Abstract
Major ampullate (dragline) spider silk is a coveted biopolymer due to its combination of strength and extensibility. The dragline silk of different spiders have distinct mechanical properties that can be qualitatively correlated to the protein sequence. This study uses amino acid analysis and carbon-13 solid-state NMR to compare the molecular composition, structure and dynamics of major ampullate dragline silk of four orb-web spider species (Nephila clavipes, Araneus gemmoides, Argiope aurantia and Argiope argentata) and one cobweb species (Latrodectus hesperus). The mobility of the protein backbone and amino acid side chains in water exposed silk fibers is shown to correlate to the proline content. This implies that regions of major ampullate spidroin 2 protein, which is the only dragline silk protein with any significant proline content, become significantly hydrated in dragline spider silk.
Keywords: solid-state NMR, spider silk, major ampullate silk, hydration dynamics, dragline silk, protein polymer
Introduction
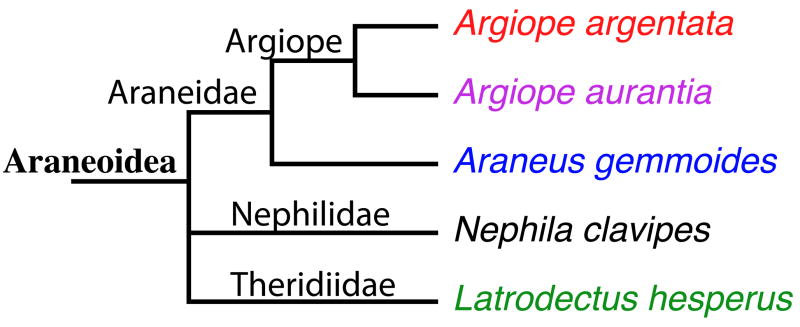
Spiders have evolved over hundreds of millions of years. The Araenoidea superfamily diverged into the araneidae and the “derived araneoids” around 125 million years ago.1 This split, which defines araneidae as orb weavers, includes Araneus gemmoides, Argiope argentata, Argiope aurantia, and groups other species such as orb weaver Nephila clavipes and cobweb weavers Latrodectus hesperus into “derived araneoids” (see Figure 1). All five species listed above have evolved to make six different types of silk fibers and an aqueous glue.2 These silks generally have the same function, including web structure (major ampullate, minor ampullate, flagelliform, pyriform, and aqueous glue), prey immobilization (aciniform) and egg case (aciniform and tubuliform).3
Figure 1.
A cladogram showing the relationship between Araneus gemmoides (A.g.), Argiope argentata (A.ar), Argiope aurantia (A.au), Latrodectus Hesperus (L.h.), and Nephila clavipes produced(N.c.) using sequenced genes indexed in Pubmed. All five spiders (Order – Araneae) are in the Superfamily – Araneoidea and produce either orb webs (Nephilidae and Araneidae) or cob webs (Theridiidae).
Although the silks of various species serve the same general purposes, the mechanical properties differ slightly for each silk of a given species, allowing them to adapt to their unique ecosystems. Of all the different silk fibers, dragline silk (major ampullate silk) has shown the greatest mechanical variation between individual species.4, 5 The mechanical property variation in dragline silks can be partially attributed to the nanostructure composite nature of Major ampullate Spidroin 1 (MaSp1) and Major ampullate Spidroin 2 (MaSp2) proteins that make up dragline fibers.6, 7 Both MaSp1 and MaSp2 have evolutionarily conserved highly repetitive motif structures found in a large class of web building spiders.8 Repetitive amino acid motifs make up the majority of the major ampullate spidrion proteins and have been the focus of numerous molecular-level structural investigations.
Solid-state NMR (ssNMR) has been instrumental in elucidating secondary structure within the highly repetitive amino acid motifs of spider and silkworm silk. For example, NMR was used to determine the amino acid repetitive motifs responsible for β-sheet crystalline domains in orb-weaving dragline spider silk9–11 and cocoon silk from Bombyx mori.12, 13 Furthermore, in spider dragline silk, ssNMR has been integral in characterizing the molecular structure of glycine-rich regions (GGX and GPGXX repetitive motifs)14, 15 and providing molecular structure and dynamic elucidation of supercontraction and the plasticizing effect of water.16–20 To date, however, very few papers have performed NMR studies on spider silks on any genus of spider other than Nephila.21–24 In this work, we compare dragline fibers from three arachnid families (four different genuses). The similarities and differences in the cross polarization and direct 13C detection NMR spectra of all five species are discussed.
Materials and Method
Spider Dragline Silks
Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes major silk were collected by forcibly silking adult female spiders at 2 cm/s.25 The spiders were anesthetized using CO2, which was done to reduce the stress of capture. Spiders were restrained and typically gained function within 2–5 minutes. Silking occurred after a spider was able to drink 20μL of water to ensure that it was awake. The silking process was monitored under a dissection microscope to ensure that only major ampullate silk was collected (no minor ampullate silk was mixed with the fiber). The spiders were fed one small cricket per silking and webs were misted with water twice daily. All silk samples have the natural abundance of 13C and 15N; no enrichment was performed on these samples. The amount of silk collected from each type of spider species was 9.1 mg of Araneus gemmoides silk, 11.5 mg of Argiope argentata silk, 6.4 mg of Argiope aurantia silk, 8.2 mg of Latrodectus hesperus silk for the dry experiments, 14.1 mg of Latrodectus hesperus silk for the wet experiments, 13.1 mg of Nephila clavipes silk for the dry experiments and 10.6 mg of Nephila clavipes dragline silk for the wet experiments.
Amino Acid Analysis (AAA)
Amino acid analysis was done using the Acquity Ultra Performance LC from Waters according to manufacturer protocols for the AccQ-Tag system. A small sample of natural silk fiber (< 1 cm) from each of the five species was hydrolyzed in 6 M HCl at 155°C for 30 minutes. Following hydrolysis, the samples were dried and then dissolved in 20mM HCl for derivatization. Each sample was derivatised with Waters pre-mixed derivatization compounds (ACQ, 6-aminoquinolyl-N-hydrozysuccinimidyl carbamate), which adds a fluorescent group to each amino acid prior to column injection. The manufacturer’s standard program for amino acid analysis was used for all identification and analysis. Known standards were run prior to the run and after each set of samples.
Solid-State NMR
Solid-state NMR spectra were collected on a Varian VNMRS 400 MHz wide-bore spectrometer equipped with a 3.2 mm triple resonance MAS probe operating in double resonance mode (1H/13C). Dry silk samples were run in standard zirconia Varian MAS rotors with Torlon caps. For wet samples, deuterated water (D2O) was added to each silk and the sample was packed in zirconia rotors that were sealed with O-ring Kel-f inserts to ensure the samples did not dehydrate. The thermal properties of spider silk indicate that the fibers will not be impacted by heating effects from MAS and/or 1H decoupling.26 1H→13C CP-MAS spectra were collected at both 5 and 10 kHz MAS with the CP condition matched to the Hartmann-Hahn condition and −1 spinning sideband of the Hartmann-Hahn profile, respectively. 13C CP-MAS NMR spectra for both wet and dry silks were collected using a 4 μs 1H 90° pulse, a 1 ms CP contact time, 100 kHz two pulse phase modulated (TPPM)27 decoupling during acquisition, 1024 data points, 12,288 scans, 100.525 MHz carbon spectrometer frequency, a 50 kHz sweep width, and a 4 s recycle delay.
Direct detection 13C{1H} MAS (DD-MAS) spectra collected with proton dipolar decoupling were obtained with 100 kHz TPPM 1H decoupling during acquisition, 1024 data points, 16,384 scans, a 50 kHz sweep width, and a recycle delay of 1 s for both wet and dry silk samples. Using a short recycle delay in the 13C{1H} DD-MAS experiments allows the mobile species to be enhanced, while saturating species with long T1 relaxation times.19 Processing parameters for 13C CP-MAS and DD-MAS spectra include baseline correction, zero-filling to 4096 points, and 25 Hz of exponential line broadening. Chemical shifts were attained utilizing an external adamantane standard setting the downfield peak at 38.56 ppm.
Results and Discussion
Amino Acid Analysis - Composition of Five Species of Spider Dragline Silk
Variations in mechanical properties of major ampullate silk from different species can be accounted for in part by the different ratios of MaSp1 and MaSp2, which can be estimated using the percentage of proline in the fiber.4, 28, 29 Table 1 provides the average mole percent of each amino acid residue for major ampullate (Ma) silk from Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes (Figure 1) spiders measured using standard amino acid analyses (AAA). These mole percentages have been shown to have large variability within a species of spider.30, 31 This is attributed in part to the apparent lack of uniformity in the spider silk fiber blending process of MaSp1 and 2 and associated inhomogeneity in spider silk fibers. The values tabulated from AAA in Table 1 are only provided for amino acids that are greater than 1 mole % in one or more species. Our results are within the range of previously reported AAA values for these species.28, 32 Clearly evident is the significant variation of both proline and serine among the species’ silk. The proline content of spider dragline silk has been shown to directly correlate to the elasticity and supercontraction effect.4, 5, 28, 29 Furthermore, proline is only present in the repetitive motifs of MaSp2 and is not found in any significant quantities in MaSp1.4, 7, 8, 28 Hence, the concentration of proline is directly dependent on the MaSp2/MaSp1 ratio, which affects the elasticity and supercontraction in spider dragline silk.
Table 1.
Amino acid analysis of major ampullate silk from Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus, and Nephila clavipes giving molar percent of the most abundant amino acids.
| Gly | Ala | Pro | Glx | Ser | Tyr | Leu | Val | Thr | Asx | Arg | Phe | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Araneus gemmoides | 42.8 | 19.4 | 11.1 | 8.2 | 7.1 | 5.2 | 1.1 | 1.1 | 1.0 | 0.9 | 0.6 | 0.5 |
| Argiope argentata | 44.6 | 19.3 | 10.2 | 9.2 | 6.8 | 5.4 | 1.0 | 0.5 | 0.5 | 0.4 | 1.1 | 0.8 |
| Argiope aurantia | 46.4 | 17.9 | 9.5 | 9.4 | 5.9 | 4.8 | 1.5 | 0.7 | 0.5 | 0.5 | 1.2 | 0.9 |
| Latrodectus hesperus | 45.7 | 31.1 | 1.5 | 8.7 | 1.1 | 4.5 | 0.7 | 0.6 | 0.7 | 0.7 | 1.5 | 0.4 |
| Nephila clavipes | 47.1 | 26.5 | 1.2 | 8.8 | 3.0 | 3.6 | 4.2 | 1.1 | 0.6 | 0.6 | 1.2 | 0.3 |
Asx = aspartate and aspartic acid, Glx = glutamine and glutamate. The variability in composition within each spider silk fiber sample was observed to be large (up to 50%) and the values presented are the average of five analysis runs on five different samples of each spider silk.
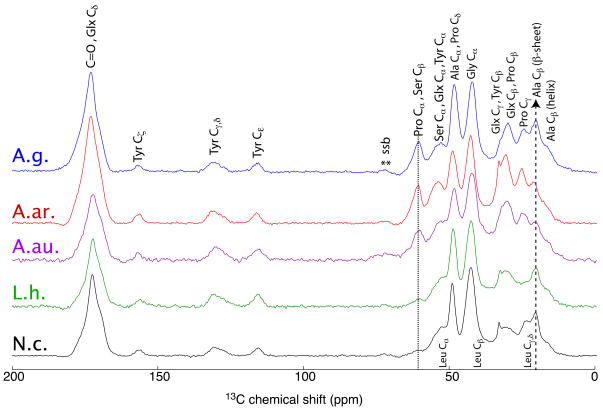
The 13C CP-MAS NMR Spectrum of Major Ampullate Silk from Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes spiders is shown in Figure 2. The spectra are scaled to the Gly-Cα resonance because this is the most abundant and conserved amino acid in all five spider silks, roughly 45.7 ± 3.8 mole % for all silks. The top to bottom order of the spectra is based on their average proline content, with Araneus gemmoides having the largest average proline content of 11.1 ± 2.5 mole % and Nephila clavipes having the smallest average proline content of 0.9 ± 0.3 mole %. The 13C CP-MAS NMR spectra (ωr = 10 kHz) are all fairly similar and contain resonances that can be assigned to Gly, Ala, Pro, Glx, Ser and Tyr; the amino acids that make up 90+ mole % of each spider silk (see Table 1). All 13C resonances assignments are based on several previous NMR studies.9, 19, 33–35 Also, on the bottom spectrum in Figure 2, Nephila clavipes silk, we have labeled resonances in the regions where Leu contributions are expected. Of the five species studied, Leu is only found in any significant amount in Nephila clavipes and is not clearly resolved from the other abundant amino acids in dragline silk. However, the shoulder at 22 ppm is in large part a contribution from the Leu Cγ and Leu Cδ.19, 36–39 The carbon resonances observed in the 13C CP-MAS NMR spectra of all five species of spiders’ dragline silk are heterogeneously broadened due to a distribution of chemical shifts that result from a continuum of structural conformations and environments. This heterogeneous distribution is additionally large in the carbonyl region (160–180ppm) as all of the amino acids in all different structural motifs and environments contribute to this region.14, 33 Also, the glutamine side chain group carboxyl contributes to this resonance.19, 23, 33 The structural and environmental heterogeneity observed is similar in all five species of spider silks and indicates that major ampullate spider silk contains a significant degree of disorder in the material.
Figure 2.
The 1H→ 13C CP-MAS NMR spectra of major dragline silk from Araneus gemmoides (A.g.), Argiope argentata (A.ar), Argiope aurantia (A.au), Latrodectus Hesperus (L.h.), and Nephila clavipes (N.c.). Spectra were collected with 10 kHz MAS and 1 ms CP contact time. The spinning side band is denoted with ssb (**).
The most notable difference in the 13C CP-MAS NMR spectra shown in Figure 2 for the five spider silks is the clear (relative to Gly) increase in a resonance centered at 25 ppm and 62 ppm.40–43 This is primarily attributed to the increase in proline content from the bottom spectrum (Nephila clavipes) to the top spectrum (Araneus gemmoides). The resonance at 62 ppm has a significant contribution from the Ser Cβ, which also increases in Agiope and Araneus silks. The broad nature of the proline and serine resonances is indicative of a polymeric material in a disordered or amorphous state.
The Ala Cβ 13C NMR resonance is commonly used to probe local structure in silk, because of its chemical shift sensitivity to various secondary structures.9, 44 There have been recent NMR studies that characterize and quantify the amount of helical and β-sheet component in spider dragline silk primarily based on the 13C Ala Cβ resonance and its carbon-carbon correlations to other amino acids.35, 45 All silks presented in figure 2 show a remarkably similar Ala Cβ resonance at 21 ppm with a shoulder at 17 ppm. The resonance at 21 ppm is indicative of Ala in a β-sheet and the shoulder at 17 ppm is a mixture of the helical, turn and random coil components. From the NMR spectra, it is clear that alanine-rich motifs (poly-A and poly-GA) primarily adopts a β-sheet structure for all five species of dragline silk.33, 45, 46 However, all five species also contain a small amount of Ala in helical, turn or random coil environments. This has been shown to primarily be Ala in the poly-GGX motif (X=Ala), which adopts a disordered 31-helical structure.19, 33, 47
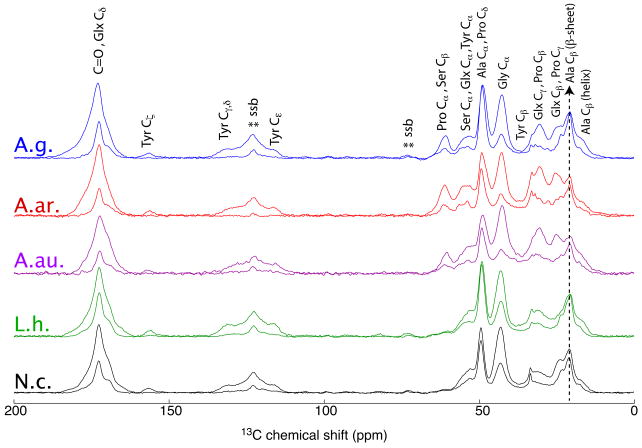
The 13C CP-MAS NMR Spectra of Wet and Dry Major Ampullate Silk from Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes spiders is shown in Figure 3. The effect of water on major ampullate silk fibers and water-induced supercontraction has been extensively studied at both functional and structural levels.3, 11, 17–19, 21, 28, 48–59 In Figure 3, 13C CP-MAS NMR spectra are shown for wet and dry dragline silks from five spider species. The same material used to collect NMR spectra for Figure 2 was used for the dry silk data shown in Figure 3. The only difference was that all spectra in Figure 3 were collected at a slower spinning speed (ωr = 5 kHz) to aid in comparison with the wet spectra.19 The 13C CP MAS NMR spectra of all wet silks show a clear loss of intensity for most of the resonances. The spectral intensity loss can be attributed to the increased protein backbone and side-chain mobility in spider silk when the material is wet.17–19 The exceptions to this loss in CP intensity are the Ala Cα and Cβ resonances at 49 and 21 ppm, respectively. These resonances only experience a minor loss in intensity between the dry and hydrated state in the five silk species. This effect has been well documented in past NMR results and is attributed to alanine located in the poly-(Ala) motif of spider silk that remain rigid in wet spider silk. These poly-(Ala) regions of spider silk are primarily in an anti-parallel β-sheet conformation, which is not solvated by water.9, 13, 18, 45
Figure 3.
The 1H→ 13C CP-MAS NMR spectra of dry (darker color) and wet (lighter color) major silk from Araneus gemmoides (A.g.), Argiope argentata (A.ar), Argiope aurantia (A.au), Latrodectus Hesperus (L.h.), and Nephila clavipes (N.c. Spectra were collected with 5 kHz MAS and 1 ms CP.) contact time. The spinning side bands are denoted with ssb (**).
The amount of proline in dragline silk has been related to (i) the amount of MaSp2 protein, (ii) an increased mobility in wet silk fibers and supercontraction effects and (iii) changes in the mechanical extensibility when hydrated.28, 60–62 The intensity loss in the 13C CP-MAS NMR spectra in Figure 3 shows a substantial difference between species, and generally track with the amount of proline. The intensity loss for wet major ampullate silk in Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes spiders is 66, 67, 71, 50, and 55 % for the Gly Cα peak at 43 ppm, respectively. The loss of NMR intensity in the Gly Cα resonance is similar in the araneidae species (Araneus and Argiope), where the derived araneoids (Nephila and Latrudectus) have a smaller intensity loss. This indicates that hydration of the glycine-rich region is a common feature in spider dragline silks from different families and that mobility is enhanced in proportion to the content of MaSp2 protein.
The Direct 13C{1H} MAS NMR Spectra of Wet and Dry Major Ampullate Silk utilizing a fast recycle delay (1 s) from Araneus gemmoides, Argiope argentata, Argiope aurantia, Latrodectus hesperus and Nephila clavipes spiders is shown in Figure 4. The wet and dry spectra are scaled to the Ala Cβ resonance at 21 ppm, which are of similar absolute intensity for all silks (wet and dry) and the dominant resonance in all dry silks. The Ala Cβ has two clear resonances at 17 ppm and 21 ppm for wet silks. The Ala Cβ resonance at 21 ppm is assigned to poly-(Ala) in a β-sheet. The line-width is similar for all wet and dry silks. This indicating that water does not hydrate the poly-(Ala) β-sheet domains in any of these silks. The resonance at 17 ppm is only well resolved in the wet silk spectra and is assigned to helical and/or random coil Ala regions of spider silk. The narrowing of this region under wet conditions indicates a significant increase in mobility of the Ala residues of helical or random coil structures within all the spider dragline silks. Conversely, the Ala Cα NMR resonance at 49 ppm does not appear in the dry silk spectra, nor is this resonance prominent in the wet silk spectra. It is common to selectively observe methyl resonances in fast recycle delay DD-MAS spectra of solid peptides or proteins due to their shorter spin lattice relaxation time (T1). A short T1 for methyl resonances is due to the inherent rotational motion of methyl groups, even if they are located in rigid structures such as β-sheets. All other resonances besides the methyl Ala Cβ are partially or fully saturated because of the long 13C T1 values of common backbone and carbonyl resonances and hence do not contribute to the dry silk DD-MAS NMR signal (and have a reduced contribution in the wet silk).
Figure 4.
The 13C DD-MAS NMR spectra of dry and wetted (lighter color) major silk from Araneus gemmoides (A.g.), Argiope argentata (A.ar), Argiope aurantia (A.au), Latrodectus Hesperus (L.h.), and Nephila clavipes (N.c.). Spectra were collected with 10 kHz MAS and 1 s recycle delay.
The glycine-rich regions of major ampullate spider silk have increased mobility when hydrated.18, 19 Hence, it is believed that water plasticizes the glycine-rich repetitive motif regions of spider dragline silk. Glycine and other amino acid residues that interact with water can be identified in 13C CP-MAS spectra of wet silk (figure 3) because they will exhibit a substantial decrease in signal intensity compared to dry spider silks. Conversely, the regions that become mobile are often enhanced when 13C direct spectra are collected with a fast recycle delay (1 s). The direct 13C MAS spectra of wetted silks show significantly enhanced resolution when compared to the dry silks or the CP-MAS spectra. This is most noticeable in the Araneus and Argiope spider silk samples, where spectra are significantly sharper (> 40% FWHM) compared to the Nephila clavipes and Latrodectus hesperus silk. This indicates that MaSp2 rich spider silks (silks high in proline content) become significantly more plasticized compared to MaSp 1 rich spider silks.
The enhanced resolution afforded by the fast-repetition direct 13C MAS spectra of wetted silks can be used to identify proline and glutamine (Gln) resonances in Argiope and Araneus dragline silk. Also, we see a significant increased resolution of the carbonyl region in these spider silks and can resolve the Gln Cδ resonance. In combination with INADEQUATE ssNMR45, the increased resolution and ability to identify Pro residues in wet dragline silk will allow the first structural and dynamics characterization of MaSp2 through the NMR chemical shift environment of Pro, which is found almost exclusively in GPGXX motifs within MaSp2.
Conclusion
Through evolution spider species have produced unique properties in silk by changing the MaSp1 to MaSp2 ratio with minor changes to the amino acid sequence. 13C CP-MAS and DD-MAS have allowed a more detailed examination into the similarities and differences of five spider species’ major ampullate dragline silk. Four orb-web spider species (Nephila clavipes, Araneus gemmoides, Argiope aurantia and Argiope argentata) and one cobweb species (Latrodectus hesperus) were studied and shown to have proline content ranging from 0.9 to 11.1 mole percent. The mobility of the protein backbone and amino acid side chains in water exposed silk fibers is shown to correlate to the proline content. This implies that regions of major ampullate spidroin 2 protein, which is the only dragline silk protein with any significant proline content, become significantly hydrated in dragline spider silk. Also, it is clear that various solid state NMR techniques can be used to discern various types of spider silk and characterize their structure and hydration dynamics.
Acknowledgments
This work was supported by grants from the US National Science Foundation (NSF-DMR 0805197), the US National Institute of Health (NIH-EB000490) and the DOD Air Force Office of Scientific Research (AFOSR). We would like to thank Dr. Brian Cherry for his help with NMR experiments and the Magnetic Resonance Research Center at Arizona State University as well as the Macromolecular Core Facility at the University of Wyoming for the use of NMR and AAA instrumentation, respectively.
References
- 1.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Science. 2001;291(5513):2603–5. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 2.Peters HM. Z Naturforsch. 1955;10:95. [Google Scholar]
- 3.Vollrath F. Scientific American. 1992;266:70–76. [Google Scholar]
- 4.Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. Biomacromolecules. 2005;6(6):3095–9. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- 5.Blackledge TA, Summers AP, Hayashi CY. Zoology (Jena) 2005;108(1):41–6. doi: 10.1016/j.zool.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Xu M, Lewis RV. Proc Natl Acad Sci U S A. 1990;87(18):7120–4. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinman M, Lewis RV. J Biol Chem. 1992;267(27):19320–19324. [PubMed] [Google Scholar]
- 8.Hayashi CY, Shipley NH, Lewis RV. Int J Biol Macromol. 1999;24(2–3):271–5. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 9.Simmons A, Ray E, Jelinski LW. Macromolecules. 1994;27(18):5235–5237. [Google Scholar]
- 10.Simmons AH, Michal CA, Jelinski LW. Science. 1996;271(5245):84–87. doi: 10.1126/science.271.5245.84. [DOI] [PubMed] [Google Scholar]
- 11.Parkhe AD, Seeley SK, Gardner K, Thompson L, Lewis RV. J Mol Recognit. 1997;10(1):1–6. doi: 10.1002/(SICI)1099-1352(199701/02)10:1<1::AID-JMR338>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 12.Asakura T, Ashida J, Yamane T, Kameda T, Nakazawa Y, Ohgo K, Komatsu K. J Mol Biol. 2001;306(2):291–305. doi: 10.1006/jmbi.2000.4394. [DOI] [PubMed] [Google Scholar]
- 13.Asakura T, Yang M, Kawase T, Nakazawa Y. Macromolecules. 2005;38:3356–3363. [Google Scholar]
- 14.van Beek JD, Hess S, Vollrath F, Meier BH. Proc Natl Acad Sci U S A. 2002;99(16):10266–71. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eles PT, Michal CA. Biomacromolecules. 2004;5(3):661–5. doi: 10.1021/bm0342685. [DOI] [PubMed] [Google Scholar]
- 16.Eles P, Michal CA. Macromolecules. 2004;37:1342–1345. [Google Scholar]
- 17.Yang Z, Liivak O, Seidel A, LaVerde G, Zax DB, Jelinski LW. J Am Chem Soc. 2000;122(37):9019–9025. [Google Scholar]
- 18.Holland GP, Lewis RV, Yarger JL. J Am Chem Soc. 2004;126(18):5867–72. doi: 10.1021/ja031930w. [DOI] [PubMed] [Google Scholar]
- 19.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Biomacromolecules. 2008;130(30):9871–77. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 20.Sapede D, Seydel T, Forsyth VT, Koza MM, Schweins R, Vollrath F, Riekel C. Macromolecules. 2005;38:8447–8453. [Google Scholar]
- 21.Bonthrone KM, Vollrath F, Hunter BK, Sanders JKM. Proc Roy Soc Lond Biol Sci. 1992;248(1322):141–144. [Google Scholar]
- 22.Hu XY, Lawrence B, Kohler K, Falick AM, Moore AMF, McMullen E, Jones PR, Vierra C. Biochemistry. 2005;44(30):10020–10027. doi: 10.1021/bi050494i. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence BA, Vierra CA, Moore AM. Biomacromolecules. 2004;5(3):689–95. doi: 10.1021/bm0342640. [DOI] [PubMed] [Google Scholar]
- 24.Bonev B, Grieve S, Herberstein ME, Kishore AI, Watts A, Separovic F. Biopolymers. 2006;82(2):134–143. doi: 10.1002/bip.20471. [DOI] [PubMed] [Google Scholar]
- 25.Work RW, Emerson PD. J Arachn. 1982;10(1):1–10. [Google Scholar]
- 26.Kaplan DL. Poly Degrad Stab. 1998;59(1–3):25–32. [Google Scholar]
- 27.Bennet AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J Chem Phys. 1995;103(16):6951–6958. [Google Scholar]
- 28.Liu Y, Sponner A, Porter D, Vollrath F. Biomacromolecules. 2008;9(1):116–21. doi: 10.1021/bm700877g. [DOI] [PubMed] [Google Scholar]
- 29.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. J Exp Biol. 1999;202(Pt 23):3295–303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 30.Lombardi SJ, Kaplan DL. J Arachn. 1990;18(3):297–306. [Google Scholar]
- 31.Work RW, Young CT. J Arachn. 1987;15:65–80. [Google Scholar]
- 32.Andersen SO. Comp Biochem Phys. 1970;35(3):705–711. [Google Scholar]
- 33.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. J Am Chem Soc. 2008;130(30):9871–7. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 34.Izdebski T, Akhenblit P, Jenkins JE, Yarger JL, Holland GP. Biomacromolecules. 2010;11(1):168–174. doi: 10.1021/bm901039e. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins JE, Creager MS, Lewis RV, Holland GP, Yarger JL. Biomacromolecules. 2010;11(1):192–200. doi: 10.1021/bm9010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jelinski LW, Blye A, Liivak O, Michal C, LaVerde G, Seidel A, Shah N, Yang Z. Int J Biol Macromol. 1999;24(2–3):197–201. doi: 10.1016/s0141-8130(98)00085-3. [DOI] [PubMed] [Google Scholar]
- 37.Michal CA, Jelinski LW. J Biomol NMR. 1998;12(2):231–41. doi: 10.1023/a:1008286004222. [DOI] [PubMed] [Google Scholar]
- 38.Taki T, Yamashita S, Satoh M, Shibata A, Yamashita T, Tabeta R, Hazime S. Chem Lett. 1981;10(12):1803–1806. [Google Scholar]
- 39.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Proc Natl Acad Sci U S A. 2002;99(26):16748–53. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kricheldorf H, Muller D. Int J Biol Macromol. 1984;6(3):145–51. [Google Scholar]
- 41.Ohgo K, Kawase T, Ashida J, Asakura T. Biomacromolecules. 2006;7(4):1210–1214. doi: 10.1021/bm0600522. [DOI] [PubMed] [Google Scholar]
- 42.Saitô H, Tabeta R, Shoji A, Ozaki T, Ando I, Miyata T. Biopolymers. 1984;23(11):2279–2297. doi: 10.1002/bip.360231111. [DOI] [PubMed] [Google Scholar]
- 43.Liivak O, Flores A, Lewis RV, Jelinski LW. Macromolecules. 1997;30:7127–7130. [Google Scholar]
- 44.Asakura T, Yang M, Kawase T. Polymer Journal. 2004;36(12):999–1003. [Google Scholar]
- 45.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Chem Comm. 2008;(43):5568–5570. doi: 10.1039/b812928b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rathore O, Sogah D. J Am Chem Soc. 2001;123:5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]
- 47.Marcotte I, van Beek J, Meier B. Macromolecules. 2007;40:1995–2001. [Google Scholar]
- 48.Bell FI, McEwen IJ, Viney C. Nature. 2002;416(6876):37. doi: 10.1038/416037a. [DOI] [PubMed] [Google Scholar]
- 49.Bonthrone KM, Vollrath F, Hunter BK, Sanders JKM. Proceedings of the Royal Society London: Biological Sciences. 1992;248(1322):141–144. [Google Scholar]
- 50.Dunford HB, Morrison JL. Can J Chem. 1955;33:904–12. [Google Scholar]
- 51.Hirai Y, Ishikuro J, Nakajima T. Polymer. 2001;42(12):5495–5499. [Google Scholar]
- 52.Jelinski LW, Blye A, Liivak O, Michal C, LaVerde G, Seidel A, Shah N, Yang Z. Int J Biol Macromol. 1999;24(2–3):197–201. doi: 10.1016/s0141-8130(98)00085-3. [DOI] [PubMed] [Google Scholar]
- 53.Savage KN, Guerette PA, Gosline JM. Biomacromolecules. 2004;5(3):675–9. doi: 10.1021/bm034270w. [DOI] [PubMed] [Google Scholar]
- 54.Sohn S, Strey HH, Gido SP. Biomacromolecules. 2004;5(3):751–7. doi: 10.1021/bm0343693. [DOI] [PubMed] [Google Scholar]
- 55.van Beek JD, Kummerlen J, Vollrath F, Meier BH. Int J Biol Macromol. 1999;24(2–3):173–8. doi: 10.1016/s0141-8130(98)00083-x. [DOI] [PubMed] [Google Scholar]
- 56.Grubb DT, Jackrel D, Jelinski LW. Polymer Preprints (American Chemical Society, Division of Polymer Chemistry) 1997;38(2):73–74. [Google Scholar]
- 57.Grubb DT, Ji G. Int J Biol Macromol. 1999;24(2–3):203–10. doi: 10.1016/s0141-8130(98)00086-5. [DOI] [PubMed] [Google Scholar]
- 58.Shao Z, vollrath F, Sirichaisit J, Young RJ. Polymer. 1999;40(10):2493–2500. [Google Scholar]
- 59.Work RW. J Exp Biol. 1985;118:379–404. [Google Scholar]
- 60.Liu Y, Shao ZZ, Vollrath F. Biomacromolecules. 2008;9(7):1782–1786. doi: 10.1021/bm7014174. [DOI] [PubMed] [Google Scholar]
- 61.Savage KN, Gosline JM. J Exp Biol. 2008;211(12):1937–1947. doi: 10.1242/jeb.014217. [DOI] [PubMed] [Google Scholar]
- 62.Savage KN, Gosline JM. J Exp Biol. 2008;211(12):1948–1957. doi: 10.1242/jeb.014225. [DOI] [PubMed] [Google Scholar]