substitution mutations affecting sporulation
The sporulation phenotype of each mutant was analyzed. Approximately 30% of the substitution mutants displayed lowered sporulation efficiency (Fig. 1C): Ten percent of them were severely reduced (0%–20% of wild type) (Fig. 1C, in purple), while the remainder of the substitutions were lowered to a lesser degree (20%–60% of wild type [Fig. 1C, in red]; 60%–80% of wild type [Fig. 1C, in orange]). Thus, although most individual substitution mutations in H3 and H4 are well tolerated in rich media, the integrity of numerous residues within each histone are important for spore differentiation.
The substitution mutants that are defective are localized largely in spatially accessible regions within the nucleosome (Figs. 1D, 2). That is, many are excluded from the α-helices of the globular domains, and are instead concentrated in the N-terminal and C-terminal tails, and in the loops between helices.
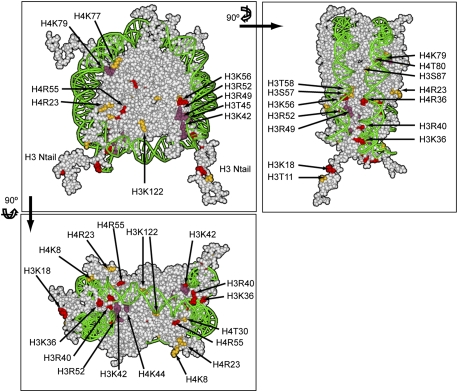
Figure 2.
Representation of the nucleosome, highlighting the residues essential for completion of sporulation. The same color-coding is used as in Figure 1. Front and two different lateral views are presented. PDB file 1KX5 was used (Davey et al. 2002), and was color-coded using Accelrys Discovery Studio Visualizer 2.5.
We found two distinct patches localized within H3 and H4 that are particularly sensitive to substitution. One patch involves a stretch of seven residues on H4 (K77–S83) (Figs. 1D, 3B), three of which are nearly blocked in spore formation (Fig. 3A), and therefore we named this patch LOS (low sporulation). The LOS patch is located on loop 2 of H4, between α-helices 2 and 3, and includes almost all residues within the loop (Figs. 1D, 2, 3B). This is especially clear when the region is enlarged (Fig. 3C), showing that the patch is localized on the edge of the nucleosome, in close proximity to contacts with DNA, as the double helix wraps around the nucleosome.
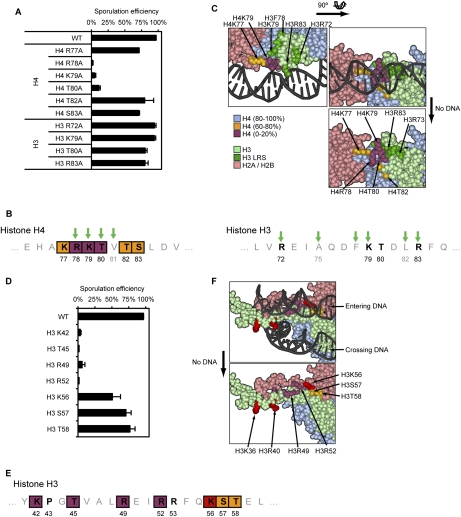
Figure 3.
Detailed analysis of the LOS and DEE patches. (A) Sporulation efficiency data for the LOS substitution mutants. (B) Graphical representation of the LOS (H4) and the H3 LRS regions. Green arrows represent identified LRS mutations (Park et al. 2002). (C) Three-dimensional representation of the LOS patch. The severity of the sporulation phenotype for each substitution is represented following the same color-coding as in Figure 1C. The H3 LRS mutants are represented in dark green. H2A/H2B, H3, and H4 are represented in light pink, light green, and light blue, respectively. (D) Sporulation efficiency data for DEE mutants. (E) Graphical representation of the DEE patch. (C) Three-dimensional representation of the DEE patch. Refer to C and Figure 1C for details on the color-coding.
The phenotypes of the LOS mutants were characterized more precisely. The sporulation repressor Rme1 is not derepressed by LOS mutations (data not shown), which likely excludes a general defect in silencing genes that repress sporulation. Induction of the sporulation master regulators Ime2 and Ime4 was variable in the mutants, but remained in the range of 50%–100% of their levels in wild-type cells (Supplemental Fig. S3A). In addition, DNA replication was similar to wild-type cells, as assessed by flow cytometry carried out on cells stained with propidium iodide (data not shown). Therefore, LOS mutations do not dramatically affect the induction of sporulation, or the replication of the DNA. However, using DAPI staining of DNA, we did not observe four normal meiotic products (data not shown), which suggests that LOS mutants may be defective in the meiotic process itself.
The LOS patch shares residues with a previously described patch called LRS (loss of rDNA silencing) (Park et al. 2002). However, the LRS mutants encompass residues located on both H3 and H4, and occupy a large patch (Fig. 3A,B [green arrows], C [dark green]), whereas only H4 mutants located in this region have defects in sporulation, and occupy a more restricted surface area (Fig. 3A,B [purple and orange], C [purple and orange]). It has been demonstrated recently that H3 LRS mutations disrupt recruitment of Sir3 to this nucleosomal interface, leading to silencing defects (Hyland et al. 2005; Norris et al. 2008). The H3 LRS mutants that lose Sir3 binding do not have a defect in sporulation in our study. Hence, we conclude that the new LOS patch is independent of the LRS phenotype, and LOS may define a new interface required for sporulation, whose function is unrelated to Sir3 recruitment.
A second patch required for sporulation occurs on the α-helix in the N terminus of H3, encompassing residues K42–T58 (Figs. 1D, 3E) in the region DNA enters and exits the nucleosome (Figs. 2, 3F), and therefore we named this patch DEE (DNA entry/exit). Substitutions in residues in one region within this patch (T42–R52) cause severe reduction of sporulation, while residues in a second region (K56–T58) are less defective (Fig. 3D,E). The sporulation repressor Rme1 remained repressed (data not shown), and the induction of master regulators Ime2 and Ime4 remained similar to wild type (Supplemental Fig. S3B; data not shown). Therefore, DEE mutants do not prevent general induction of sporulation. The patch includes many residues involved in histone interaction with DNA (Figs. 2, 3), and it is thus unclear why some of these mutants are fully competent for mitotic growth, but nearly fail to form spores.
The DEE patch includes H3K56, an acetylated residue involved in nucleosome assembly during replication and DNA repair. Indeed, as shown previously (Recht et al. 2006), we detect a transient peak in H3K56ac during meiosis, with similar timing to H3S10ph (Fig. 4A,B), which also increases at that time (Wei et al. 1998; Krishnamoorthy et al. 2006). We tested several substitutions, and found that K56A and K56R both lowered sporulation efficiency, while K56Q, which mimics the neutralization of charge caused by acetylation, is closer to wild type in sporulation efficiency (Fig. 4C). This is in contrast to DNA damage conditions, where a K56Q strain is defective similarly to K56R (Schneider et al. 2006; Driscoll et al. 2007; Han et al. 2007), and thus there may be a distinct role for K56 acetylation within the entire DEE patch during sporulation.
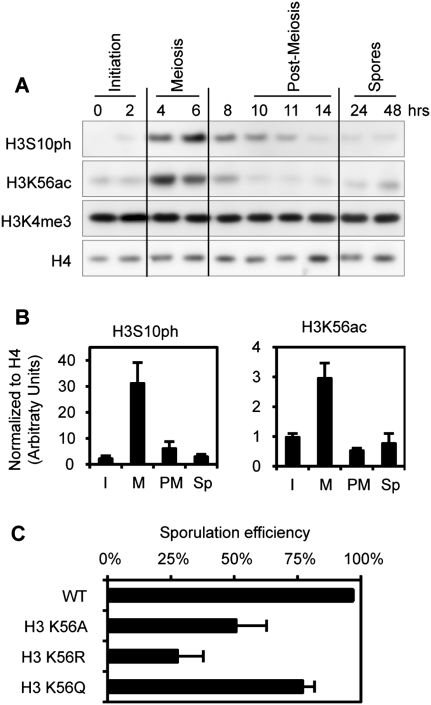
Figure 4.
Dynamics of meiosis-specific histone modifications during sporulation. (A) H3S10ph and H3K56ac were detected by Western blot during sporulation. (B) Quantification of the Western blots, including images presented in A. The time course has been broken into stages, as indicated in A. (I) Initiation; (M) meiosis; (PM) post-meiosis; (Sp) spores. Quantification was performed using two to three biological independent replicates (depending on the time points). (C) Sporulation efficiency data for different H3K56 mutants. H3K56R mimics the unmodified form of H3K56, while H3K56Q mimics the acetylated form of H3K56.
Analysis of modifications of H3 during gametogenesis
Histone tails are modified during all major cellular processes. Surprisingly, despite an abundant literature describing the occurrence of many modifications, our data show that substitution of 70% of the known modified residues does not show defects during either vegetative growth in rich media or sporulation (Fig. 5; Dai et al. 2008; Nakanishi et al. 2008). For example, H3K4me3 is a key modification that occurs over start sites of transcription during induction (for review, see Berger 2007). In spite of its ubiquity, we detected no loss of vegetative growth or sporulation (Fig. 5) in an H3K4A strain lacking K4 methylation, and, furthermore, the level of H3K4me3 was unchanged during a sporulation time course (Fig. 4A). Thus, the absence of many individual modifications on the H3 and H4 tail does not appear to severely affect chromatin function during most of the yeast life cell cycle.
Figure 5.
Analysis of mutants located in the N-terminal tails of H3 and H4. (A,B) Detailed sporulation efficiency data for histones H3 and H4. (C,D) Graphical representation of the residues essential for sporulation completion (same color-coding as in Fig. 1).
However, substitution of certain residues within the N-terminal tails does lower sporulation efficiency (Fig. 5), and we carried out further analysis of some of these sites. H3K18A and H3K36A mutations decrease sporulation efficiency to 30% and 60%, respectively (Fig. 5A,C). This may be due to absence of H3K18ac or H3K36me; however, no clear changes were detected in the global levels of the K18ac or K36me modifications over the sporulation time course (data not shown). Moreover, a mutation of H3K18 does not significantly affect the expression of the master regulators Ime2 and Ime4 (Supplemental Fig. S3C).
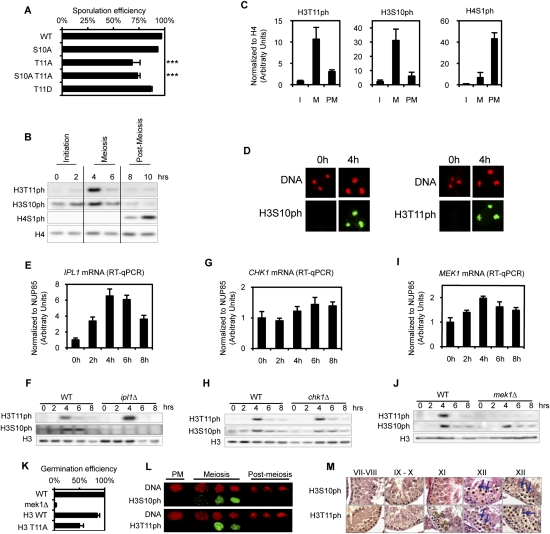
Within the H3 and H4 tails, we found that H3T11A substitution had a significant effect, reducing sporulation efficiency (Fig. 5A). We tested multiple isolates of this substitution, and found that the level was ∼65% of wild type (Fig. 6A). It is striking that H3S10A substitution did not reduce sporulation efficiency, and that the introduction of the double substitution (H3S10A H3T11A) did not grow more poorly than the single H3T11A substitution, suggesting that H3S10ph is not a key modification during sporulation. We determined that H3T11 is phosphorylated during sporulation, and, as expected, the signal is eliminated in the H3T11A strain (Fig. 6B; Supplemental Fig. S4). A H3T11D strain, as a potential mimic of phosphorylation, behaved more similarly to wild type (Fig. 6A).
Figure 6.
H3T11ph is a new meiotic mark in yeast and mammals. (A) Sporulation efficiency data for selected mutants. P-values are 10−7 and 10−3 for H3T11A and H3S10AT11A mutants, respectively (asterisks). (B) Detection of H3T11ph during sporulation by Western blot. (C) Quantification of the Western blots, including images presented in B. The time course has been broken into stages, as indicated in B. (I) Initiation; (M) meiosis; (PM) post-meiosis. Quantification was performed using two to three biological independent replicates (depending on the time points). (D) In situ detection of H3S10ph and H3T11ph by immunofluorescence during sporulation. (E,G,I) Quantifications of mRNA abundance of IPL1, CHK1, and MEK1, respectively. Data were obtained from four independent biological replicates, and normalized to NUP85 (see the Materials and Methods). (F) Ipl1 is essential for H3S10ph deposition. Depletion of Ipl1 during meiosis (using a conditional strain; see the text for details) leads to a loss of H3S10ph, but does not affect H3T11ph. Western blot analysis using indicated antibodies. (H) Chk1 is not required for H3S10ph or H3T11ph deposition. Western analysis of the histone modifications in a chk1Δ strain compared with the wild-type strain. (J) Mek1 is essential for H3T11ph deposition. Western analysis of the histone modifications in a mek1Δ strain compared with the wild-type strain. (K) Germination efficiency. Spores were dissected and then germinated on YPD plates. More than 50 spores were analyzed using two independent isolates of each strain. (L) Analyses of H3S10ph and H3T11ph during mouse spermatogenesis using immunofluorescence. (M) Immunohistochemistry using H3S10ph and H3T11ph antibodies during mouse spermatogenesis.
The H3T11ph modification specifically increased during meiosis, with similar timing as H3S10ph and prior to post-meiotic H4S1ph, as we observed previously (Fig. 6B,C; Krishnamoorthy et al. 2006). Immunofluorescence experiments confirm this pattern, and also show a homogeneous distribution for H3S10ph and H3T11ph during meiosis (Fig. 6D).
The H3T11A substitution mutation slightly lowered both H3S10ph and H4S1ph, but quantification indicated that the decrease in H3S10ph and H4S1ph signal is commensurate with the reduction in sporulation observed in a H3T11A mutant (Supplemental Fig. S5; data not shown), indicating that these phosphorylation events are independent. Taken together, it appears that phosphorylation of H3T11 has a more important role during meiosis than phosphorylation of H3S10.
We investigated the kinase that is required for H3T11ph. Our analysis of published data identified three kinases whose transcription is specifically induced during meiosis (Fig. 6E,G,I). The first is Ipl1, the ortholog of Aurora kinase, which phosphorylates H3S10ph during meiosis (Hsu et al. 2000). IPL1 is an essential gene, and cannot be deleted. Thus, we used a strain in which IPL1 is expressed under the control of the CLB2 promoter, which is active during vegetative growth, but is rapidly repressed upon induction of sporulation (Monje-Casas et al. 2007). This allowed analysis of the abundance of H3S10ph and H3T11ph during meiosis in the absence of Ipl1. We found reduced H3S10ph, but no effect on H3T11ph (Fig. 6F), confirming that Ipl1 is responsible for H3S10ph deposition during meiosis, but excluding a role in H3T11 phosphorylation. The second potential kinase is yChk1, the ortholog of mammalian Chk1, which phosphorylates H3T11ph in mammalian cells and regulates gene activation during DNA damage-induced transcriptional repression (Shimada et al. 2008). CHK1 is not essential, and its deletion did not decrease either H3S10ph or H3T11ph (Fig. 6H). The third potential kinase is Mek1, and, strikingly, we found that deletion of MEK1 completely abolished H3T11ph, but had little effect on H3S10ph (Fig. 6J). These data implicate the meiotic Mek1 kinase specifically in phosphorylation of H3T11. It is interesting that H3S10 and H3T11 are adjacent residues and are both involved in meiosis, but are phosphorylated by distinct kinases.
Mek1 promotes interchromosomal recombination, and its kinase activity is required for the formation of viable spores, as mek1Δ spores do not germinate (Fig. 6K; Rockmill and Roeder 1991; Wan et al. 2004). We tested the viability of H3T11A mutant spores. The wild-type shuffle strain (H3 WT) germinates efficiently (>80%) (Fig. 6K); however, only 50% of H3T11A spores were able to germinate (Fig. 6K). These results suggest that the H3T11A substitution partially phenocopies the deletion of mek1, and that H3T11 phosphorylation is required for the maturation of fully functional spores. We found that the H3T11A spores are not affected in chromatin compaction (see below; Supplemental Fig. S5A), further suggesting that their lowered viability stems from a defect during meiosis.
H3S10 phosphorylation occurs during both yeast sporulation and mammalian gametogenesis (Wei et al. 1998; Krishnamoorthy et al. 2006). We therefore investigated whether H3T11ph similarly occurs during spermatogenesis. By immunodetection of mouse germ cells from staged tubules (Fig. 6L) and on paraffin testis sections (Fig. 6M), we found that H3S10ph and H3T11ph both occur during meiosis, and that both strongly mark condensed meiotic chromosomes of spermatocytes from stage XI and XII tubules (Fig. 6M, see arrows).
Analysis of modifications of H4 during gametogenesis
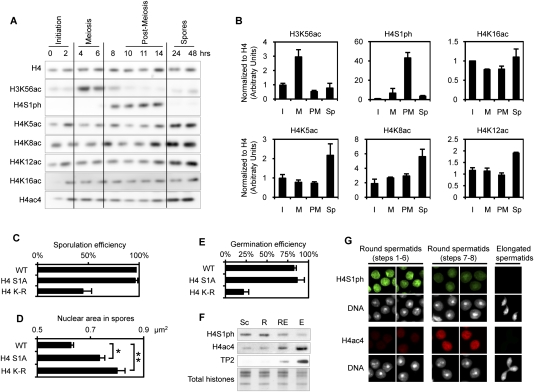
We also found, among H4 tail substitutions, a modest effect of K8A on sporulation efficiency (Fig. 5B). There is redundancy between K5, K8, and K12 acetylation during vegetative growth, in regulation of transcription and DNA repair, among other processes (Dion et al. 2005). We examined whether these H4 acetylations increase during sporulation, and found that all three (K5ac, K8ac, and K12ac) increase late in the process, after meiotic and post-meiotic phosphorylation events on the H3 and H4 tails, as described above (Fig. 7A). For example, H4K5ac, H4K8ac, and H4K12ac all begin to increase at ∼14 h, after H4S1ph has peaked and begun to decline (Fig. 7A,B). The acetylations then remain at this elevated level—even at the end of the process, when the spore reprogramming is complete (Fig. 7A,B). It is notable that H4K16ac, which has a distinct role in chromatin (Shogren-Knaak et al. 2006), did not change significantly over the time course (Fig. 7A,B).
Figure 7.
Dynamics of H4 modifications during post-meiotic maturation of the gamete. (A) The phosphorylation and acetylation status of H4 during sporulation analyzed by Western blot. The H3K56ac and H4S1ph pattern confirms that the cells are sporulating synchronously. (B) Quantification of the Western blots, including images presented in A. Quantification was performed using two to three biological independent replicates (depending on the time points). (C) Sporulation efficiency data for H4S1A and H4K5/8/12R mutants. (D) Average nuclear size of fully mature spores of wild-type (WT), H4S1A, and H4K5/8/12R strains. The increase in size is statistically significant for both mutants: P-values are 10−4 and 10−11 for H4S1A and H4K–R mutants, respectively (asterisks). (E) Germination efficiency. Spores were dissected and then germinated on YPD plates. More than 80 spores were analyzed using two independent isolates of each strain. (F) Phosphorylation and acetylation status of H4 during mouse spermatogenesis, analyzed by Western blot. H4S1ph is detected during post-meiosis and disappears, while H4 is hyperacetylated during spermatid elongation. (G) The phosphorylation and acetylation status of H4 during mouse spermatogenesis, analyzed by immunofluorescence on staged cells. The results confirm the data presented in E.
Since K5ac, K8ac, and K12ac displayed similar kinetics during late sporulation, we tested whether these modifications work synergistically, via simultaneous substitution of the three residues to arginine. The triple H4K5/8/12R substitution mutant displayed <50% sporulation efficiency compared with wild-type H4 (Fig. 7C). Furthermore, the rise in acetylation on H4 follows the transient rise of post-meiotic H4S1ph (Fig. 7A), which is involved in chromatin compaction (Krishnamoorthy et al. 2006). Thus, we investigated whether elimination of the acetylation in the H4K5/8/12R strain altered compaction, using DAPI staining of DNA to measure nuclear size. We examined a large number of spore nuclei (>200), and, as a control, confirmed that H4S1A substitution leads to a significant increase in median nuclear size, from 0.6 μm2 to 0.73 μm2 (P < 10−4) (Fig. 7D,E; Krishnamoorthy et al. 2006). Using the same approach, we found that the H4K5/8/12R triple substitution correlates with an even larger median nuclear size, to 0.8 μm2 (t-test, P < 10−11) (Fig. 7D; Supplemental Fig. 5). Thus, H4 modifications that increase following the end of meiosis appear to be involved in nuclear compaction.
We investigated whether H4 modifications are important for spore viability during germination. We found that H4S1A spores are viable, but spores expressing the H4K5/8/12R substitution mutant are nearly completely inviable (Fig. 7E). This suggests that H4 acetylation has an important function in the compaction of the chromatin in spores, and subsequently plays a key role during germination.
We showed previously that H4S1ph is conserved in mammalian spermatogenesis, and in fact persists later in the process relative to H3S10ph (Krishnamoorthy et al. 2006), similar to our findings during sporulation. This led us to investigate how these marks, and H4ac, behave during spermatogenesis in the mouse. As published previously, we found, using both Western and immunofluorescence of staged sperm, that H4K5/8/12ac increases in later stages of round spermatids (Fig. 7F,G; Hazzouri et al. 2000). We then compared the hyperacetylation to the kinetics of H4S1ph, and found that the dramatic increase in acetylation occurs only following the decline of H4S1ph in early stages of round spermatids (Fig. 7F,G). It is striking that these modifications have a similar relative temporal relationship during sporulation and mammalian spermatogenesis, suggesting that histone H4 phosphorylation and acetylation dynamics during the final differentiation stages to generate mature gametes has been conserved through evolution.
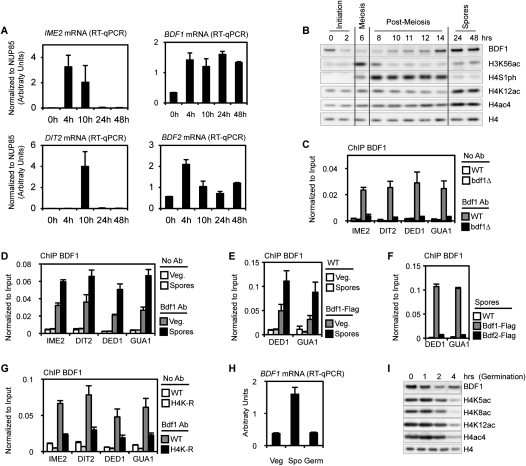
The bromodomain-containing protein Brdt has been shown to compact hyperacetylated chromatin (Pivot-Pajot et al. 2003). The orthologous proteins yBdf1 and yBdf2 also bind to acetylated H4 (Ladurner et al. 2003), and we investigated whether they may have a similar function during spore maturation. First, we analyzed their relative expression levels during sporulation, by RT–qPCR. IME2 (early sporulation, at 4 h) and DIT2 (mid-sporulation, at 10 h) transcript levels demonstrate that the cells are synchronous (Fig. 8A). BDF1 and BDF2 mRNA were both induced during meiosis (4 h after induction of sporulation). However, only BDF1 mRNA was maintained at a higher level during the remainder of the differentiation program and in fully mature spores, until germination is induced and the spores return to vegetative growth (see below; Fig. 8A,H).
Figure 8.
Bdf1 is highly expressed and enriched on acetylated chromatin in mature spores. (A) Quantifications of mRNA abundance of IME2, DIT2, BDF1, and BDF2. Data were obtained from three independent biological replicates, and were normalized to NUP85 (see the Materials and Methods). IME2 and DIT2 profiles confirm that the cells were sporulating synchronously. (B) Bdf1 abundance and acetylation status of H4 during sporulation analyzed by Western blot. The H3K56ac and H4S1ph pattern confirms that the cells were sporulating synchronously. Bdf1 accumulates in spores concomitantly with H4ac (quantification is presented in Supplemental Fig. 7B). (C) Bdf1 antibody is specific by ChIP. Wild-type (WT) and bdf1Δ cells were used to assess the specificity of Bdf1 antibody by ChIP. (D) Bdf1 is enriched on the chromatin of mature spores. ChIP was performed using the Bdf1 antibody in vegetative cells before sporulation induction (Veg.) or in mature spores (24 h after sporulation induction), and was analyzed on four different loci. (E) ChIP experiments using a Flag antibody in Bdf1-Flag strains. Results are consistent with data for Bdf1 antibody (Fig. 7D). (F) Bdf1, but not Bdf2, is recruited to the chromatin of mature spores. ChIP experiments were performed in Bdf1-Flag or Bdf2-Flag spores. (G) Bdf1 recruitment in mature spores is dependent on H4 acetylation. ChIP experiments were performed in wild-type and H4K-R strains in fully mature spores. (H) Quantification of BDF1 mRNA in vegetative cells (before sporulation induction, Veg), in fully mature spores (48 h after induction, Spo), and during germination. The level of BDF1 mRNA is specifically enriched in fully mature spores. (I) Bdf1 abundance analyzed by Western blot during germination. The Bdf1 level is reduced to vegetative levels 3–4 h after induction of germination. This profile is highly similar to the decrease of acetylated H4, as detected using several antibodies. The timing of this decrease corresponds to the re-entry into vegetative growth (Herman and Rine 1997; Joseph-Strauss et al. 2007; data not shown).
Next, the level of Bdf1 was analyzed by Western blot (Chua and Roeder 1995). The specificity of the Bdf1 antibody was tested by ourselves and others (Chua and Roeder 1995; Ladurner et al. 2003): First, we found that the signal was eliminated in a bdf1Δ strain (Chua and Roeder 1995; Ladurner et al. 2003), and second, in a Bdf1-Flag epitope-tagged strain, both a Flag antibody and the Bdf1 antibody immunoprecipitated the slightly larger Bdf1-Flag (Supplemental Fig. S7A). We then found, using Western blot experiments, that Bdf1 is highly expressed in fully mature spores, concurrent with the hyperacetylation of H4 (Fig. 8B; Supplemental Fig. S7B). This pattern was confirmed using the Flag-Bdf1 strain (data not shown). This increase in Bdf1 levels during late sporulation suggests that Bdf1 mediates the final compaction of the spore nuclei, similarly to the putative function of Brdt in spermatogenesis (Govin et al. 2004; Moriniere et al. 2009). It remains unclear why BDF1 mRNA is induced during meiosis at 4 h of sporulation, whereas a dramatic increase of Bdf1 protein is observed only at the 14-h mark in the process.
The role of Bdf1 during chromatin compaction in fully mature spores could not be assessed in a bdf1Δ strain, because the deletion strain does not complete the sporulation program and does not form tetrads (Chua and Roeder 1995; our data). Nevertheless, chromatin immunoprecipitation (ChIP) experiments strongly suggest an important role for Bdf1 in fully mature spores. First, we determined that the Bdf1 antibody was able to chromatin-immunoprecipitate the protein, as the Bdf1 Western signal was lost in a bdf1Δ strain (Fig. 8C). The amount of Bdf1 bound to the chromatin was then determined in vegetative cells versus mature spores at different locations. A recent study reported that Bdf1 is enriched on the 5′end of each ORF (Koerber et al. 2009); therefore, primers were designed on the +1 and +2 nucleosomes of a variety of genes having typical transcription patterns (sporulation- or germination-specific). We found Bdf1 enriched on the chromatin of mature spores compared with vegetative cells (Fig. 8D). This result was confirmed using both the Bdf1 antibody (Fig. 8D) and a Flag antibody (Fig. 8E) in the Bdf1-Flag strain (only a selection of genes is shown in Fig. 8E). More than 15 different loci were tested, and the enrichment of Bdf1 appeared to be homogeneous and independent of the transcription status of each gene. Indeed, IME2 and DIT2, which are specifically expressed during sporulation, show the same level of recruitment as DED1 and GUA1, genes that are slightly or greatly induced during germination (Fig. 8A [mRNA levels], D [ChIP data]; Supplemental Fig. S7C,D). We then attempted to chromatin-immunoprecipitate H4Kac in fully mature spores (48 h after induction), but were unable to detect the modification, although we previously detected H4ac at 11 h after induction of sporulation using ChIP (Govin et al. 2010). We therefore hypothesize that the high level of chromatin compaction at the late stage of maturation may block ChIP detection of H4ac. We also found that Bdf1, and not Bdf2, appears to have a role in late sporulation based on findings (1) of low levels of BDF2 mRNA levels in mature spores (Fig. 8A), and (2) that Bdf2 was not recruited to the spore chromatin, using ChIP of a Flag-tagged strain (Fig. 8F).
Importantly, recruitment of Bdf1 to spore chromatin was dependent on the acetylation status of H4, because the Bdf1 ChIP signal was abolished in H4K–R mutant spores (Fig. 8G). Finally, we investigated the levels of H4ac and Bdf1 when spores return to vegetative growth; i.e., as the spores begin to germinate. BDF1 mRNA levels drop to those detected before induction of sporulation (Fig. 8H). Bdf1 protein levels also were reduced, and concomitantly with the decrease of H4 acetylation. The timing of the decrease in both H4 acetylation and Bdf1 perfectly matches the return to growth of the cells, which divide 4 h after the induction of germination (Herman and Rine 1997; Joseph-Strauss et al. 2007; data not shown). Taken together, these data provide strong evidence for a role of H4 hyperacetylation to recruit Bdf1 to compact chromatin specifically during late sporulation.
Discussion
The yeast S. cerevisiae has long provided a highly informative model for chromatin mechanisms that are evolutionarily conserved. In this study, we extend the utility of the yeast model to identify numerous novel nucleosome regions and histone H3 and H4 alterations that underlie chromatin reorganization during sporulation. These include both new patches on the surfaces of the nucleosome and new post-translational modifications. We initially determined the sporulation efficiency of the substitutions relative to wild-type histones in a simple endpoint assay, and then examined the stage of the process blocked by the substitution mutations. This analysis identified a new meiotic modification, H3T11ph, and shed light on a new pattern of H4 modifications during the final stages of spore maturation. Finally, we investigated whether these modifications are conserved through evolution to spermatogenesis, a process having numerous features in common with sporulation, including ortholog proteins able to read and interpret these modifications.
Importantly, in this first large-scale mutational study, we analyzed in detail only a small number of modifications; we note that the first three modifications addressed for mechanism (H3T11ph and H4K5/8/12ac, along with H4S1ph in our previous study) (Krishnamoorthy et al. 2006) are conserved during mouse spermatogenesis. Thus, an important conclusion is that our broad analysis of yeast sporulation will usher in further discovery of specific modifications and novel chromatin-modulating proteins that regulate spermatogenesis.
It has been revealed in S. cerevisiae that substitutions of single amino acids in histones do not, in general, have a dramatic effect on mitotic growth (Dai et al. 2008; Nakanishi et al. 2008). However, our results show that, in striking contrast, there are numerous single substitutions that have significant deleterious effects on meiosis and sporulation.
We identify two surface patches on H3 and H4, each comprising a group of substitutions that profoundly lower sporulation efficiency. These patches are likely to provide surfaces involved in protein interactions. The substitutions in the LOS patch have a strong negative effect on sporulation, affecting a stage following early steps, such as induction of the master regulators Ime1 and Ime2, and DNA synthesis. Our data suggest that the inflection point follows DNA synthesis, but is before meiotic divisions. LOS mutants do not show any mitotic defects, but cannot complete meiosis, suggesting that the main function of the LOS patch is meiotic-specific.
Interestingly, loss of telomere silencing has been linked to defects in meiosis completion in Schizosaccharomyces pombe (Nimmo et al. 1998), but no clear functional mechanism has been proposed. The LOS patch in H4 occurs within a larger patch including both H3 and H4, called the LRS region, which regulates heterochromatic silencing of telomeres via direct interaction with the Sir3 component of the Sir2/Sir3/Sir4 silencing complex. However, it appears highly unlikely that the LOS phenotype is directly related to a loss of Sir3 recruitment, because some of the key residues in H3 within the LRS region that interact with Sir3 are not in the LOS group of residues (Norris et al. 2008). Furthermore, a deletion of Sir3 only delays meiosis completion, but does not greatly affect sporulation efficiency (Trelles-Sticken et al. 2003). Taken together, this suggests that the LOS patch involves a new Sir3-independent pathway critical for meiosis completion.
One speculative possibility involves assembly of a telomeric cluster, called a bouquet, which forms at the beginning of meiosis and is critical for chromosomal pairing and successful meiotic divisions (Siderakis and Tarsounas 2007). The LOS patch could be involved in this process, which has been shown to be independent of Sir3 (Trelles-Sticken et al. 2003), but involves other silencing telomeric proteins (Chikashige and Hiraoka 2001).
The second patch is within the H3 amino tail region that interacts with the DNA emerging from the linker region, as the DNA initially associates with the histone octamer. This patch (the DEE) may be involved in new nucleosome assembly following replication, since the patch includes H3K56, and acetylation at this residue is thought to be involved in nucleosome assembly during both DNA replication and DNA repair. Because both K56R (mimicking loss of acetylation) and K56Q (mimicking acetylation) are defective in these latter processes, it is thought that the rapid cycling of acetylation is important for assembly (Schneider et al. 2006; Driscoll et al. 2007; Han et al. 2007). Since we find that only K56R (and K56A), but not K56Q, is defective during sporulation, it may be that acetylation provides a surface for more stable association with an interacting protein, binding of which may also be affected by the neighboring residues in the DEE patch.
Identification of new histone modifications during sporulation
We did not detect global alteration of many well-characterized transcription-linked histone modifications during sporulation, nor did substitution mutations in the residues affect the process (e.g., H3K4me3 and H3K9/K14ac; H3K4A, H3K9A, or H3K14A). While we observed a modest defect in sporulation efficiency of the H3K36A strain, there was no global change in K36me levels during the course of sporulation. One implication is that modifications that regulate transcription of the cascade of genes during sporulation, even of the master regulators of sporulation, do not individually have a key role, and this includes H3K4me3 and H3 amino tail acetylations. A second conclusion is that the modified residues that do show a mutagenetic effect during sporulation, and that show specific temporal patterns of modification during sporulation, are likely to exert a broad functional role over the genome. These likely regulate meiotic replication and/or recombination, and stages of compaction in the post-meiotic sporulation process, as described below.
Meiotic replication and recombination
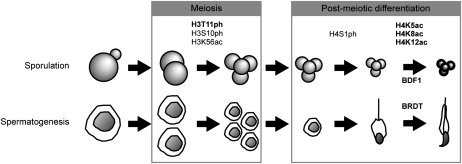
Our study represents the first detailed analysis of histone post-translational modifications through the entire course of sporulation, including within fully mature spores (summarized in Fig. 9). We show one group of modifications that is specific to meiosis. H3T11ph is emblematic of these, as it is detected for the first time in S. cerevisiae, increases during meiosis, and has a significant phenotype as H3T11A substitution (Fig. 9). H3S10ph and H3K56ac are two other modifications that increase during meiosis (Fig. 9; Krishnamoorthy et al. 2006; Recht et al. 2006; this study). It appears that H3T11ph is carried out by the Mek1 kinase, since, of several nuclear kinase candidates that are induced during the meiotic phase (Chk1, Ipl1, and Mek1), only mutation in MEK1 strongly reduces H3T11ph. Mek1 is known to regulate programmed recombination during meiosis, which is critical for continuity of the sporulation process and spore viability (Rockmill and Roeder 1991; Wan et al. 2004). Prevention of H3T11 phosphorylation by introduction of an H3T11A mutation also decreases sporulation efficiency and spore viability, which suggests that H3T11ph may be directly regulating recombination downstream from Mek1.
Figure 9.
Summary of conserved histone modifications during sporulation and spermatogenesis. Sporulation and spermatogenesis are represented as a series of steps, including meiosis and post-meiotic differentiation. Histone marks associated with specific steps are indicated. Bold is used to emphasize histone marks and patterns identified in the present study. Bdf1 and its ortholog Brdt are involved in the final compaction of the chromatin.
Post-meiotic chromatin compaction
We showed previously that H4S1ph increases just following the end of meiosis, and is involved in chromosome compaction (Fig. 9; Krishnamoorthy et al. 2006). In this study, we found that H4K5/8/12ac increases very late in meiosis, but, strikingly, H4K16ac does not show a similar increase (Fig. 9). Thus, it appears that the acetylation of K5/8/12 is involved in chromatin compaction, and H4K16ac, which, in contrast, is able on its own to decompact chromatin in vitro (Shogren-Knaak et al. 2006), is not similarly involved in this process. It is interesting that multiply acetylated H4 is an association surface for bromodomain proteins during mitosis (Dey et al. 2003) and spermatogenesis (Pivot-Pajot et al. 2003; Moriniere et al. 2009). Indeed, Brdt is specifically expressed during spermatogenesis, and contains two bromodomains (Moriniere et al. 2009). Moreover, Brdt has been shown to be able to compact acetylated chromatin (Pivot-Pajot et al. 2003; Moriniere et al. 2009). We show that the orthologous protein yeast Bdf1 appears to have a similar role during sporulation, to promote chromatin compaction through its specific recruitment to spore chromatin by acetylated histones.
Finally, an additional important conclusion from our study is that there is evolutionary conservation of histone post-translational modifications during the process of gametogenesis, extending from yeast sporulation to mammalian spermatogenesis (Fig. 9). Here we discovered a role of H3T11ph during meiosis, and extended these observations to meiosis during spermatogenesis; in both cases, there is a coincidence with H3S10ph. Furthermore, we found a remarkable late sporulation hyperacetylation of H4 following the decline in H4S1ph, and we demonstrate a similar relationship in spermatogenesis, where H4 acetylation surges after H4S1ph levels fall (Fig. 9). Thus, our study shows that systematic, discovery-based approaches to dissect chromatin regulation during yeast sporulation can enhance understanding of orthologous processes in mammalian spermatogenesis.
Materials and methods
Antibodies
H3 and H3K4me3 antibodies were obtained from Abcam (Ab1791 lot 147540 and Ab8580 lot 151301). H4ac4, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H3K56ac, and H3T11ph antibodies were obtained from Millipore-Upstate Biotechnologies (08-866 lot 31992, 07-595 lot 30417, 07-328 lot 30399, 07-327 lot 07-595, 07-329 lot 26818, 07-677 lot 30328, and 05-789 lot 26854). H4 and H3S10ph antibodies were obtained from Active Motif (39269 lot 11908001 and 39274 lot 13008001). H4S1ph was a gift from David Allis at Rockefeller, and was described previously (Barber et al. 2004). Bdf1 antibody was a gift from Shirleen Roeder (Yale University).
Yeast strains
All strains are in the SK1 background. The yeast wild-type diploid used for sporulation has the following genotype: MATa/MATα leu2∷hisG/leu2∷hisG trp1∷hisG/trp1∷hisG lys2-SK1/lys2-SK1 his4-N/his4-G ura3-SK1/ura3-SK1 ho∷LYS2/ho∷LYS2.
A histone shuffling strain was created in the SK1 background by deletion of endogenous copies of the histone H3 and H4 genes using conventional deletion methods, creating the strain yJG52 (MATa/MATα leu2∷hisG/leu2∷hisG trp1∷hisG/trp1∷hisG lys2-SK1/lys2-SK1 his4-N/his4-G ura3-SK1/ura3-SK1 ho∷LYS2/ho∷LYS2 hhf1-hht1∷LEU2/hhf1-hht1∷LEU2 hhf2-hht2∷trp1∷KanMX3/hhf2-hht2∷trp1∷KanMX3). Histone genes are provided on plasmid pDM1 (HHF2-HHT2 CEN-ARS1 URA3). This pDM1 plasmid was replaced by pRM204 plasmid (HHF2-HHT2 CEN-ARS1 TRP1), allowing the introduction of desired mutations into yeast strains. Most of the mutated plasmids were provided by Ali Shilatifard (Nakanishi et al. 2008). Other mutations were introduced using QuickChange kit (Stratagene, 200518-5). The presence of the desired mutations, as well as the disappearance of the wild-type version, was verified by sequencing of the histone genes present in the new yeast strains, and, where indicated, by Western blot.
Sporulation induction and spore analysis
For sporulation, diploid yeast in the SK1 background were grown in YPA to an OD of 0.5–0.8. Growth rates were tracked for each mutant by monitoring the OD600. Yeast were then washed in water, and transferred into sporulation media (K Acetate 2%) at OD 2 and supplemented for auxotrophic amino acids. Samples were collected at the indicated times, washed twice in water, and stored at −80°C. Sporulation efficiency was typically assessed after 24 h of induction. Sporulation efficiency is defined by the number of original cells that formed tetrads, and is usually >98% in a wild-type SK1 background.
Whole-cell extracts and Western blots were performed as described previously (Krishnamoorthy et al. 2006). Quantification of Western blot images was performed using two to three biological independent replicates, depending on the time points, even in cases in which images from only one replicate are shown. Measurements of the spore nuclei were performed as described previously (Krishnamoorthy et al. 2006). t-tests were performed to determine statistical significance.
mRNA quantification
RNA was purified using Qiagen RNAeasy Purification kit according to the manufacturer's instructions. RT was performed using TaqMan Reverse Transcription (ABI). cDNA were quantified using standard procedures on a 7900HT Fast-Real-Time PCR (ABI). During sporulation, cDNA levels were normalized to NUP85, whose mRNA levels appear to be constant in three independent genome-wide transcriptome analyses (Chu et al. 1998; Primig et al. 2000; Friedlander et al. 2006). During germination (Fig. 8H; Supplemental Fig. 7C,D), cDNA levels were normalized to a combination of three different genes—MEF1, SVP26, and ATP10—whose mRNA levels appear to be constant in a genome-wide transcriptome analysis carried out during germination (Joseph-Strauss et al. 2007) using Genorm (Vandesompele et al. 2002). Primers are presented in Supplemental Table 3. Three to four independent biological replicates were analyzed.
ChIP
ChIP analyses were performed as described (Govin et al. 2010). The only minor difference is that cross-linking was done with 1% EGS for 15 min, and then 1% formaldehyde for 10 min before quenching for 5 min with 125 mM glycine.
Mouse spermatogenesis
Immunofluorescence and immunohistochemistry
Immunofluorescence experiments were performed on germ cells from staged microdissected tubules, which were prepared as described by Kotaja et al. (2004). The detailed protocol for immunofluorescence was described (Govin et al. 2007). The protocol of immunohistochemistry experiments on testis paraffin sections was described (Faure et al. 2003).
Cell fractionation and analysis
Cells were fractioned on a 2%–4% BSA gradient, and were analyzed as described previously (Pivot-Pajot et al. 2003).
Acknowledgments
We thank former and present members of the Berger laboratory for valuable discussions. The research in the Berger laboratory was supported by research grant GM55360 from the National Institutes of Health. J.G. has been supported by the Excellence Biomedical Program of the “Fondation pour la Recherche Médicale Française,” and by the Philippe Foundation. S.K.'s laboratory is supported by ANR EpiSperm and empreinte, as well as ARC and INCa programs.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1954910.
Supplemental material is available at http://www.genesdev.org.
References
- Balhorn R 1982. A model for the structure of chromatin in mammalian sperm. J Cell Biol 93: 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CM, Turner FB, Wang Y, Hagstrom K, Taverna SD, Mollah S, Ueberheide B, Meyer BJ, Hunt DF, Cheung P, et al. 2004. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 112: 360–371 [DOI] [PubMed] [Google Scholar]
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11:1618–1623 [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM 2001. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet 28: 82–86 [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Chua P, Roeder GS 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol 15: 3685–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD 2008. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol 319: 1097–1113 [DOI] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci 100: 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci 102: 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AK, Pivot-Pajot C, Kerjean A, Hazzouri M, Pelletier R, Peoc'h M, Sele B, Khochbin S, Rousseaux S 2003. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod 9: 757–763 [DOI] [PubMed] [Google Scholar]
- Friedlander G, Joseph-Strauss D, Carmi M, Zenvirth D, Simchen G, Barkai N 2006. Modulation of the transcription regulatory program in yeast cells committed to sporulation. Genome Biol 7: R20 doi: 10.1186/gb-2006-7-3-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S 2009. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J 277: 599–604 [DOI] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem 271: 3459–3469 [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Schug J, Krishnamoorthy T, Dorsey J, Khochbin S, Berger SL 2010. Genome-wide mapping of histone H4 serine-1 phosphorylation during sporulation in Saccharomyces cerevisiae. Nucleic Acids Res doi: 10.1093/nar/gkq218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S, Nix D, Zhang H, Purwar J, Carrell D, Cairns B 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu R-M, Zhang Z 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: Involvement of histone deacetylases. Eur J Cell Biol 79: 950–960 [DOI] [PubMed] [Google Scholar]
- Herman PK, Rine J 1997. Yeast spore germination: A requirement for Ras protein activity during re-entry into the cell cycle. EMBO J 16: 6171–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291 [DOI] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N 2007. Spore germination in Saccharomyces cerevisiae: Global gene expression patterns and cell cycle landmarks. Genome Biol 8: R241 doi: 10.1186/gb-2007-8-11-r241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F 2003. Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol 162: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589 [DOI] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell 35: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch J-L, Davidson I, Parvinen M, Sassone-Corsi P 2004. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods 1: 249–254 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy T, Chen X, Govin J, Cheung WL, Dorsey J, Schindler K, Winter E, Allis CD, Guacci V, Khochbin S, et al. 2006. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev 20: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner AG, Inouye C, Jain R, Tjian R 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell 11: 365–376 [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci 102: 2808–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37: 179–183 [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461: 664–668 [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 15: 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828 [DOI] [PubMed] [Google Scholar]
- Norris A, Bianchet MA, Boeke JD 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet 4: e1000301 doi: 10.1371/journal.pgen.1000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Cosgrove MS, Youngman E, Wolberger C, Boeke JD 2002. A core nucleosome surface crucial for transcriptional silencing. Nat Genet 32: 273–279 [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S 2003. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23: 5354–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE 2000. The core meiotic transcriptome in budding yeasts. Nat Genet 26: 415–423 [DOI] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci 103: 6988–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS 1991. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev 5: 2392–2404 [DOI] [PubMed] [Google Scholar]
- Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A 2006. Rtt109 is required for proper H3K56 acetylation. J Biol Chem 281: 37270–37274 [DOI] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M 2008. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132: 221–232 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Siderakis M, Tarsounas M 2007. Telomere regulation and function during meiosis. Chromosome Res 15: 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al. 2005. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol 25: 7107–7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Loidl J, Scherthan H 2003. Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J Cell Sci 116: 2431–2442 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F 2002. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, De Los Santos T, Zhang C, Shokat K, Hollingsworth NM 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell 15: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci 95: 7480–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]