Abstract
Eukaryotic translation initiation begins with ribosomal recruitment of aminoacylated initiator tRNA (Met-tRNAMeti) by eukaryotic initiation factor eIF2. In cooperation with eIF3, eIF1, and eIF1A, Met-tRNAMeti/eIF2/GTP binds to 40S subunits yielding 43S preinitiation complexes that attach to the 5′-terminal region of mRNAs and then scan to the initiation codon to form 48S initiation complexes with established codon–anticodon base-pairing. Stress-activated phosphorylation of eIF2α reduces the level of active eIF2, globally inhibiting translation. However, translation of several viral mRNAs, including Sindbis virus (SV) 26S mRNA and mRNAs containing hepatitis C virus (HCV)-like IRESs, is wholly or partially resistant to inhibition by eIF2 phosphorylation, despite requiring Met-tRNAMeti. Here we report the identification of related proteins that individually (Ligatin) or together (the oncogene MCT-1 and DENR, which are homologous to N-terminal and C-terminal regions of Ligatin, respectively) promote efficient eIF2-independent recruitment of Met-tRNAMeti to 40S/mRNA complexes, if attachment of 40S subunits to the mRNA places the initiation codon directly in the P site, as on HCV-like IRESs and, as we show here, SV 26S mRNA. In addition to their role in initiation, Ligatin and MCT-1/DENR can promote release of deacylated tRNA and mRNA from recycled 40S subunits after ABCE1-mediated dissociation of post-termination ribosomes.
Keywords: Ligatin, MCT-1, translation initiation, ribosomal recycling, HCV, Sindbis virus
During the first stage of protein synthesis, initiation, the P site of the small ribosomal subunit is occupied by aminoacylated initiator tRNA (Met-tRNAMeti), whereas, at the last stage, ribosomal recycling, it contains deacylated elongator tRNA, which remains bound to the ribosome after the hydrolysis of peptidyl-tRNA and release of peptide that occur during termination. In both bacteria and eukaryotes, occupancy of the P site during initiation and ribosomal recycling is regulated by initiation factors. However, the mechanisms of ribosomal attachment of Met-tRNAMeti during initiation, and the factors involved in this process, differ considerably between the two kingdoms.
In bacteria, 30S ribosomal subunits bind to mRNA directly via the Shine-Dalgarno (SD) interaction, which places the initiation codon into the 30S subunit's P site. fMet-tRNAfMeti binds to 30S subunits that are associated with all three initiation factors (IF1, IF2, and IF3) and (usually) mRNA, and the ribosome-bound IF2 accelerates its attachment by either direct interaction with the CCA-end of fMet-tRNAfMeti, or inducing favorable conformational changes in the 30S subunit (Milon et al. 2010). During the next stage of initiation, IF2 also promotes association of the 50S ribosomal subunit with the 30S initiation complex (Milon et al. 2008). IF3, in cooperation with IF1, plays the key role in monitoring the fidelity of initiation codon and initiator tRNA selection, accelerating dissociation of incorrect tRNA/mRNA complexes (Petrelli et al. 2001; Milon et al. 2008). In IF3's absence, initiation complexes containing mismatched codon–anticodon interactions can form with fMet-tRNAfMeti on non-AUG codons, and elongator tRNAs also bind readily to 30S/mRNA complexes if they contain cognate P-site codons (Hartz et al. 1989). IF3 and IF1 bind to the platform and to the area of the ribosomal A site, respectively, and act by inducing conformational changes in 30S subunits (Carter et al. 2001; Dallas and Noller 2001). IF3 also promotes dissociation of deacylated elongator tRNA from the P site of 30S subunits after EF-G/RRF-mediated splitting of post-termination ribosomes into subunits (Peske et al. 2005).
Initial ribosomal attachment to mRNA and initiation codon selection in eukaryotes (Jackson et al. 2010) differ fundamentally from these processes in bacteria. In eukaryotes, 40S subunits do not normally bind mRNAs directly, and this step is mediated by initiation factors (eIFs) instead of by the SD interaction. Moreover, in contrast to 30S subunits that attach directly to the initiation codon, the initiation codon in eukaryotes is selected during ribosomal scanning after factor-mediated attachment of 40S subunits to the capped 5′-proximal region of mRNA. Another key difference is that, in eukaryotes, it is not mRNA-bound ribosomal subunits that recruit Met-tRNAMeti, but Met-tRNAMeti instead binds first to 40S subunits via its dedicated “carrier” eIF2 (a eukaryote-specific heterotrimeric factor that interacts specifically with the methionine moiety and forms an eIF2/GTP/Met-tRNAMeti ternary complex), after which tRNA-bound 40S subunits attach to mRNAs. In outline, the mechanism of initiation on most eukaryotic mRNAs is as follows. In cooperation with eIF3, eIF1, and eIF1A, eIF2/GTP/Met-tRNAMeti binds a 40S subunit, yielding a 43S preinitiation complex. Attachment of 43S complexes to the 5′-terminal region of mRNA is mediated by eIF4F, eIF4A, and eIF4B, which unwind the cap-proximal region of mRNA and likely promote attachment via the eIF3/eIF4G interaction. 43S complexes then scan downstream to the initiation codon, where they stop to form 48S initiation complexes with established codon–anticodon base-pairing. Importantly, scanning through stable RNA secondary structures also requires DHX29, a DExH-box protein, which binds directly to 40S subunits, likely at the entrance of the mRNA-binding channel (Pisareva et al. 2008). In cooperation with eIF1A, eIF1 plays a key role in maintaining the fidelity of initiation codon selection, enabling 43S complexes to discriminate against non-AUG triplets, and AUG triplets that have suboptimal nucleotide context or are located within 8 nucleotides (nt) of the 5′ end of mRNA (Pestova and Kolupaeva 2002; Pisarev et al. 2006). Like their functional (IF3) and functional/sequence (IF1) bacterial homologs, eIF1 and eIF1A also bind to the platform and to the A-site area of the 40S subunit, respectively (Lomakin et al. 2003; Yu et al. 2009), and act by inducing conformational changes in 40S subunits that involve opening of the “latch” in the entry part of the mRNA-binding channel and establishment of a new head–body connection on the 40S subunit's solvent side (Passmore et al. 2007). Following start codon recognition and eIF5-induced hydrolysis of eIF2-bound GTP, eIF5B, an IF2 homolog, promotes joining of a 60S subunit. Thus, eIF5B has retained the function of its IF2 homolog in subunit joining, but does not promote ribosomal attachment of Met-tRNAMeti during scanning-mediated initiation.
Importantly, like its bacterial functional analog, IF3, eIF1 also plays a key role in releasing P-site deacylated tRNAs from recycled 40S subunits after ABCE1/eRF1-mediated dissociation of post-termination ribosomal complexes into subunits (Pisarev et al. 2007, 2010). However, in contrast to IF3, which alone promotes efficient tRNA dissociation, efficient release of tRNA by eIF1 also requires eIF1A and eIF3.
In mammals, four stress-activated eIF2α kinases reduce the level of active eIF2—and, consequently, the global level of translation—by preventing recycling of eIF2-GDP to the active eIF2-GTP (Jackson et al. 2010). However, translation of several viral proteins is wholly or partially resistant to inhibition by phosphorylation of eIF2α, despite requiring Met-tRNAMeti. These include Sindbis virus (SV) and Semliki Forest virus structural proteins (McInerney et al. 2005; Ventoso et al. 2006), rotavirus proteins (Montero et al. 2008), and the hepatitis C virus (HCV) polyprotein (Robert et al. 2006). A mechanism for eIF2-independent initiation on HCV-like IRESs has been proposed recently (Pestova et al. 2008; Terenin et al. 2008). Initiation on HCV-like IRESs (including the classical swine fever virus [CSFV] IRES) is based on their direct interaction with the 40S subunit and eIF3 (Pestova et al. 1998). Binding of 40S subunits positions the initiation codon in the ribosomal P site, and, as a result, under conditions of eIF2 sufficiency, eIF2/GTP/Met-tRNAMeti-containing 43S complexes attach to initiation codons of these IRESs directly, without prior scanning (Pestova et al. 1998). Ribosomal attachment to HCV-like IRESs is thus similar to the analogous process in bacteria, in that it also places the initiation codon in the P site, and recent studies have shown that efficient recruitment of Met-tRNAMeti to 40S/eIF3/IRES complexes can be mediated by eIF5B (Pestova et al. 2008; Terenin et al. 2008). Thus, on mRNAs on which some aspects of ribosomal attachment resemble the analogous process in bacteria, eIF5B was able to substitute for eIF2 and, like its IF2 homolog, promote ribosomal recruitment of initiator tRNA.
The structural diversity of the 5′-untranslated regions (UTRs) of viral mRNAs that are efficiently translated under conditions of eIF2 phosphorylation suggests the existence of multiple distinct eIF2-independent mechanisms for ribosomal recruitment of Met-tRNAMeti. Here we report the identification and characterization of three related mammalian proteins as factors that have a dual function, and that, singly (Ligatin) or in combination (MCT-1 and DENR), can (1) promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits, and (2) recruit Met-tRNAMeti to 40S/mRNA complexes, and thus mediate formation of 48S complexes if the process of ribosomal attachment to mRNA places the initiation codon directly in the 40S subunit's P site. Thus, on some specific mRNAs, these conserved eukaryotic proteins have a potential to substitute for the canonical initiation factor eIF2 in circumstances when its activity is down-regulated.
Results
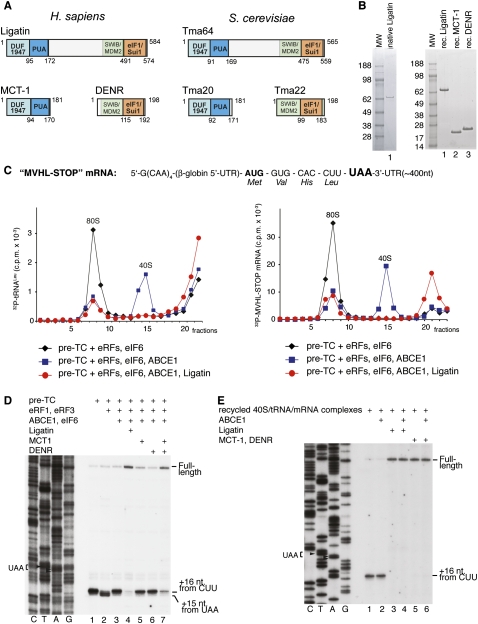
During purification from rabbit reticulocyte lysate (RRL) of ABCE1, which dissociates eukaryotic post-termination complexes (post-TCs) into free 60S subunits and tRNA/mRNA-associated 40S subunits (Pisarev et al. 2010), we also obtained a ribosomal salt wash fraction that induced release of deacylated tRNA and mRNA from recycled 40S subunits. Isolation of its active component yielded an ∼65-kDa protein, which was identified as Ligatin (Supplemental Table S1). Ligatin occurs in all eukaryotes, and its Saccharomyces cerevisiae ortholog is Tma64 (Fleischer et al. 2006). All eukaryotes also encode interacting pairs of proteins that correspond to N-terminal and C-terminal regions of Ligatin, such as human MCT-1 and S. cerevisiae Tma20, and human DENR and S. cerevisiae Tma22, respectively (Fig. 1A; Deyo et al. 1998; Prosniak et al. 1998; Fleischer et al. 2006). Ligatin contains N-terminal DUF1947 and PUA domains that also occur in MCT-1 and Tma20, and C-terminal SWIB/MDM2 and SUI1/eIF1 domains that also occur in DENR and Tma22. Ligatin, MCT-1/DENR, and Tma20/Tma22 have been implicated in translation on the basis of bioinformatic, proteomic, and overexpression/silencing analyses (e.g., Aravind and Koonin 1999; Fleischer et al. 2006; Reinert et al. 2006), but their function has remained obscure. We therefore investigated the activities of Ligatin and MCT-1/DENR in initiation and post-termination ribosomal recycling using an in vitro reconstituted system. Native Ligatin and bacterially expressed His-tagged Ligatin, MCT-1, and DENR (Fig. 1B) were used in these studies. The activities of native and recombinant Ligatin were identical in all assays described below.
Figure 1.
Ligatin and MCT-1/DENR promote release of tRNA and mRNA from recycled 40S subunits. (A) Domain organization of Homo sapiens Ligatin, MCT-1, and DENR and their S. cerevisiae orthologs Tma64, Tma20, and Tma22. (B) Purified native Ligatin (left panel) and recombinant Ligatin, MCT-1, and DENR (right panel) resolved by SDS-PAGE. (C) Ribosomal association of [32P]tRNALeu (left panel) and [32P]MVHL-STOP mRNA (right panel) after incubation of pre-TCs assembled on MVHL-STOP mRNA with eRFs, ABCE1, eIF6, and Ligatin, assayed by SDG centrifugation. (D) Toe-print analysis of ribosomal complexes, obtained by incubating pre-TCs assembled on MVHL-STOP mRNA, with combinations of eRFs, ABCE1, eIF6, Ligatin, MCT-1, and DENR. (E) Toe-printing analysis of SDG-purified recycled tRNA/mRNA-associated 40S subunits, obtained from pre-TCs assembled on MVHL-STOP mRNA, after their incubation with ABCE1, Ligatin, MCT-1, and DENR, as indicated. Lanes C, T, A, and G depict corresponding DNA sequences. Positions of the stop codon, full-length cDNA, and toe prints corresponding to ribosomal complexes are indicated.
Ligatin and MCT-1/DENR promote release of P-site deacylated tRNA and mRNA from recycled 40S subunits
Dissociation of deacylated tRNA and mRNA from recycled 40S subunits by Ligatin and MCT-1/DENR was studied using pre-TCs assembled from 40S and 60S subunits; eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, eIF4G, eIF5, and eIF5B; elongation factors eEF1H and eEF2; Met-tRNAMeti; Val-tRNAVal; His-tRNAHis; and Leu-tRNALeu on MVHL-STOP mRNA encoding a MVHL tetrapeptide followed by a UAA stop codon (Fig. 1C), and purified by sucrose density gradient (SDG) centrifugation. As reported (Pisarev et al. 2010), tRNA and mRNA remained bound to recycled 40S subunits after incubation of pre-TCs with eRF1/eRF3, ABCE1, and eIF6, whereas inclusion of Ligatin led to their near-complete release (Fig. 1C). In toe-printing experiments, incubation of pre-TCs with eRF1/eRF3, ABCE1, eIF6, and Ligatin yielded mostly full-length cDNA, indicating that Ligatin can induce efficient tRNA/mRNA release from recycled 40S subunits even in conditions that do not involve SDG centrifugation (Fig. 1D, lane 4). In combination but not individually, MCT-1 and DENR also promoted tRNA/mRNA release, albeit with slightly lower efficiency (Fig. 1D, lanes 5–7). To verify if Ligatin and MCT-1/DENR can promote tRNA/mRNA release in the absence of ABCE1, pre-TCs were incubated with eRF1/eRF3, ABCE1, and eIF6, and subjected to SDG centrifugation to isolate recycled tRNA/mRNA/40S subunit complexes. These purified complexes did not contain ABCE1, since it does not associate stably with ribosomal complexes in the presence of nucleoside triphosphates (NTPs) (Pisarev et al. 2010). In toe-printing experiments, incubation of 40S/tRNA/mRNA complexes with Ligatin or MCT-1/DENR yielded mostly full-length cDNA, whether or not ABCE1 was present (Fig. 1E), confirming that Ligatin and MCT-1/DENR alone could promote tRNA/mRNA release from recycled 40S subunits. Importantly, Ligatin promoted release of not only elongator, but also initiator tRNAMeti from 40S subunits obtained after ABCE1-mediated dissociation of pre-TCs formed on M-STOP or MMM-STOP mRNAs in the presence of only Met-tRNAMeti (data not shown).
Ribosomal association of Ligatin, MCT1, and DENR
We next investigated the ability of Ligatin, MCT-1, and DENR to bind to ribosomal subunits and 80S ribosomes using SDG centrifugation. Ligatin bound stably and near-stoichiometrically to 40S subunits and 80S ribosomes, whereas binding to 60S subunits was much less efficient (Fig. 2A, lanes 1–3). Consistently, preincubation of 40S subunits with Ligatin did not impair their association with 60S subunits (Fig. 2B). Binding of Ligatin to 40S subunits was inhibited by their preincubation with eIF1, and even more so with eIF1 and eIF1A (Fig. 2A, lanes 4,5). MCT-1 also efficiently and stably bound to 40S subunits and 80S ribosomes, but not to 60S subunits (Fig. 2C). Unfortunately, currently available antibodies did not have sufficient affinity to reliably monitor the ribosomal association of DENR (data not shown).
Figure 2.
Ribosomal association of Ligatin, MCT-1, and DENR. Association of Ligatin (A) and MCT-1 (C) with 40S subunits, 60S subunits, and 80S ribosomes in the presence/absence of eIF1 and eIF1A, as indicated. Ribosomal peak fractions obtained by SDG centrifugation were analyzed by SDS-PAGE followed by fluorescent SYPRO staining and Western blotting using Ligatin antibodies (A) and Western blotting using MCT-1 antibodies (C). (B) Influence of Ligatin on ribosomal subunit association assayed by SDG centrifugation.
Activities of Ligatin, MCT-1, and DENR in 48S complex formation by 5′-end-dependent scanning
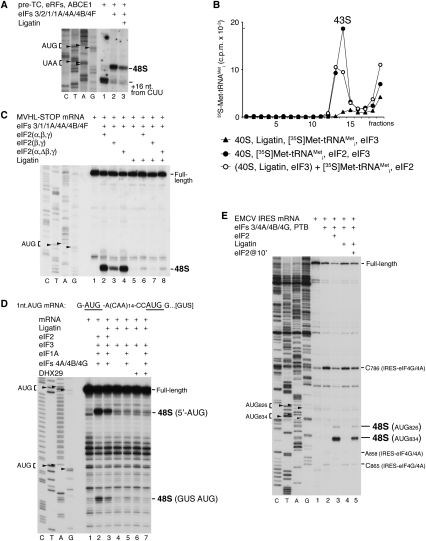
Incubation of pre-TCs assembled on MVHL-STOP mRNA with eRF1/eRF3, ABCE1, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and Met-tRNAMeti led to formation of 48S complexes on the same mRNA (Fig. 3A, lane 2). However, initiation was substantially reduced if pre-TCs were preincubated with eRF1/eRF3, ABCE1, and Ligatin before addition of eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and Met-tRNAMeti (Fig. 3A, lane 3). Thus, even though Ligatin promoted release of tRNA and mRNA from recycled 40S subunits, it also impaired the ability of these 40S subunits to participate in the next round of initiation. To investigate the mechanism of inhibition, we assayed Ligatin's influence on 43S complex formation (Fig. 3B). Ligatin did not promote attachment of Met-tRNAMeti to 40S subunits (Fig. 3B, triangles), and preincubation of 40S subunits with Ligatin even moderately inhibited eIF2-mediated recruitment of Met-tRNAMeti (Fig. 3B, cf. filled and open circles). Ligatin did not influence association of eIF3 with 40S subunit in the presence of ssRNA (Kolupaeva et al. 2005; data not shown), which suggests that Ligatin likely impaired tRNA recruitment through a direct effect on eIF2, rather than by inhibiting ribosomal association of eIF3. Thus, inhibition by Ligatin of 48S complex formation on MVHL-STOP mRNA could be due at least in part to its influence on eIF2-mediated ribosomal recruitment of Met-tRNAMeti. We next determined whether Ligatin sterically interfered with ribosomal binding of eIF2α or eIF2β, by using eIF2 lacking the α subunit or containing small substoichiometric amounts of N-terminally truncated β subunit (Fig. 3C). As reported (Pisarev et al. 2006), 48S complex formation was reduced by omission of eIF2α but not eIF2β (Fig. 3C, lanes 2–4). In the absence of eIF2, Ligatin did not support 48S complex formation, and also inhibited 48S complex assembly mediated by all forms of eIF2 (Fig. 3C, lanes 5–8), which indicates that inhibition was not due to a steric clash between Ligatin and eIF2α or eIF2β.
Figure 3.
Activity of Ligatin in 48S complex formation on mRNAs translated by the 5′-end-dependent scanning mechanism and on the EMCV IRES. (A) Toe-printing analysis of 48S complex formation on MVHL-STOP mRNA after incubation of pre-TCs, assembled on this mRNA, with eRFs, ABCE1, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, and eIF4G in the presence/absence of Ligatin. Positions of initiation and stop codons and assembled ribosomal complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences. (B) Influence of Ligatin on eIF2-mediated 43S complex formation, assayed by SDG centrifugation. (C–E) Toe-printing analysis of 48S complex formation on MVHL-STOP mRNA (C), GUS mRNA with an unstructured 5′-UTR containing an AUG triplet 1 nt from the 5′ end followed by 14 CAA repeats (D), and EMCV IRES in the presence of 40S subunits, Met-tRNAMeti, Ligatin, eIF2, and other factors (E), as indicated. Positions of initiation codons and assembled ribosomal complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences.
Next, we tested Ligatin's activity in 48S complex formation on an AUG triplet located only 1 nt from the 5′ end of mRNA, using GUS mRNA with an unstructured 5′-UTR containing a 5′-terminal AUG triplet followed by 14 CAA repeats (Fig. 3D). Ligatin's activity was tested in the presence of different combinations of eIFs with or without DHX29 (which strongly stimulated 48S complex formation by Ligatin on SV 26S mRNA, as discussed below). In the absence of eIF1, eIF2 promoted efficient 48S complex formation on the 5′-terminal AUG triplet, whereas initiation on the downstream GUS AUG codon was much weaker (Fig. 3D, lane 2). Again, Ligatin inhibited eIF2-mediated 48S complex formation on both AUG triplets (Fig. 3D, lane 3), and, in the absence of eIF2, mediated assembly of only a marginal amount of 48S complexes on the 5′-terminal AUG triplet (Fig. 3D, lanes 4–7). Thus, Ligatin could not promote efficient 48S complex formation on a near-leaderless mRNA.
Taken together, these data indicate that Ligatin cannot promote initiation on mRNAs translated by the 5′-end-dependent scanning mechanism, and even inhibits eIF2-mediated 48S complex formation.
Activities of Ligatin, MCT-1, and DENR in IRES-mediated 48S complex formation
The fact that MCT-1 has been implicated in translation of specific mRNAs, initiation on which might proceed by noncanonical mechanism(s) (see the Discussion), prompted us to investigate whether Ligatin and MCT-1/DENR could promote 48S complex formation on mRNAs translated by IRES-mediated initiation.
Different groups of structurally related viral IRESs use distinct initiation mechanisms that are based on noncanonical-specific interactions of IRESs with canonical components of the translational apparatus. Initiation on the encephalomyocarditis virus (EMCV) IRES is based on specific binding of its J–K domain upstream of the initiation codon with eIF4G, which likely mediates direct attachment of 43S complexes to the initiation codon by interaction with 43S-bound eIF3 (Pestova et al. 1996), and is enhanced by the pyrimidine tract-binding protein (PTB). 48S complexes form at AUG834, and infrequently at AUG826 (Fig. 3E, lane 3). Ligatin could not substitute for eIF2 on this IRES (Fig. 3E, lane 4)—irrespective of whether DHX29, eIF1, or eIF1A was included or PTB or other factors were omitted (data not shown)—and, again, it moderately inhibited 48S complex formation mediated by eIF2 (Fig. 3E, lane 5).
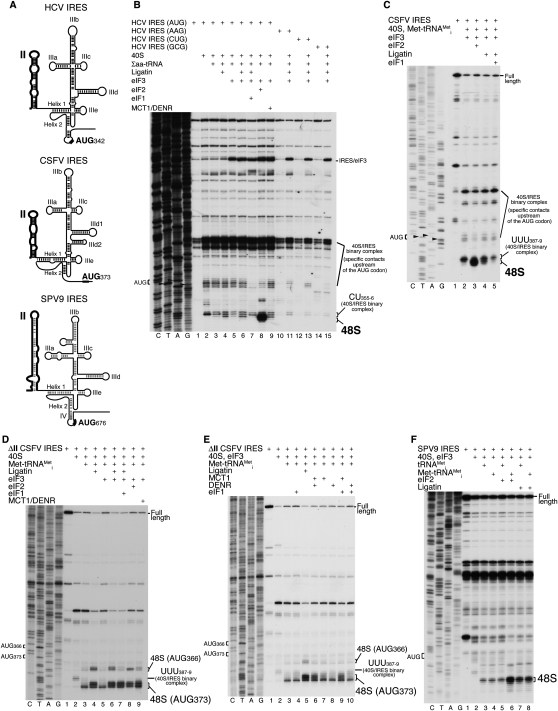
Initiation on HCV and the related CSFV and simian picornavirus type 9 (SPV9) IRESs (Fig. 4A) is based on their direct interaction with 40S subunits and eIF3 (Pestova et al. 1998; de Breyne et al. 2008). Binding of 40S subunits to these IRESs positions the initiation codon directly in the P site, and, as a result, 48S complexes assemble without prior scanning. Moreover, on HCV and CSFV IRESs, eIF5B can substitute for eIF2 in recruiting Met-tRNAMeti to IRES/40S complexes (Pestova et al. 2008; Terenin et al. 2008), and, on the SPV9 IRES, Met-tRNAMeti can even bind 40S/IRES complexes directly in the absence of all eIFs, albeit with lower efficiency (de Breyne et al. 2008). Binding of 40S subunits to HCV-like IRESs yields two sets of toe prints: One (CU355–356 on HCV, and UUU387–389 on CSFV IRESs) corresponds to the 40S subunit's leading edge, and the second corresponds to specific 40S/IRES contacts upstream of the initiation codon, outside the mRNA-binding cleft (Fig. 4B,C, lanes 2; Pestova et al. 1998). eIF2-mediated attachment of Met-tRNAMeti to 40S/IRES complexes on HCV and CSFV IRESs resulting in 48S complex formation causes a 1- to 2-nt forward shift of the toe print corresponding to the 40S subunit's leading edge (Fig. 4B [lane 8], C [lane 3]). Replacing eIF2 by Ligatin resulted in a much lower level of 48S complex formation (Fig. 4B [lane 6], C [lane 4]). Ligatin's ability to promote recruitment of Met-tRNAMeti to 40S/IRES complexes was not enhanced by eIF3 (e.g., Fig. 4B, lanes 4,6). Like 48S complexes formed with eIF2 or eIF5B (Pestova et al. 2008), 48S complexes formed with Ligatin were susceptible to dissociation by eIF1 (Fig. 4B [lane 7], C [lane 5]). MCT-1 and DENR together also promoted 48S complex formation about as efficiently as Ligatin (Fig. 4B, lane 9). To determine if Ligatin could recruit cognate elongator tRNAs to non-AUG codons, we used mutant HCV IRESs in which the initiation codon was substituted by AAG, CUG, or GCG triplets (Reynolds et al. 1995). As reported (Pestova et al. 1998), mutant IRESs efficiently interacted with 40S subunits, yielding characteristic toe prints at CU355–356, but no toe prints indicative of 48S complex formation were observed (Fig. 4B, lanes 11,13,15). Thus, Ligatin did not recruit elongator tRNAs to ribosomal complexes assembled on HCV IRESs with mutated initiation codons.
Figure 4.
Activity of Ligatin, MCT-1, and DENR in 48S complex formation on HCV-like IRESs. (A) Secondary structures of HCV, CSFV, and SPV9 IRESs, with domain II in bold. (B–F) Toe-printing analysis of 48S complex formation on HCV (B), CSFV (C), ΔII CSFV, (D,E) and SPV9 (F) IRESs in the presence of 40S subunits, aminoacylated and deacylated tRNAMeti, Ligatin, MCT-1, DENR, eIF2, and other eIFs as indicated. Initiation codons and positions of assembled ribosomal complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences.
HCV-like IRESs comprise two major domains: II and III (Fig. 4A). Although domain II does not determine IRES/40S subunit affinity, it induces conformational changes in 40S subunits involving rotation of the head and opening of the mRNA channel entry “latch” (Spahn et al. 2001) that are likely required to accommodate mRNA in the mRNA-binding channel, and for efficient ribosomal subunit joining (Locker et al. 2007; Pestova et al. 2008). Deletion of CSFV domain II (ΔII CSFV IRES) eliminates the sensitivity of 48S complexes formed on the IRES to dissociation by eIF1, and leads to weak factor-independent binding of Met-tRNAMeti to 40S/IRES complexes (Pestova et al. 2008). Since conformational changes in 40S subunits induced by domain II might influence the ability of Ligatin and MCT-1/DENR to promote Met-tRNAMeti recruitment to 40S/IRES complexes, we investigated whether they could do so efficiently on the ΔII CSFV IRES. Although, as reported (Pestova et al. 2008), weak binding of Met-tRNAMeti to binary 40S/ΔII CSFV IRES complexes occurred in the absence of factors (Fig. 4D, lane 3), Ligatin and MCT-1/DENR strongly stimulated this process in an eIF3-independent manner (Fig. 4D, lanes 4,6,9). Importantly, in contrast to the wild-type IRES, their activity on ΔII CSFV IRES was very similar to that of eIF2 (Fig. 4D, lane 8). Individually, MCT-1 and DENR did not promote efficient attachment of Met-tRNAMeti, but MCT-1 promoted relatively efficient Met-tRNAMeti binding in the presence of eIF1 (Fig. 4E, lanes 7–9). Like 48S complexes assembled on the ΔII CSFV IRES with eIF2 or eIF5B (Pestova et al. 2008), complexes assembled with Ligatin were resistant to eIF1-induced dissociation (Fig. 4D, lane 7). Interestingly, the toe print of 48S complexes assembled in the absence of factors (Fig. 4D, lane 3) shifted ∼2 nt downstream relative to toe prints of 48S complexes assembled with eIF2, Ligatin, or MCT-1/DENR (Fig. 4D, lanes 4,8,9), but the reason for this is not clear.
The SPV9 IRES has an HCV-like domain III, but its domain II lacks the conserved apex, and its predicted structure differs significantly from HCV and CSFV domain II (Fig. 4A; Hellen and de Breyne 2007). The conformational changes induced in 40S subunits by this IRES that depend on domain II might thus differ, at least in magnitude, from those induced by HCV and CSFV IRESs. Consistently, as with the ΔII CSFV IRES, Met-tRNAMeti bound directly to 40S/SPV9 IRES binary complexes in the absence of eIFs, and 48S complexes formed with eIF2 resisted eIF1-induced dissociation (de Breyne et al. 2008). Ligatin strongly stimulated Met-tRNAMeti binding to 40S/SPV9 IRES complexes, and, importantly, in contrast to eIF2 (which promoted recruitment of only aminoacylated tRNAMeti), did not discriminate between aminoacylated and deacylated tRNAMeti (Fig. 4F, lanes 5–8).
These results indicate that Ligatin and MCT-1/DENR can promote attachment of Met-tRNAMeti to ribosomal complexes assembled on mRNAs that place their initiation codon directly in the P site; e.g., HCV-like IRESs. The Met-tRNAMeti recruitment activity of Ligatin and MCT-1/DENR is strongly enhanced by deletion of IRES domain II, likely by elimination of the conformational changes in 40S subunits that it induces.
The mechanism of 48S complex formation on SV 26S mRNA
The ability of Ligatin and MCT-1/DENR to promote eIF2-independent 48S complex formation on HCV-like IRESs suggests that they might also contribute to initiation on other mRNAs when the level of active eIF2 is reduced by phosphorylation. Infection by alphaviruses such as SV and Semliki Forest virus leads to robust phosphorylation of eIF2α and shutoff of host protein synthesis, but their 26S subgenomic mRNAs continue to be translated efficiently (McInerney et al. 2005; Ventoso et al. 2006). The 5′ ends of their coding regions contain hairpins that enhance translation, determine initiation codon recognition, and confer resistance to inhibition by phosphorylation of eIF2α (Frolov and Schlesinger 1996; McInerney et al. 2005; Ventoso et al. 2006). At its natural location ∼25 nt downstream from the initiation codon, this enhancer can promote initiation at correctly located non-AUG codons, whereas its displacement or disruption impairs initiation codon selection, so that initiation occurs downstream (Frolov and Schlesinger 1996; Sanz et al. 2009).
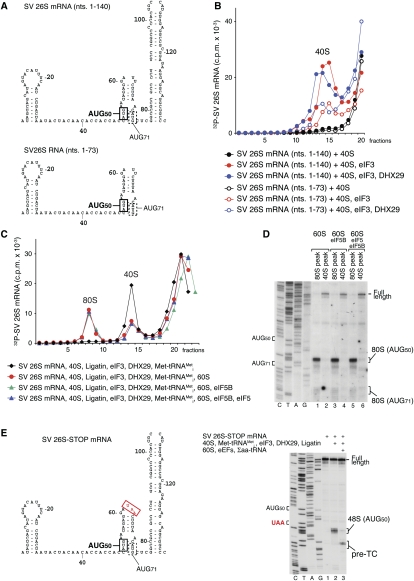
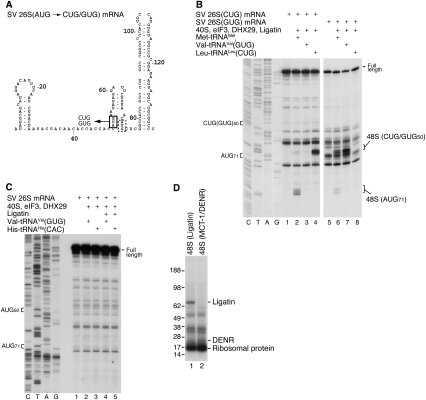
The initiation codon in SV 26S mRNA is at AUG50, but initiation can occur at the in-frame AUG71 if the enhancer stem (nucleotides 77–139) (Fig. 5A) is disrupted (Frolov and Schlesinger 1996; Sanz et al. 2009). We investigated the ability of Ligatin and MCT-1/DENR to promote 48S complex formation on SV 26S mRNA, and compared it with that of eIF2. Interestingly, eIF2-mediated 48S complex formation on AUG50 was strongly stimulated by DHX29 (Fig. 5B, cf. lanes 5–7 and 8–10) and by the simultaneous presence of eIF1 and eIF1A, which also eliminated weak 48S complex formation on AUG71 (Fig. 5B, lane 10), but not by eIF4A, eIF 4B, and eIF4G (Fig. 5B, cf. lanes 8,14). 48S complexes did not form in eIF3's absence with any combination of other factors (data not shown). Thus, efficient eIF2-mediated 48S complex formation on AUG50 required eIF3, DHX29, eIF1, and eIF1A.
Figure 5.
eIF2- and Ligatin-mediated 48S complex formation on SV 26S mRNA. (A) Secondary structure of a fragment of SV 26S mRNA comprising the 5′-UTR and the adjacent enhancer stem. (B–F) Toe-printing analysis of 48S complex formation on SV 26S mRNA in the presence of 40S subunits, aminoacylated and deacylated tRNAMeti, Ligatin, MCT-1, DENR, eIF2, and other eIFs, as indicated. AUG50 and AUG71 triplets and positions of assembled 48S complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences.
48S complex formation on SV 26S mRNA with Ligatin was as efficient as with eIF2, but depended even more strongly on DHX29 (Fig. 5C, cf. lanes 5–7 and 8–10), and was inhibited rather than stimulated by eIF1 and eIF1A (Fig. 5C, lanes 8,10). Interestingly, in the absence of eIF1 and eIF1A, fewer 48S complexes formed on AUG71 with Ligatin than with eIF2 (Fig. 5B,C, lanes 8). Like eIF2, Ligatin did not require eIF4A, eIF4B, and eIF4G to promote 48S complex formation (Fig. 5C, lanes 8,14), but absolutely depended on eIF3 (data not shown). Thus, efficient Ligatin-mediated 48S complex formation required only eIF3 and DHX29. As with the SPV9 IRES, Ligatin did not discriminate between recruitment of aminoacylated and deacylated tRNAMeti during initiation on SV 26S mRNA (Fig. 5D). In the presence of eIF3 and DHX29, MCT-1/DENR also mediated 48S complex formation on SV 26S mRNA, albeit less efficiently (Fig. 5E, lanes 2,3).
Importantly, low-level 48S complex formation occurred on SV 26S mRNA without eIF2 or Ligatin if eIF3 and DHX29 were present (e.g., Fig. 5D [lane 3], E [lane 1], F [lanes 2,7]). Thus, low-level eIF2-independent binding of Met-tRNAMeti occurred on all mRNAs on which Ligatin and MCT-1/DENR promoted efficient 48S complex formation (ΔII CSFV and SPV9 IRESs; SV 26S mRNA).
Although eIF5B promotes binding of Met-tRNAMeti to 40S/IRES complexes formed on HCV-like IRESs (Pestova et al. 2008; Terenin et al. 2008), it could not mediate 48S complex formation on SV 26S mRNA at either 1 or 5 mM free Mg2+ (Fig. 5F, lanes 4–6,9–11). However, the elevated Mg2+ concentration enhanced Ligatin-dependent, eIF2-dependent, and eIF2/Ligatin-independent 48S complex formation (Fig. 5F, lanes 2,3,7,8; data not shown).
Enhanced 48S complex formation at elevated Mg2+; its lack of dependence on eIF4A, eIF4B, and eIF4G; and the fact that omitting eIF1 did not lead to 48S complex formation on near-cognate start codons preceding AUG50, all suggested that initiation on SV 26S mRNA does not involve conventional 5′-end-dependent scanning, and raised the question of how it is recruited to 40S subunits. Using SDG centrifugation, we found that a nucleotide 1–140 fragment (Fig. 6A) comprising the 5′-UTR and the enhancer stem bound efficiently to eIF3-associated 40S subunits (Fig. 6B, red filled circles). Deletion of the stem (nucleotides 1–73 fragment) (Fig. 6A) strongly reduced binding (Fig. 6B, red open circles). DHX29 did not influence the efficiency of SV 26S mRNA binding to eIF3-associated 40S subunits (Fig. 6B, blue filled and open circles), suggesting that the requirement for it in initiation likely involves correct fitting of 26S mRNA into the 40S subunit's mRNA-binding cleft, rather than its initial ribosomal recruitment.
Figure 6.
Attachment of SV 26S mRNA to 40S subunits, and ribosomal subunit joining on 48S complexes assembled on SV 26S mRNA in the presence of Ligatin. (A) Secondary structure of 73-nt-long and 140-nt-long fragments of SV 26S mRNA. (B) Association of 73-nt-long and 140-nt-long SV 26S mRNA fragments with 40S subunits, depending on the presence of eIF3 and DHX29, assayed by SDG centrifugation. (C) Joining of 60S subunits to 48S complexes assembled on SV 26S RNA with Ligatin, eIF3, and DHX29, depending on the presence/absence of eIF5 and eIF5B, assayed by SDG centrifugation. (D) Toe-printing analysis of 80S- and 40S-containing fractions shown in C. (E) Elongation competence of 80S complexes assembled with Ligatin on SV 26S mRNA containing a UAA stop codon four triplets downstream from the initiation codon (left panel), assayed by toe-printing (right panel). (D,E) Lanes C, T, A, and G depict corresponding cDNA sequences. Positions of initiation and stop codons, full-length cDNA, and toe prints corresponding to ribosomal complexes are indicated.
We next investigated factor requirements for joining of 60S subunits to 48S complexes formed on SV 26S mRNA with Ligatin, eIF3, and DHX29. Joining was direct and did not require eIF5 or eIF5B (Fig. 6C). Irrespective of eIF5/eIF5B's presence or absence, toe-printing analysis of peak fractions corresponding to 80S complexes yielded strong stops 15–17 nt downstream from AUG50 that are characteristic of correctly assembled 80S initiation complexes (Fig. 6D, lanes 1,3,5). Synthesis of only full-length cDNA in reaction mixtures containing peak fractions corresponding to residual 40S ribosomal complexes (Fig. 6D, lanes 2,4,6) indicated that they were not 48S complexes and likely lacked Met-tRNAMeti, and thus that subunit joining on 48S complexes formed with Ligatin was very efficient. Incubation with 60S subunits, eEF1H, eEF2, and aa-tRNAs of 48S complexes formed with Ligatin on SV26S mRNA containing a stop codon four triplets downstream from the initiation codon yielded pre-TCs, indicating that 80S complexes formed on SV 26S mRNA with Ligatin were elongation-competent (Fig. 6E).
To investigate if Ligatin-mediated initiation on SV mRNA is specific for AUG codons and Met-tRNAMeti, we used mutant SV 26S mRNAs with substitutions of AUG50 to CUG(Leu) or GUG(Val) (Fig. 7A). In the presence of Met-tRNAMeti, Ligatin promoted weak 48S complex formation on AUG71 of both mRNAs, and moderately efficient 48S complex formation on the near-cognate GUG, but not the CUG codon (Fig. 7B, lanes 2,6). Efficient 48S complex formation on the CUG and GUG triplets occurred in the presence of cognate elongator Leu-tRNALeu and Val-tRNAVal (Fig. 7B, lanes 4,7), indicating that Ligatin could promote recruitment of noninitiator tRNAs to cognate codons during 48S complex formation. However, Ligatin did not recruit His-tRNAHis to any of the three cognate CAC codons immediately preceding AUG50 (Fig. 7C), which suggests that, during Ligatin-mediated initiation, the SV 26S initiation codon is defined by its relative position in the mRNA rather than by its sequence. Although, in contrast to SV 26S mRNA, Ligatin did not promote appreciable recruitment of elongator tRNAs to cognate P-site codons in the case of ribosomal complexes formed on the HCV IRES (Fig. 4B), it should be noted that the overall activity of Ligatin in initiation on it was low.
Figure 7.
Ligatin-mediated 48S complex formation on SV 26S mRNA with noninitiator tRNAs, and interaction of Ligatin and MCT-1/DENR with tRNAMeti on the 40S subunit. (A) Mutant SV 26S mRNAs with AUG50 replaced by CUG or GUG. (B) Analysis of Ligatin's ability to promote 48S complex formation on mutant SV 26S mRNAs (shown in A) in the presence of tRNAMeti and cognate Leu-tRNALeu and Val-tRNAVal, assayed by toe-printing. (C) Analysis of Ligatin's ability to promote 48S complex formation on CAC codons preceding AUG50 of wild-type SV 26S mRNA in the presence of cognate His-tRNAHis, assayed by toe-printing. (B,C) Lanes C, T, A, and G depict corresponding cDNA sequences. Positions of AUG, CUC, and GUG codons, full-length cDNA, and toe prints corresponding to ribosomal complexes are indicated. (D) UV-cross-linking of Ligatin and MCT-1/DENR with tRNAMeti in 48S complexes assembled on ΔII CSFV IRES with 32P-labeled tRNAMeti containing 4-thioU and purified by SDG centrifugation, assayed by SDS-PAGE and autoradiography. The positions of Ligatin, DENR, cross-linked ribosomal protein, and molecular weight markers are indicated.
The results described above are consistent with a mechanism of initiation on SV 26S mRNA, in which its binding to eIF3/DHX29-associated 40S subunits places AUG50 directly into the P site, after which recruitment of Met-tRNAMeti to the AUG-containing P site could be promoted by eIF2, Ligatin, or, less efficiently, MCT-1/DENR. Thus, data concerning 48S complex formation on HCV-like IRESs and SV 26S mRNA indicate that Ligatin and MCT-1/DENR can recruit Met-tRNAMeti to 40S/mRNA complexes if the process of mRNA binding to the ribosome places the initiation codon directly in the P site.
Interaction of Ligatin and MCT-1/DENR with tRNAMeti on the 40S subunit
Ligatin and MCT-1/DENR might promote ribosomal recruitment of Met-tRNAMeti by binding it directly, or by inducing favorable conformational changes in 40S subunits. To shed light on how they recruit Met-tRNAMeti, we used UV-cross-linking to investigate if they contact tRNA on the 40S subunit. tRNAMeti was in vitro transcribed with 32P-labeled ATP, CTP, GTP, and 4-thioU, which can be cross-linked to proteins by low-energy irradiation yielding zero-length cross-links, representing direct contact between tRNA and other translational components. 48S complexes were formed on ΔII CSFV IRES with 40S subunits, Met-tRNAMeti, and Ligatin or MCT-1/DENR; purified by SDG centrifugation; and subjected to irradiation. In both cases, tRNA cross-linked strongly to a ≤17-kDa ribosomal protein (Fig. 7D). Cross-linking to Ligatin was inefficient (Fig. 7D, lane 1), and cross-linking to DENR was even weaker (Fig. 7D, lane 2). Thus, these data do not support extensive direct contact between Met-tRNAMeti and Ligatin or MCT-1/DENR on 40S subunits.
Discussion
We identified roles for Ligatin and the related interacting proteins MCT-1 and DENR in ribosome recycling and initiation, two linked stages of translation. Ligatin—and, to a slightly lesser extent, MCT-1 and DENR together (but not individually)—promoted release of deacylated tRNA and mRNA from recycled 40S subunits following dissociation of post-TCs by ABCE1, and could substitute for eIF1, eIF1A, and eIF3 in this process. Ligatin—and, to a lesser extent, MCT-1 and DENR—also promoted efficient eIF2-independent recruitment of Met-tRNAMeti to the P site of 40S/mRNA complexes, but only if ribosomal attachment to the mRNA placed the initiation codon directly in the P site. In contrast to eIF2, Ligatin also promoted binding of deacylated initiator tRNA as well as elongator tRNAs to 40S/mRNA complexes if the P site was occupied by a cognate codon. However, it did not mediate 48S complex formation on mRNAs translated by the 5′-end-dependent scanning mechanism, on a near-leaderless mRNA, and on the EMCV IRES, and even inhibited eIF2-dependent initiation.
The mechanism of Ligatin- or MCT-1/DENR-mediated 48S complex formation on HCV-like IRESs differed from another eIF2-independent mechanism of initiation on these IRESs based on recruitment of Met-tRNAMeti to 40S/IRES complexes by eIF5B (Pestova et al. 2008; Terenin et al. 2008). Although, like Ligatin and MCT-1/DENR, eIF5B alone mediated only low-efficiency recruitment of Met-tRNAMeti to 40S/mRNA binary complexes on wild-type HCV and CSFV IRESs, its activity was strongly stimulated by eIF3, so that 48S complex assembly by eIF2- and eIF5B/eIF3-mediated attachment of Met-tRNAMeti to 40S/IRES complexes was equally efficient. In contrast, the weak activity of Ligatin and MCT-1/DENR in promoting 48S complex formation on these IRESs was not enhanced by eIF3. However, Ligatin and MCT-1/DENR stimulated very efficient eIF3-independent attachment of Met-tRNAMeti to 40S/mRNA binary complexes formed on a CSFV IRES variant lacking domain II and on the SPV9 IRES, in which domain II is highly divergent. Interestingly, low-level factor-independent binding of Met-tRNAMeti to 40S/mRNA complexes has been observed on these IRESs, and eIF5B was also able to mediate efficient 48S complex formation on ΔII CSFV IRES in the absence of eIF3 (de Breyne et al. 2008; Pestova et al. 2008). Thus, conformational changes caused by binding of HCV-like IRESs to 40S subunits that depend on domain II (Spahn et al. 2001) most likely destabilize P-site binding of initiator tRNA, which could be suppressed by eIF3 in the case of eIF5B, but not Ligatin or MCT-1/DENR, possibly because eIF5B has the potential to interact directly with the CCA end of Met-tRNAMeti. Elimination of these conformational changes by deletion of domain II (in the case of ΔII CSFV IRES), or a possible reduction in their magnitude (in the case of the SPV9 IRES), strongly enhanced recruitment of Met-tRNAMeti by Ligatin, MCT-1/DENR, and eIF5B alone. Although the activity of Ligatin in 48S complex formation on wild-type HCV and CSFV IRESs was lower than that of eIF2, it may nevertheless be sufficient to contribute to their partial resistance to inhibition by eIF2 phosphorylation (Robert et al. 2006), and might even contribute significantly to initiation in virus-infected cells on more divergent HCV-like IRESs (Hellen and de Breyne 2007).
Analysis of 48S complex formation on SV 26S mRNA revealed a novel mechanism for initiation that, as on HCV-like IRESs, involves ribosomal attachment to mRNA in a manner that places the initiation codon directly in the P site. Efficient assembly of 48S complexes on SV 26S mRNA requires only eIF3, DHX29, and eIF2 or Ligatin. The 5′-terminal 140 nt of this mRNA comprising the 5′-UTR and the enhancer stem associate stably with eIF3-bound 40S subunits, in what is likely the first stage of its ribosomal recruitment, and, significantly, the enhancer stem is essential for this step. However, eIF2 or Ligatin cannot promote efficient recruitment of Met-tRNAMeti to mRNA/40S/eIF3 complexes unless DHX29 is also present. Since DHX29 did not influence the level of SV 26S mRNA binding to eIF3-associated 40S subunits, its role in initiation might involve correct positioning of mRNA in the mRNA-binding cleft of the 40S subunit. The ability of SV 26S mRNA to initiate translation at AUG50, even if it is preceded by an AUG triplet in good context (ACCAUGG) (Van Duijn et al. 1988); enhancement rather than inhibition of 48S complex formation by elevating Mg2+ concentrations; its independence from eIF4A, eIF4B, and eIF4G; the lack of 48S complex formation on near-cognate start codons preceding AUG50 in the absence of eIF1; and the fact that low-level 48S complex formation occurred on AUG50 even without eIF2 or Ligatin (this study) all point to a mechanism in which binding of SV 26S mRNA to eIF3/DHX29/40S complexes places AUG50 directly in the P site without prior scanning. This mechanism would account for the requirement for the enhancer stem for accurate and efficient initiation on SV 26S mRNA (Frolov and Schlesinger 1996; Sanz et al. 2009), which we found is needed for stable binding of this mRNA to eIF3/40S complexes, and is also consistent with several unusual aspects of the translation of alphavirus 26S mRNAs, such as its lack of dependence on intact eIF4G (Sanz et al. 2009) and low requirement for eIF4E and eIF4B (van Steeg et al. 1981). Ligatin's ability to promote recruitment of Met-tRNAMeti to 40S/eIF3/DHX29/mRNA complexes at a level similar to that of eIF2 accounts for the efficient translation of alphavirus 26S mRNAs in virus-infected cells at times when cellular translation is impaired due to eIF2α phosphorylation. The fact that initiation on SV 26S mRNA can occur even from noncognate codons, such as GCG and UGU if they replace AUG50 (Sanz et al. 2009), can now be explained by Ligatin's ability to promote recruitment of elongator tRNAs to cognate P-site codons. Importantly, in contrast to 48S complexes formed on SV 26S mRNA with eIF2, joining of 60S subunits to 48S complexes formed with Ligatin occurred directly and did not require additional factors.
In addition to the exact role of DHX29 and the nature of the enhancer stem's interactions with components of the translation apparatus, another important unresolved question concerning this mechanism of initiation is whether ribosomal attachment to SV 26S mRNA is end-dependent or results from a form of internal ribosomal entry. The sequence of the 26S subgenomic mRNA is contained entirely within the genomic 42S mRNA, and there is some controversy whether the AUG triplet corresponding to AUG50 of 26S mRNA is used at all during translation of 42S mRNA (for a discussion, see Bonatti et al. 1980). Thus, if the translation-enhancing region of 26S mRNAs has any activity in promoting end-independent ribosomal attachment, it must be highly dependent on its context.
Although Ligatin and eIF5B were equally efficient in promoting recruitment of Met-tRNAMeti to 40S/mRNA complexes on ΔII CSFV and SPV9 IRESs, in contrast to Ligatin, eIF5B could not replace eIF2 during initiation on SV 26S mRNA. This likely reflects differences in the conformation of 40S subunits in binary 40S/IRES complexes and in 40S/eIF3/DHX29 complexes formed on SV 26S mRNA.
Interestingly, the ability to recruit Met-tRNAMeti to 40S subunits in a codon-dependent manner in the presence of AUG triplets has been reported previously for eIF2A, a protein that is unrelated to Ligatin or MCT-1/DENR (Merrick and Anderson 1975), and silencing of eIF2A reduced the ability of SV 26S mRNA to translate under conditions of eIF2 phosphorylation (Ventoso et al. 2006). In our experiments, eIF2A was unable to substitute for Ligatin in initiation on SV 26S mRNA (MA Skabkin and CUT Hellen, unpubl.), and the activity of eIF2A in initiation on mRNAs remains to be demonstrated in in vitro studies.
Ligatin and MCT-1 could bind stably to individual 40S subunits (which is consistent with the presence of MCT-1 and yeast Tma64 and Tma20 in 40S-containing fractions of polysome gradients) (Fleischer et al. 2006; Reinert et al. 2006), but not to 60S subunits. Ligatin and MCT-1 also interacted efficiently with individual 80S ribosomes. Ligatin (and the MCT-1/DENR pair) contains several domains with the potential to interact with the 40S subunit, including the PUA domain in Ligatin and MCT-1 (which commonly binds dsRNA in tRNA or rRNA) (Perez-Arellano et al. 2007), the intimately associated DUF1947 domain (first identified in archaeal MCT-1 homologs such as Pyrococcus horikoshii PHO734.1) (Miyazono et al. 2008), and the Sui1 domain in Ligatin and DENR (also found in eIF1, which binds to the platform of the 40S subunit). Many PUA domains bind dsRNA through either the major or the minor groove, but others—including the 60S-associated Nip7 (Coltri et al. 2007), MCT-1, and Ligatin—lack most or all of the conserved basic residues involved in these interactions, and their potential contacts with rRNA must thus involve different residues and may have an altered specificity. The DUF1947 domain in P. horikoshii PHO734.1 has an exposed electropositive cluster—also present in MCT-1 and Ligatin—that could mediate an interaction with the ribosome. Although eIF1 (particularly in the presence of eIF1A) impaired binding of Ligatin to 40S subunits (Fig. 2A), and could substitute for DENR in supporting MCT-1-mediated binding of Met-tRNAMeti to the ΔII CSFV IRES (Fig. 4E), it is nevertheless premature to conclude that the Sui domains of Ligatin and DENR bind to the same site as eIF1, since binding of eIF1 and eIF1A induces conformational changes in the 40S subunit (Passmore et al. 2007), which might influence Ligatin's ribosomal association.
The mechanism of action of Ligatin and MCT-1/DENR in promoting release and recruitment of tRNA could be direct (involving interaction with tRNA) and/or indirect (by induction of conformational changes in the 40S subunit). The low efficiency of UV-cross-linking of Ligatin to Met-tRNAMeti in 48S complexes assembled on the ΔII-CSFV IRES, the lack of obvious discrimination against deacylated or noninitiator tRNA in 48S complex formation on SPV9 and SV 26S mRNAs, the efficient dissociation of tRNAMeti from recycled 40S subunits, and the fact that these proteins could (depending on circumstances) promote both release and recruitment of P-site tRNA are more consistent with an indirect mode of action in which the interactions of Ligatin and MCT-1/DENR with 40S subunits induce conformational changes that would regulate entry and egress of tRNA from the P site. However, since the ribosomal positions of Ligatin and MCT-1/DENR are not currently known, a direct mode of action cannot strictly be excluded. Although the activities of Ligatin and MCT-1/DENR in mediating release of deacylated tRNA from the P site of recycled 40S subunits and promoting recruitment of aminoacylated initiator tRNA to the P site might seem contradictory, they likely reflect the contrasting outcomes of fundamentally similar interactions of these proteins with the 40S subunit (and possibly, but less likely, also with tRNA) in the context of conformationally distinct 40S ribosomal complexes, which are moreover associated with mRNAs with substantially different affinities. Thus, whereas, during recycling, Ligatin and MCT-1/DENR promote dissociation of deacylated tRNA from 40S subunits that are not bound to other factors and do not interact specifically with associated mRNAs, tRNA recruitment by Ligatin and MCT-1/DENR has so far been found to occur only on 40S subunits specifically associated with HCV-like IRESs that are known to induce conformational changes in 40S subunits, and on 40S subunits specifically associated with SV 26S mRNA and also bound to eIF3 and DHX29, which are thought to induce conformational changes in 40S subunits. Moreover, the orientation of deacylated tRNA on recycled 40S subunits might differ from that of initiator tRNA, and the association of the former with 40S subunits might be incompatible with potential conformational changes induced by Ligatin or MCT-1/DENR.
The apparently strict dependence of Ligatin and MCT-1/DENR on direct P-site placement of the initiation codon during initiation on viral mRNAs raises the question of what role they might play in translation of cellular mRNAs. MCT-1 is an oncogene that is overexpressed in some types of lymphoma; its overexpression can transform mammary epithelial cells, suppress apoptosis, override cell cycle control, and stimulate cell proliferation (Prosniak et al. 1998; Dai et al. 2009 and references therein). Overexpression and gene silencing studies indicate that MCT-1 enhances translation of a subset of cellular mRNAs (including cyclin D1, Dp-1, cIAP2, E2F1, and Bcl2L2), whereas overexpression of its PUA domain up-regulates translation of some proteins but down-regulates that of others (Reinert et al. 2006; Mazan-Mamczarz et al. 2009). Bidirectional control of cellular translation by Ligatin and MCT-1/DENR would be consistent with the observations that, while Ligatin promoted 48S complex formation on specific mRNAs, it also impaired eIF2-mediated initiation by the canonical scanning mechanism. 40S subunits could potentially acquire Ligatin or MCT-1/DENR during ribosomal recycling, but it is not yet known how the affinity of these factors to recycled 40S subunits compares with that of eIF3, eIF1, and eIF1A, and thus whether they would be able to compete effectively for recycled 40S subunits. However, even if Ligatin and MCT-1/DENR can do so, their cellular abundance may, like that of their yeast homologs, Tma64 and Tma20 (Ghaemmaghami et al. 2003), be several-fold lower than that of eIFs. Many mRNAs whose translation is enhanced by MCT-1 have 5′-UTRs containing stable secondary structures and/or conserved upstream ORFs (uORFs), and might thus function as IRESs or be translated as a consequence of reinitiation after translation of a uORF. Another intriguing observation is that translation of cIAP2, which is enhanced by MCT-1 overexpression, is also up-regulated during endoplasmic reticulum stress in conditions of eIF2 phosphorylation by PERK kinase (Hamanaka et al. 2009). It is likely that translation of specific cellular mRNAs can also be promoted by Ligatin. The focus of future research will therefore be to characterize the mechanisms of initiation on cellular mRNAs whose translation is regulated by MCT-1 and, possibly, Ligatin.
Materials and methods
Plasmid construction; purification of ribosomal subunits, initiation factors, elongation factors, termination factors, aminoacyl-tRNA synthetases, recombinant Ligatin, MCT-1, and DENR; preparation of mRNAs and tRNAs; and aminoacylation of tRNAs are described in the Supplemental Material, which also contains detailed protocols for all experimental procedures.
Purification of native Ligatin
Ligatin was purified from RRL on the basis of activity in promoting release of tRNA and mRNA from recycled 40S subunits obtained after ABCE1-mediated dissociation of post-TCs formed on MVHL-STOP mRNA (Pisarev et al. 2010). Purification involved preparation of ribosomal salt wash, fractionation by ammonium sulfate precipitation, chromatography on DEAE-cellulose and phosphocellulose, and fast protein liquid chromatography (FPLC) on MonoS, MonoQ, hydroxyapatite, and Superdex 75 columns.
tRNA/mRNA release from recycled 40S subunits
Release of tRNA and mRNA was assayed by SDG centrifugation and toe-printing using pre-TCs assembled on MVHL-STOP mRNA with 32P-labeled or unlabeled mRNA and Leu-tRNALeu (Pisarev et al. 2010).
Analysis of ribosomal binding of Ligatin, MCT-1, and DENR
Ligatin and MCT-1 were incubated with 40S subunits, 60S subunits, and 80S ribosomes in the presence/absence of eIF1 and eIF1A, and were subjected to centrifugation through 10%–30% SDGs. Fractions that corresponded to ribosomal complexes were analyzed by SDS-PAGE with subsequent fluorescent SYPRO staining or Western blotting.
Toe-printing analysis of 48S complex formation
48S complexes were assembled on MVHL-STOP, 1nt-AUG-CAA-GUS, wild-type SV 26S, SV 26S-STOP, SV 26S(CUG), and SV 26S(GUG) mRNAs; and mRNAs containing the EMCV IRES, wild-type and AUG → AAG/CUG/GCG HCV IRESs, wild-type and ΔII CSFV IRESs, and SPV9 IRES in the presence of 40S subunits and factors, as indicated in Figures 3 (C and D), 4, 5, 6E, and 7 (B andC), and were analyzed by primer extension using AMV reverse transcriptase and 32P-labeled primers (Pestova et al. 1996).
UV-cross-linking of tRNAMeti to translational components in 48S complexes formed with Ligatin and MCT-1/DENR
48S complexes were assembled on ΔII CSFV IRES mRNA with 40S subunits, 32P-labeled tRNAMeti containing 4-thioU, and Ligatin or MCT-1/DENR, and were purified by centrifugation through 10%–30% SDGs. 48S-containing fractions were irradiated at 360 nm, subjected to a second round of SDG centrifugation, and treated with RNases A, T1, and V1, and cross-linked proteins were analyzed by SDS-PAGE (Pisarev et al. 2006).
Acknowledgments
We thank W. Merrick for a generous gift of eIF2 lacking the α subunit. This work was supported by NIH grants GM59660 and GM80623 to T.V.P. and AI51340 to C.U.T.H.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1957510.
Supplemental material is available at http://www.genesdev.org.
References
- Aravind L, Koonin EV 1999. Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol 48: 291–302 [DOI] [PubMed] [Google Scholar]
- Bonatti S, Sonenberg N, Shatkin AJ, Cancedda R 1980. Restricted initiation of protein synthesis on the potentially polycistronic Sindbis virus 42 S RNA. J Biol Chem 255: 11473–11477 [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Coltri PP, Guimarães BG, Granato DC, Luz JS, Teixeira EC, Oliveira CC, Zanchin NI 2007. Structural insights into the interaction of the Nip7 PUA domain with polyuridine RNA. Biochemistry 46: 14177–14187 [DOI] [PubMed] [Google Scholar]
- Dai B, Zhao XF, Hagner P, Shapiro P, Mazan-Mamczarz K, Zhao S, Natkunam Y, Gartenhaus RB 2009. Extracellular signal-regulated kinase positively regulates the oncogenic activity of MCT-1 in diffuse large B-cell lymphoma. Cancer Res 69: 7835–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Noller HF 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864 [DOI] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Pestova TV, Hellen CU 2008. Factor requirements for translation initiation on the simian picornavirus internal ribosomal entry site. RNA 14: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyo JE, Chiao PJ, Tainsky MA 1998. drp, a novel protein expressed at high cell density but not during growth arrest. DNA Cell Biol 17: 437–447 [DOI] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev 20: 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S 1996. Translation of Sindbis virus mRNA: Analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol 70: 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA 2009. PERK-dependent regulation of IAP translation during ER stress. Oncogene 28: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L 1989. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev 3: 1899–1912 [DOI] [PubMed] [Google Scholar]
- Hellen CU, de Breyne S 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol 81: 5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner P, Dai B, Corl S, Liu Z, Gartenhaus RB 2009. Targeted suppression of MCT-1 attenuates the malignant phenotype through a translational mechanism. Leuk Res 33: 474–482 [DOI] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 16: 3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Anderson WF 1975. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 250: 1197–1206 [PubMed] [Google Scholar]
- Milon P, Konevega AL, Gualerzi CO, Rodnina MV 2008. Kinetic checkpoint at a late step in translation initiation. Mol Cell 30: 712–720 [DOI] [PubMed] [Google Scholar]
- Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO 2010. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep 11: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Nishimura Y, Sawano Y, Makino T, Tanokura M 2008. Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3. Proteins 73: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Montero H, Rojas M, Arias CF, López S 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J Virol 82: 1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pérez-Arellano I, Gallego J, Cervera J 2007. The PUA domain—a structural and functional overview. FEBS J 274: 4972–4984 [DOI] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W 2005. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU 1996. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 16: 6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J 20: 4560–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV 2006. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosniak M, Dierov J, Okami K, Tilton B, Jameson B, Sawaya BE, Gartenhaus RB 1998. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. Cancer Res 58: 4233–4237 [PubMed] [Google Scholar]
- Reinert LS, Shi B, Nandi S, Mazan-Mamczarz K, Vitolo M, Bachman KE, He H, Gartenhaus RB 2006. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res 66: 8994–9001 [DOI] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14: 6010–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell 17: 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Castelló A, Ventoso I, Berlanga JJ, Carrasco L 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS ONE 4: e4772 doi: 10.1371/journal.pone.0004772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 15: 836–841 [DOI] [PubMed] [Google Scholar]
- van Duijn LP, Holsappel S, Kasperaitis M, Bunschoten H, Konings D, Voorma HO 1988. Secondary structure and expression in vivo and in vitro of messenger RNAs into which upstream AUG codons have been inserted. Eur J Biochem 172: 59–66 [DOI] [PubMed] [Google Scholar]
- van Steeg H, van Grinsven M, van Mansfeld F, Voorma HO, Benne R 1981. Initiation of protein synthesis in neuroblastoma cells infected by Semliki Forest Virus. A decreased requirement of late viral mRNA for eIF-4B and cap binding protein. FEBS Lett 129: 62–66 [DOI] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV 2009. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res 37: 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]