Abstract
An extensive search for isoflurane binding sites in the nicotinic acetylcholine receptor (nAChR) and the proton gated ion channel from Gloebacter violaceus (GLIC) has been carried out based on molecular dynamics (MD) simulations in fully hydrated lipid membrane environments. Isoflurane introduced into the aqueous phase readily partitions into the lipid membrane and the membrane-bound protein. Specifically, isoflurane binds persistently to three classes of sites in the nAChR transmembrane domain: (i) An isoflurane dimer occludes the pore, contacting residues identified by previous mutagenesis studies; analogous behavior is observed in GLIC. (ii) Several nAChR subunit interfaces are also occupied, in a site suggested by photoaffinity labeling and thought to positively modulate the receptor; these sites are not occupied in GLIC. (iii) Isoflurane binds to the subunit centers of both nAChR α chains and one of the GLIC chains, in a site that has had little experimental targeting. Interpreted in the context of existing structural and physiological data, the present MD results support a multisite model for the mechanism of receptor-channel modulation by anesthetics.
Keywords: anesthesia, cys-loop receptor, ligand-gated ion channel
Despite efforts reaching back over a century, the molecular mechanism through which certain small molecules (general anesthetics) cause reversible immobilization and amnesia remains unclear. Known general anesthetics fall into several diverse classes, but the dominant effects of nearly all general anesthetics are believed to reflect modulation of ion channels in the central nervous system (1–3). An understanding of the mechanisms by which general anesthetics modulate such channels is therefore not only essential for medical progress, but can also serve to illuminate underlying behavior of ion channels and their larger role in the biological processes of mobilization and consciousness. Particular attention has focused on the anesthetic-sensitive Cys-loop superfamily of ligand-gated ion channels, including cation channels such as the nicotinic acetylcholine receptor (nAChR) and serotonin receptors, as well as anion channels such as the γ-aminobutyric acid class A (GABAA) receptor and the glycine receptor. Recently a prokaryotic cation channel of this superfamily, the proton gated ion channel from Gloebacter violaceus (GLIC), has demonstrated sensitivity to both intravenous and inhaled anesthetics at subclinical concentrations (4).
High-resolution crystal structures have demonstrated that anesthetics do bind directly to proteins (5–8), whereas more indirect means such as mutagenesis and photolabeling have indicated that general anesthetics bind to Cys-loop receptors in particular (1–3). Obtaining high-resolution structures of Cys-loop receptors even in the absence of anesthetic has proven to be a challenge, however, and structures have only been solved for prokaryotic pentameric ion channels (9–11), including GLIC in a putatively open state (3EHZ, 3EAM). For the most part, these crystal structures reveal a family of proteins that is consistent with the earlier 4-Å cryo-EM structure of nAChR from Torpedo solved by Unwin and coworkers (2BG9) (12, 13).
An intriguing difference between the nAChR structure reported in 2BG9 and the prokaryotic structures are the large gaps in protein density in the extracellular half of the nAChR transmembrane domain (TMD). The high-resolution prokaryotic structures do not display such gaps (Fig. S1). Recently (14), we proposed that such gaps are occupied by cholesterol, which is essential for nAChR function (15). Because cholesterol is not found in prokaryotic membranes, this hypothesis provides an alternate explanation to the source of differences between the two structures. Furthermore, as we demonstrate in the present paper, even with docked cholesterol, there is ample space in the nAChR TMD for binding of multiple isoflurane molecules. The collapse of the nAChR TMD observed in simulations of a cholesterol-free model (14) results in a structure that presumably would not offer as many binding sites for anesthetics as observed here. Such a collapse, however, is inconsistent with structural information obtained on nAChR in native membranes (13), and consequently no cholesterol-free nAChR models were considered in this study.
In the absence of detailed structural information, most experimental efforts to determine binding sites for anesthetics in Cys-loop receptors have depended on techniques such as electrophysiology, mutagenesis, and photolabeling of various anesthetics and alcohols to the GABAA, glycine, and nACh receptors. At clinical concentrations, most volatile anesthetics positively modulate the GABAA receptor but negatively modulate the nAChR, indicating that some binding sites do not overlap (16). In general, experiments suggest multiple binding sites (17, 18) for anesthetics and alcohols on both the nAChR and GABAA receptor: Potential sites have been identified in the TMD, at subunit interfaces (1, 3, 16, 19–24) in the nAChR pore, (16, 25–29), and at various positions in the agonist-binding domain (22). The multitude of potential sites and mechanisms has particularly complicated interpretation of ion current measurements, because of the possibility of competing effects. Mutagenesis and photolabeling studies provide an incomplete picture of anesthetic binding sites, because the choice of mutations or selective reactivity of the photolabel prevent the whole receptor from being explored, and the hydrophobic regions to which anesthetics bind are difficult to isolate. In addition, such methods typically identify regions of the amino acid sequence, from which spatial location of binding sites is indirectly inferred. If multiple residues are identified, it is often not clear whether they form multiple binding sites, or simply represent multiple sides of the same binding site.
Complementing experimental approaches, computational methods can serve to directly illuminate microscopic features of anesthetic-ion channel interactions. Structure-based docking is a common technique used to find ligand binding sites on a protein of known structure. Tang and coworkers (30) reported several mostly superficial sites for halothane detected using structure-based docking to their model of the α4β2 nAChR in an open conformation; binding free energy calculations revealed that halothane bound with low affinity to most sites, with the exception of a deeper TM site suggested by experiments. Molecular dynamics (MD) computation-based “flooding” of the receptor (in which a high concentration of anesthetic is placed in the surrounding water and allowed to partition into lipid and protein binding sites over the course of an MD simulation trajectory) is a more expensive alternative to structure-based docking which holds several advantages for investigating volatile anesthetics and Cys-loop receptors. In the MD approach, nearly all protein degrees of freedom are unrestrained, resulting in a fully flexible, dynamic, and physical dock that is especially appropriate for the highly mobile nAChR (and the corresponding medium-resolution structure). Unlike docking calculations, the MD approach naturally includes explicit water, which is shown in ref. 8 to be an essential component of some anesthetic binding sites. The membrane lipid environment can also be included, along with the cholesterol that we proposed (14) is embedded in the nAChR TM-gaps. Furthermore, the MD approach accounts for interactions between anesthetic molecules, and can therefore identify multiply occupied sites.
Flooding of a protein by anesthetics has been reported previously by members of our group (31, 32); here we present MD simulations on a much increased scale involving isoflurane partitioning into two nearly complete and fully solvated pentameric ligand-gated ion channels: nAChR and GLIC. In order to achieve full partitioning of isoflurane into deeply buried protein sites, this method requires substantially longer computation times than those reached by previous simulations of Cys-loop receptors. Such simulations typically involve about 200,000 atoms. The two systems presented here were simulated for 0.4 μs each. The nAChR from Torpedo was used for the simulations for the most direct comparison with experimental data and to reduce errors caused by homology modeling.
We find that isoflurane binding is remarkably consistent between GLIC and nAChR, with the exception of sites deep within the TMD. We divide the binding sites we observe in the nAChR into eight classes, four of which are found in the TMD (pore, intersubunit, intrasubunit, and annular), and four of which are found in the extracellular domain (interfacial loops, agonist site, β-sandwich, and α1 helix). Binding to internal sites in the TMD (intersubunit and intrasubunit sites) is much reduced in the tightly packed GLIC structure, whereas other classes of sites are similarly occupied in the two receptors. In Results, we address the three sites with the strongest implications for channel function, and remaining sites are addressed in the SI Text. The data presented here only provide direct information regarding the location of binding sites, and not their effect on the receptor structure and dynamics. Furthermore, the present paper reports only on the location of sites of at least moderate affinity as demonstrated by persistent occupancy. Quantitative measurements of affinity that reliably include even relatively fast protein dynamics (such as rotation of side-chain dihedral angles) would require significantly more computational time, as demonstrated in ref. 33.
Results
By the final frames of the simulation, there were between zero and one isoflurane molecules in the aqueous phase surrounding the nAChR, corresponding to a concentration ranging between 0 and 1 mM; this range includes the EC50 for anesthesia as well as the concentrations used in most experiments cited here. Isoflurane surrounding the GLIC simulation equilibrated to a concentration of 4 mM. As demonstrated in Fig. S2, fewer isoflurane molecules partitioned to the GLIC surface than the nAChR surface.
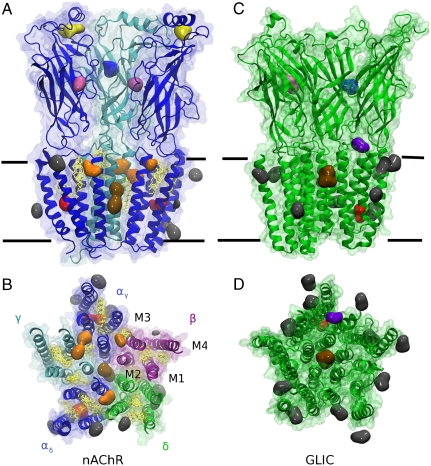
Binding sites were identified by constructing an isoflurane density map, averaged over frames from the last 100 ns of the simulation. Regions of high density therefore reflect sites in which isoflurane was persistently bound, and are shown for both nAChR and GLIC in Fig. 1. A few sites (depicted as small blobs) were occupied for less than half of the production period. Contact residues in the TMD are reported in Fig. S3.
Fig. 1.
Regions of persistent occupation by isoflurane. (A and B) Isoflurane binding sites in the nAChR. Protein is colored by subunit: α, blue; β, purple; δ, green; γ, cyan. Embedded cholesterol is yellow. Colored blobs represent an isoflurane density isosurface averaged over the last 100 ns of the simulation; large blobs represent occupation over at least most of that period, whereas a few much smaller blobs represent occupation for less than half of that period. (A) Side view of the αδ, γ, and αγ subunits, as well as isoflurane sites contacting those subunits. Isoflurane binding sites in the TMD are colored as follows: superficial/annular sites (gray), intrasubunit sites (red), intersubunit sites (orange), and pore site (brown). Isoflurane binding sites in the LBD are pink, blue, and yellow, corresponding with binding to the agonist site, beta sandwiches, and α1 helices, respectively. (B) View of the nAchR TM domain, looking down on the membrane from the extracellular region. (C and D) Isoflurane binding sites in GLIC. Protein is green and blobs are colored as in A–C, with the addition of isoflurane in the loop site (purple). (C) Side view of three chains of GLIC. (D) TMD of GLIC, with isoflurane in loop site, intrasubunit site, pore site, and annular sites. Binding to LBD for both proteins is shown in Figs. S4–S6
Pore.
An isoflurane dimer (colored brown in Fig. 1) occupies the nAChR pore for the last 300 ns of the simulation. Each molecule in the dimer shields the other from water on one side, so that in narrow regions of the water-filled pore each binding site assumes qualities of a hydrophobic-polar interface, like those known (5–8) to bind volatile anesthetics.
The two isoflurane molecules are mobile within the nAChR pore, with contact residues ranging from those in 6′ (a serine ring) to those in 16′ (a hydrophobic ring). Mutagenesis studies (28, 29) have indicated that mutation of α∶S10′ (α∶S252) to a hydrophobic residue increases sensitivity to isoflurane; our results are highly consistent with that work, because we find both that the 10′ position is frequently occupied by isoflurane (Fig. 2) and that hydrophobic contacts are preferred by isoflurane bound to the pore (Fig. S3) Smaller isoflurane density peaks, associated with the second molecule in the dimer, are observed between 6′ and 10′ and between 13′ and 16′. Exchange between the two isoflurane molecules is frequent, despite the presence of low-probability regions between the discrete sites.
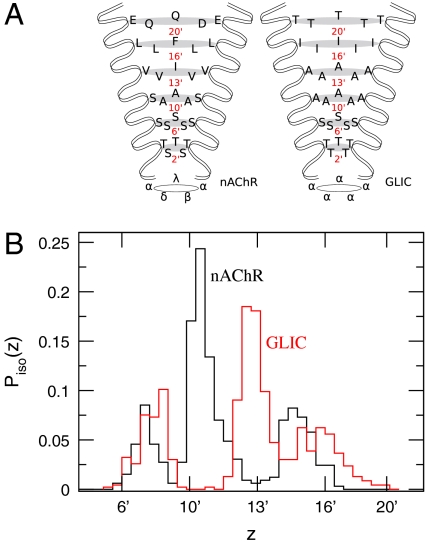
Fig. 2.
Distribution of isoflurane density in ion channel pore. (A) Standard prime numbering scheme for both nAChR (2) and GLIC (11). Figure is based on that of ref. 2. (B) Probability to find at least one isoflurane molecule with height z in the pore. Both curves include combined data from each isoflurane molecule in the dimer.
Binding to an analogous site (also by an isoflurane dimer) is observed in simulations of the GLIC channel (Fig. 1). Isoflurane bound to the GLIC channel (Fig. 2) also follows a strongly peaked trimodal distribution with the smaller peaks in similar locations as for nAChR. The most probable contact residue for isoflurane in the GLIC pore is 13′, rather than 10′ as in the nAChR. This discrepancy is consistent with differences in pore radius as a function of z: In nAChR, the pore is relatively wide at 10′, whereas a similarly sized opening occurs at 13′ in GLIC (11). Isoflurane bound to the GLIC pore generally displays fewer hydrogen bonds with pore residues than does isoflurane bound to the nAChR pore (Fig. S7), which is likely due to the higher hydrophobicity of the GLIC pore and the absence of serine and threonine residues found in nAChR. Although there is presently no experimental data regarding the location of binding sites for anesthetics on GLIC, binding of anesthetics to the pore provides the simplest explanation for the inhibitory effect reported in ref. 4.
The results of several experiments with differing methodologies are consistent with a binding site for anesthetics in the nAChR pore. In addition to the mutagenesis studies (28, 29) already mentioned, photolabeling of nAChR with (4-[3-(trifluoromethyl)-3H-diazirin-3-yl]benzyl 1-(1-phenylethyl)-1H-imidazole-5-carboxylate) (TDBzl-etomidate) (16) yields labeling of pore-lining residues (α∶S252). Furthermore, single channel recordings of the nAChR under control conditions indicate that openings of the channel are isolated. After exposure to isoflurane, the openings occur in clusters (or bursts) and are separated by brief closures. The duration of the bursts is nearly equivalent to the open time in the control system (25–27). One interpretation of this “flickering” effect is that major conformational changes occur on relatively similar time scales in the absence and presence of isoflurane, but that isoflurane binds and unbinds quickly to the pore, blocking ion flow and dramatically shortening the effective open time. We see binding of isoflurane to the pores of both the nAChR and GLIC structures, which were solved under conditions resulting in a functionally closed and open conformation, respectively.
Intersubunit.
Four isoflurane molecules (colored orange in Fig. 1) bind to three sites in the subunit interfaces of the nAChR TM domain, below the M2–M3 loop. The αδ–δ and αγ–γ interfaces are both occupied persistently by either one or two isoflurane molecules; in the ligand binding domain (LBD) these interfaces correspond to agonist-binding sites. In addition, one isoflurane molecule binds loosely to the αγ–β interface without occupying a well-defined binding site. A sample frame showing the multiply occupied site is shown in more detail in Fig. 3. Isoflurane hydrogen bonds to nonbulk water and (in the δ and γ subunits) asparagine residues located on M1 (see Fig. S3). Isoflurane is not observed to bind to subunit interfaces in the TMD of GLIC over the course of the 400 ns simulation. Reduced binding to the TMD of GLIC is likely due to the increased packing density of GLIC relative to nAChR (Fig. S1).
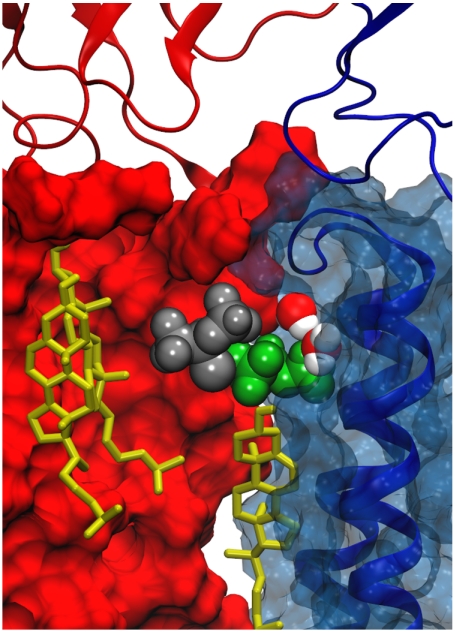
Fig. 3.
Sample frame showing the αγ–γ interface in the TMD. The γ subunit is red and the αγ subunit is blue, with cartoon overlay showing M2–M3 loop. Cholesterol is yellow, water is colored by atom type, and the two isoflurane molecules are silver or green.
Intersubunit sites for isoflurane in the nAChR overlap with what we termed the “B” hypothetical binding sites for cholesterol in ref. 14. The present simulations demonstrate that binding of cholesterol to the B site does not preclude the binding of one or more volatile anesthetics to the anesthetic site. Isoflurane rests near the cholesterol hydroxyl, although hydrogen bonding between isoflurane and cholesterol in the B sites is minimal. As shown in Fig. 3, isoflurane can bind between cholesterol and the transmembrane-LBD interface, disrupting contacts between cholesterol and the M2–M3 loop.
Significant experimental evidence is consistent with a site for anesthetics at subunit interfaces. Photolabeling of the nAChR reveals a halothane binding site at the αδ–δ interface (22) and a TDBzl-etomidate binding site at αγ–γ (16), the occupation of which is proposed to positively modulate the nAChR. Moreover, residues in the GABAA and glycine receptors have been identified that contribute to positive modulation of those receptors, using photolabeling and mutagenesis (1, 3, 19–24); some of these residues are thought to lie at the subunit interfaces in a site analogous to the intersubunit sites we observe, according to experimentally confirmed (34) homology models based on the nAChR. A similar site is occupied in simulations of halothane interacting with the nAChR (30, 35). The site is highly suggestive as an allosteric modulation site, as the M2–M3 loop is thought to act in transduction of a ligand binding signal to the pore (12).
Intrasubunit.
Isoflurane binds to both α subunit interiors (sites colored red in Fig. 1) in the intracellular half of the nAChR TMD, beneath cholesterol occupying “C” sites (14). Isoflurane is also observed to bind in the same region of one GLIC subunit. All three isoflurane molecules bound to these sites share a homologous pair of contact residues: α∶T229 and α∶V230 in nAChR and S212 and W213 in GLIC (Fig. S3). Additional contact residues are found but are not consistently detected across the three instances of binding to this class of site. In particular, serine residues forming the nAChR site frequently serve as hydrogen bonding partners.
Given that isoflurane only binds to such sites in α subunits of the nAChR, it is tempting to rule out sites in the other chains. The unexpected asymmetry of this mode of interaction across GLIC subunits suggests that the isoflurane concentration is too low to consistently bind to all five sites, but it is possible that isoflurane has a higher affinity for α subunits of the nAChR than for GLIC. (Alternatively, it is possible that fivefold symmetry of binding to GLIC would be seen if the simulation were run longer, but it seems unlikely because most isoflurane has partitioned into the membrane by the end of the simulation.) In GABAA receptors, mutation of only one or two subunits greatly reduces anesthetic sensitivity, suggesting that fivefold binding is not necessary for clinical effects (19). Supposing exactly two isoflurane molecules binding to five intrasubunit sites with equal affinity, there is only a 10% chance that both would bind to α subunits. A preference for α subunits appears to stem from the replacement of α∶T229 with hydrophobic residues in β, δ, and γ chains; because there is little solvation by water in these regions (Fig. S7), polar residues offer the only likely hydrogen bond acceptors.
A binding site in this region of the nAChR has not been conclusively pinpointed experimentally, to our knowledge. With embedded cholesterol included, the site is likely too small for occupancy by etomidate or neurosteroids, so it is not surprising that it is not labeled by TDBzl-etomidate (16). A photolabeling study of halothane binding to the nAChR (22) primarily detected labeling of aromatic residues, which are not found in the vicinity of the nAChR intrasubunit sites. A site for ethanol in the center of the subunit of GABAA has been considered in the absence of information regarding the location of various critical residues (19), but the contact residues (Fig. S3) we find for isoflurane in this site are not homologous to any previously tested (using the alignment prescribed in ref. 36).
Discussion
The simulations presented here constitute an extensive search for anesthetic binding sites on a Cys-loop receptor; the method allows for an unbiased search in which no preliminary information regarding site location is required. Multiple binding sites for isoflurane on nAChR are found. Importantly, isoflurane binds as a dimer to the pores of both nAChR and GLIC, in a configuration that would clearly obstruct ion flow. Given confirmation of this presumably inhibitory site (which would dominate the effect of isoflurane at concentrations for which it was occupied), as well as the positive effect of many anesthetics on other Cys-loop receptors, the presence of a positively modulating site elsewhere in the nAChR as suggested by ref. 16 seems increasingly likely. Our work is consistent with the results of many studies indicating that anesthetics bind to intersubunit sites in the TM domain, below the M2–M3 loop. Such positive modulation could therefore occur through increased correlations between the conformation of the LBD and the TMD. We find additional indirect evidence that binding to this site positively modulates the receptor, because it is not found in GLIC, which is significantly more sensitive than nAChR to inhibition by anesthetics (4).
Our MD simulations raise the possibility that isoflurane and other small anesthetics partition into the cytoplasmic half of Cys-loop receptor α subunit TM domains, in addition to sites at the subunit interface. Because such sites lie in the core of the domain, in the same plane that contains the hydrophobic constriction of the pore, and in the same subunit that confers sensitivity to agonist, their occupancy may have a detectable or even physiologically significant effect on function. Such effects could potentially be through allosteric means, in which the occupancy of the site stabilizes the closed or open conformation. Because of the sites’ location behind M2 helices, however, it is also possible that occupancy directly hinders either tilting or rotation of M2 helices, which is likely required for pore opening. Fig. S3 lists contact residues that are conserved across the three observed instances of binding, which could be targeted in mutagenesis studies.
Materials and Methods
System Setup.
Systems nAChR and GLIC were set up similarly. The nAChR coordinates were taken from 2BG9, missing β8–β9 loops were modeled using MODELLER (37), as in ref. 14, and cholesterol was inserted as in the full occupancy system of that reference. Unlike the simulations reported in ref. 14, the vestibule domain helices (MA) were removed for efficiency, because their behavior probably cannot be modeled realistically without the missing 100-residue M3–MA loop, and their removal significantly decreases the amount of water required for simulation. Starting cholesterol coordinates were optimized using a self-implemented Monte Carlo algorithm that minimized van der Waals clashes while applying a global biasing potential encouraging fivefold symmetry. The GLIC protein structure was taken from the 3EAM Protein Data Bank entry and prepared in a similar manner as by Bocquet et al. (11). In particular, acid dissociation constants were taken directly from the results given in ref. 11 and used to protonate appropriate amino acids to describe the protein at pH 4.6.
Both proteins were placed in 1-palmitoyl-2-oleoyl-sn-glycerol-phosphatidylcholine bilayers originally made using the MEMBRANE plug-in of Visual Molecular Dynamics (VMD) (38) and equilibrated using the procedure in ref. 39. The systems were solvated using the SOLVATE plug-in of VMD (38) and NaCl was added to a 0.15 M concentration. One thousand steps of minimization were conducted and then all atoms of the protein were constrained for 2 ns while the membrane healed around the protein. Subsequently, isoflurane was inserted randomly into the water surrounding the protein, with an isoflurane-to-lipid ratio of about 1∶3 for nAChR and about 1∶2 for GLIC. Both systems comprised approximately 200,000 atoms.
Simulation Details.
The simulations used the CHARMM22-CMAP force field with torsional cross-terms (40, 41) for proteins, CHARMM27 (42) for phospholipids, ions, and water, the Cournia et al. model for cholesterol (43), and parameters developed in our group for isoflurane (33). Minimization and dynamics were conducted with the NAMD2.7b1 package (44). Periodic boundary conditions were applied, with particle-mesh Ewald long-range electrostatics and a cutoff of 1.2 nm for Lennard–Jones potentials, with a smooth switching function starting at 1.0 nm. Simulations were conducted at a constant temperature of 300 K and pressure of 1 bar. Bonds involving hydrogen atoms were constrained to their equilibrium length using the SHAKE/RATTLE algorithm. Multiple-timestep integration was carried out using r-RESPA, with a base timestep of 2 fs and a secondary timestep of 4 fs for long-range interactions.
The systems were simulated for 15 ns with Cα atoms restrained with a force constant of 1 kcal/mol/Å, followed by 400 ns with no restraints except 10 pseudobonds between the intracellular ends of the five nAChR M4 helices to mimic the missing vestibule domain.
Acknowledgments
This work was supported by the National Institutes of Health through Grant GM055876 and by the National Science Foundation through TeraGrid resources provided by the National Institute for Computational Sciences.
Supplementary Material
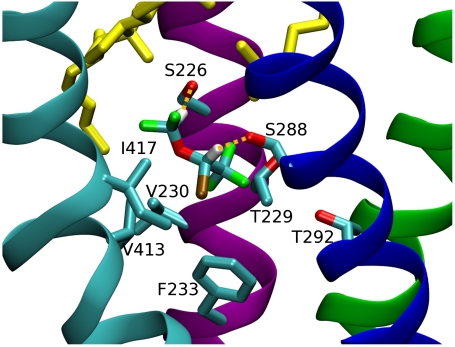
Fig. 4.
Sample frame showing isoflurane bound to the center of the TMD of the αδ subunit. M1 is purple, M2 is green, M3 is blue, and M4 is cyan; cholesterol is yellow, isoflurane is colored by atom type. Hydrogen bonds are represented by yellow dashed lines.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008534107/-/DCSupplemental.
References
- 1.Krasowski MD, Harrison NL. General anaesthetic actions on ligand-gated ion channels. Cell Mol Life Sci. 1999;55:1278–1303. doi: 10.1007/s000180050371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller KW. The nature of sites of general anaesthetic action. Br J Anaesth. 2002;89:17–31. doi: 10.1093/bja/aef167. [DOI] [PubMed] [Google Scholar]
- 3.Hemmings HC, et al. Emerging molecular mechanisms of general anesthetic action. Trends Pharmacol Sci. 2005;26:503–510. doi: 10.1016/j.tips.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Weng Y, Yang L, Corringer P-J, Sonner JM. Anesthetic sensitivity of the Gloebacter violaceus proton-gated ion channel. Anesth Analg. 2010;110:59–63. doi: 10.1213/ANE.0b013e3181c4bc69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks NP, Jenkins A, Conti E, Lieb WR, Brick P. Structural basis for the inhibition of firefly luciferase by a general anesthetic. Biophys J. 1998;75:2205–2211. doi: 10.1016/S0006-3495(98)77664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya AA, Curry S, Franks NP. Binding of the general anesthetics propofol and halothane to human serum albumin. High resolution crystal structures. J Biol Chem. 2000;275:38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 7.Liu R, Loll PJ, Eckenhoff RG. Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein. FASEB J. 2005;19:567–576. doi: 10.1096/fj.04-3171com. [DOI] [PubMed] [Google Scholar]
- 8.Vedula LS, et al. A unitary anesthetic binding site at high resolution. J Biol Chem. 2009;284:24176–24184. doi: 10.1074/jbc.M109.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 10.Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 11.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 12.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 13.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Brannigan G, Hénin J, Law R, Eckenhoff R, Klein ML. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 2000;57:1577–1592. doi: 10.1007/PL00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nirthanan S, Garcia G, Chiara DC, Husain SS, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for tdbzl-etomidate, a photoreactive positive allosteric effector. J Biol Chem. 2008;283:22051–22062. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckenhoff RG. An inhalational anesthetic binding domain in the nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 1996;93:2807–2810. doi: 10.1073/pnas.93.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Seto T, Tang P, Firestone L. Nmr study of volatile anesthetic binding to nicotinic acetylcholine receptors. Biophys J. 2000;78:746–751. doi: 10.1016/S0006-3495(00)76632-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihic SJ, et al. Sites of alcohol and volatile anaesthetic action on gaba(a) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 20.Belelli D, Pistis M, Peters JA, Lambert JJ. The interaction of general anaesthetics and neurosteroids with gaba(a) and glycine receptors. Neurochem Int. 1999;34:447–452. doi: 10.1016/s0197-0186(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 21.Yamakura T, Bertaccini E, Trudell JR, Harris RA. Anesthetics and ion channels: Molecular models and sites of action. Annu Rev Pharmacol Toxicol. 2001;41:23–51. doi: 10.1146/annurev.pharmtox.41.1.23. [DOI] [PubMed] [Google Scholar]
- 22.Chiara DC, Dangott LJ, Eckenhoff RG, Cohen JB. Identification of nicotinic acetylcholine receptor amino acids photolabeled by the volatile anesthetic halothane. Biochemistry. 2003;42:13457–13467. doi: 10.1021/bi0351561. [DOI] [PubMed] [Google Scholar]
- 23.Schofield CM, Trudell JR, Harrison NL. Alanine-scanning mutagenesis in the signature disulfide loop of the glycine receptor alpha 1 subunit: Critical residues for activation and modulation. Biochemistry. 2004;43:10058–10063. doi: 10.1021/bi036159g. [DOI] [PubMed] [Google Scholar]
- 24.Li G-D, et al. Identification of a gabaa receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dilger JP, Brett RS, Lesko LA. Effects of isoflurane on acetylcholine receptor channels: 1. Single-channel currents. Mol Pharmacol. 1992;41:127–133. [PubMed] [Google Scholar]
- 26.Dilger JP, Brett RS, Mody HI. The effects of isoflurane on acetylcholine receptor channels: 2. Currents elicited by rapid perfusion of acetylcholine. Mol Pharmacol. 1993;44:1056–1063. [PubMed] [Google Scholar]
- 27.Dilger JP, Vidal AM, Mody HI, Liu Y. Evidence for direct actions of general anesthetics on an ion channel protein. A new look at a unified mechanism of action. Anesthesiology. 1994;81:431–442. doi: 10.1097/00000542-199408000-00022. [DOI] [PubMed] [Google Scholar]
- 28.Forman SA, Miller KW, Yellen G. A discrete site for general anesthetics on a postsynaptic receptor. Mol Pharmacol. 1995;48:574–581. [PubMed] [Google Scholar]
- 29.Wenningmann I, Barann M, Vidal AM, Dilger JP. The effects of isoflurane on acetylcholine receptor channels: 3. Effects of conservative polar-to-nonpolar mutations within the channel pore. Mol Pharmacol. 2001;60:584–594. [PubMed] [Google Scholar]
- 30.Liu LT, Willenbring D, Xu Y, Tang P. General anesthetic binding to neuronal α4β2 nicotinic acetylcholine receptor and its effect on global dynamics. J Phys Chem B. 2009;113:12581–12589. doi: 10.1021/jp9039513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vemparala S, Saiz L, Eckenhoff RG, Klein ML. Partitioning of anesthetics into a lipid bilayer and their interaction with membrane-bound peptide bundles. Biophys J. 2006;91:2815–2825. doi: 10.1529/biophysj.106.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vemparala S, Domene C, Klein ML. Interaction of anesthetics with open and closed conformations of a potassium channel studied via molecular dynamics and normal mode analysis. Biophys J. 2008;94:4260–4269. doi: 10.1529/biophysj.107.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hénin J, Brannigan G, Dailey WP, Eckenhoff R, Klein ML. An atomistic model for simulations of the general anesthetic isoflurane. J Phys Chem B. 2010;114:604–612. doi: 10.1021/jp9088035. [DOI] [PubMed] [Google Scholar]
- 34.Bali M, Jansen M, Akabas MH. Gaba-induced intersubunit conformational movement in the gabaa receptor alpha 1m1-beta 2m3 transmembrane subunit interface: Experimental basis for homology modeling of an intravenous anesthetic binding site. J Neurosci. 2009;29:3083–3092. doi: 10.1523/JNEUROSCI.6090-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu LT, Haddadian EJ, Willenbring D, Xu Y, Tang P. Higher susceptibility to halothane modulation in open- than in closed-channel alpha4beta2 nachr revealed by molecular dynamics simulations. J Phys Chem B. 2010;114:626–632. doi: 10.1021/jp908944e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen M, Akabas MH. State-dependent cross-linking of the M2 and M3 segments: Functional basis for the alignment of GABAA and acetylcholine receptor M3 segments. J Neurosci. 2006;26:4492–4499. doi: 10.1523/JNEUROSCI.0224-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiser A, Sali A. Modloop: Automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:27–28,. 33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Law RJ, Henchman RH, McCammon JA. A gating mechanism proposed from a simulation of a human α7 nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 41.MacKerell AD, Feig M, Brooks CL. Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 42.Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 43.Cournia Z, Smith JC, Ullmann GM. A molecular mechanics force field for biologically important sterols. J Comput Chem. 2005;26:1383–1399. doi: 10.1002/jcc.20277. [DOI] [PubMed] [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with namd. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.