Abstract
Evolutionary relationships may exist among very diverse groups of proteins even though they perform different functions and display little sequence similarity. The tailed bacteriophages present a uniquely amenable system for identifying such groups because of their huge diversity yet conserved genome structures. In this work, we used structural, functional, and genomic context comparisons to conclude that the head–tail connector protein and tail tube protein of bacteriophage λ diverged from a common ancestral protein. Further comparisons of tertiary and quaternary structures indicate that the baseplate hub and tail terminator proteins of bacteriophage may also be part of this same family. We propose that all of these proteins evolved from a single ancestral tail tube protein fold, and that gene duplication followed by differentiation led to the specialized roles of these proteins seen in bacteriophages today. Although this type of evolutionary mechanism has been proposed for other systems, our work provides an evolutionary mechanism for a group of proteins with different functions that bear no sequence similarity. Our data also indicate that the addition of a structural element at the N terminus of the λ head–tail connector protein endows it with a distinctive protein interaction capability compared with many of its putative homologues.
Keywords: macromolecular assembly, protein evolution, unstructured protein
As the proteins existing in nature arose through diversification from a small primordial set, many evolutionarily related groups of proteins must exist that no longer share a common function or detectable sequence identity. However, the identification of such related groups is challenging because sequence similarity is the major criterion for establishing evolutionary connections between proteins. Comparative analysis of bacteriophage (phage) genomes provides a unique opportunity for tracing distant evolutionary relationships. Phage proteins are tremendously diverse and many that are clearly related through evolution bear no sequence similarity. Nevertheless, the conserved genome organization among highly diverged phages allows functional and evolutionary connections to be made even in the absence of sequence similarity (1–3). Furthermore, the sequences of tens of thousands of phage and prophage proteins are present in the databases, providing a superb resource for bioinformatic and evolutionary studies. The advantages of phage-based investigations are exemplified by studies on phage Cro proteins, which provided a description of one of the few clearly documented cases of protein fold evolution (4). In the work presented here, we investigated two phage λ virion proteins that bear no detectable sequence similarity and perform different functions, yet possess the same fold. This structural similarity prompted us to address the question of whether these proteins arose from a common ancestral protein.
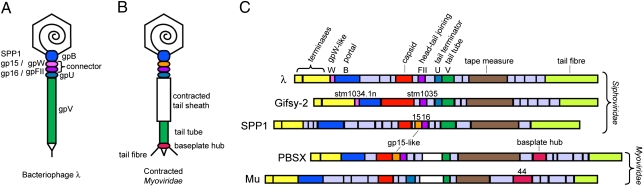
Phage λ is a member of a large and diverse group of viruses known as the Siphoviridae. These viruses possess a dsDNA genome encased within an icosahedral head that is attached to a long, noncontractile tail. The head and tail are attached to one another at a unique vertex of the head by a complex known as the connector (Fig. 1A). Upon infection, DNA exits the head through the connector and passes down the tail into the cell. The portion of the connector that is inserted into the head is composed of a dodecameric ring of the product of gene B (gpB), also known as the portal protein. The bottom surface of the connector (Fig. 1A), which interacts with the tail, is composed of gpFII (5). Another protein, gpW, is required for the stabilization of the DNA within the head and for the addition of gpFII (6, 7), suggesting that it may be positioned in the connector between gpB and gpFII. Bacillus subtilis phage SPP1 gp16, a protein with the same structure, function, and genomic position as gpFII (2) (Fig. 1 A and C), has been shown by cryoelectron microscopy (cryoEM) to form a 12-membered ring within the connector (8, 9). Although the number of molecules of gpFII in assembled phage particles has been estimated at 5 to 10 (5), their arrangement within the connector has not been determined. The structure of gpFII is also similar to XkdH from B. subtilis prophage PBSX (2, 9), and the genome position of the gene encoding this protein suggests that it performs the same function as gpFII (2). Hundreds of homologues of gpFII, SPP1 gp16, and XkdH have been identified in diverse phage and prophage genomes, indicating they are members of a large conserved family of connector proteins (2).
Fig. 1.
(A) Schematic diagram of bacteriophage λ. The λ connector is composed of rings of proteins gpW and gpFII whereas the tail tube is composed of rings of gpV, and capped by gpU. In bacteriophage SPP1, the protein rings of gpW and gpFII are occupied by gp15 and gp16, respectively (indicated beside the λ proteins). (B) Schematic diagram of a contracted Myovirus. (C) Genome maps of bacteriophage discussed in this study. Groups of genes that encode proteins of similar function are colored the same. Genes that are colored gray are unrelated. The genes encoding proteins discussed in this study are named.
The tail tube of phage λ is composed primarily of gpV. From a monomeric unassembled form, gpV forms hexameric rings upon assembly that stack to form the tail tube (10). The tube is capped by a single hexameric ring of gpU, the tail terminator protein, which forms the interface for binding to the connector (11–13). We recently solved the structure of monomeric gpV and found that it possesses the same tertiary structure as a tail tube protein from a contractile-tailed phage that is unrelated by sequence. This observation combined with conservation of genomic position implied a common evolution for the tube proteins of contractile and noncontractile phage tails (3). Structural comparison indicated that this family of tail tube proteins also includes the proteins that comprise the tube of the bacterial type VI secretion system (3).
The work described in this article was motivated by our discovery that, despite their different functions, λ gpFII and gpV display the same fold. In addition, they both possess large unstructured regions at corresponding positions in their structures. As described later, we have used structural comparisons, mutagenesis, and functional studies to provide evidence that these proteins assemble in the same manner within phage particles, and use their unstructured regions for related purposes. We propose that gpFII, gpV, and other tail proteins with the same tertiary structure were all derived from a single primordial tail tube protein.
Results and Discussion
The λ Head-Tail Joining Protein gpFII Is Similar in Structure to the Tail Tube Protein gpV.
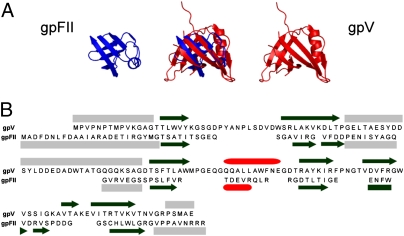
Comparison of the structures of gpV, gpFII,† and putative homologues of gpFII from phage SPP1 and prophage PBSX led to the surprising conclusion that the structure of gpV is very similar to those of these head–tail joining proteins (Fig. 2A). For example, the structure of gpV could be overlaid onto that of gpFII with an rmsd of 2.4 Å over 46 residues (gpFII has 68 structured residues in total; Fig. 2A). The secondary structure topology of these proteins is remarkably similar with equivalent connectivity, and gpV, gpFII, and gp16 of SPP1 possess unstructured regions in the same positions (Fig. 2B and Fig. S2B; see Fig. S3C for further data on protein overlays discussed here). Particularly noticeable are the large unstructured loops between the second and third strands of each structure (strands 2 and 3 of gpV, 2 and 2′ of gp16, and 3 and 4 of gpFII). Residues within these regions of gpV and gp16 have been shown to be crucial for function (3, 9).
Fig. 2.
Structural comparison of gpFII and gpV. (A) The structural overlay of gpFII and gpV is displayed in the center and the individual monomers are displayed on either side. These structures overlay with an rmsd of 2.4 Å over 46 residues. (B) The sequences of gpFII and gpV are aligned according to the structural alignment. The secondary structure of gpV is indicated above its sequence and that of gpFII is shown below its sequence (β-strands are represented as black arrows, α-helices as red cylinders, and unstructured regions as gray boxes). Unstructured regions of gpFII (Fig. S2A) and gpV (3) were delineated by measuring {1H}-15N heteronuclear NOEs.
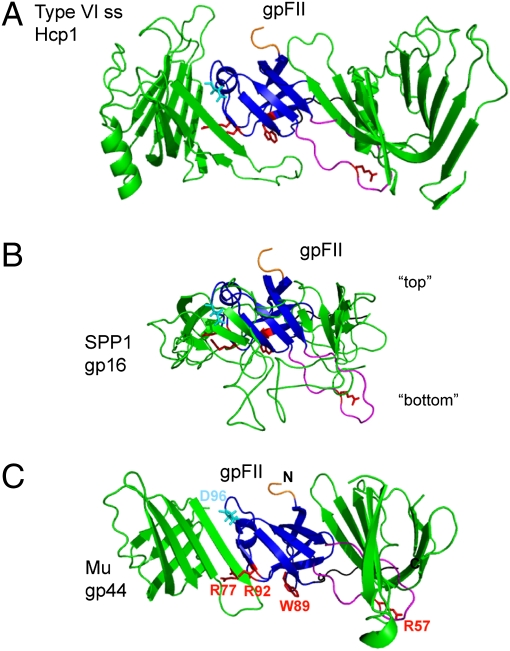
Our previous work indicated that gpV is structurally and evolutionarily related to Hcp1, which is believed to form the tube structure for the Type VI secretion system (3). As Hcp1 crystallized as a hexameric ring similar to the dimensions of the hexameric rings of gpV in the λ tail, we built a hexameric model of gpV using the Hcp1 structure as a guide. Both mutagenesis and bioinformatic data supported the validity of this model as an accurate representation of the arrangement of gpV within the λ tail (3). As the structure of gpFII is very similar to gpV, we could similarly overlay gpFII with Hcp1 (rmsd of 3.0 Å over 71 residues); thus, determining how gpFII would be oriented if it formed a ring similar to that formed by Hcp1 (Fig. 3A). In this model, the large N-terminal unstructured region (residues 1–24) of gpFII protrudes from one side of the ring whereas much of the central β3–β4 unstructured region (residues 46–62) is positioned on the opposite side.
Fig. 3.
Models of the arrangement of gpFII within phage particles. Models were made by structurally aligning gpFII to (A) Hcp1 in the hexameric form found in its crystal structure (PDB accession no. 1Y12), (B) the head–tail joining protein of phage SPP1 (gp16) as it was modeled into cryoEM density of the whole connector of SPP1, and (C) the phage Mu baseplate hub (gp44) pseudohexameric ring (PDB accession no. 1WRU). The view shown is of the β-strands that line the inside of these ring-like structures. In each view, gpFII was overlaid with the center monomer, which is not seen, and two flanking monomers are shown. The other half of each ring is not shown. Residues discussed in this work are highlighted on each model as follows: N terminus (truncated) in orange, β3–β4 unstructured region in magenta, C-terminal unstructured region in black, Asp96 in cyan (a reference point for orientation), and Arg57, Arg77, Trp89, and Arg92 in red. In B, “top” refers to the surface of this connector protein that binds to the head and “bottom” to the surface that binds tails.
Recently, a cryoEM-based pseudoatomic model was constructed of phage SPP1 gp16 as it is arranged within the connector of this phage (9). Overlaying gpFII with a single molecule of gp16 within this pseudoatomic model places gpFII in an orientation that is almost identical to that seen when it was overlaid with Hcp1 (Fig. 3B). From this overlay, we can predict that the surface of the putative gpFII ring from which the N-terminal unstructured region protrudes is the “top” surface, which would interact with the head, and the surface containing the β3–β4 unstructured region is the “bottom” surface, which would interact with the tail. These structural overlays imply that the orientation of gpFII and gp16 within the assembled phage head is the same as gpV when it is assembled into the tail. It should be noted that gp16 was modeled as a dodecamer in the SPP1 tail whereas gpV and Hcp1 form hexameric rings. The number of gpFII molecules in the assembled head is not known, but our proposed orientation of gpFII within the head could apply equally whether gpFII forms a hexamer or dodecamer.
Site-Directed Mutagenesis Experiments Support Our Model of gpFII Oligomerization.
To validate the predicted arrangement of gpFII subunits within the mature λ particle, we tested the functional properties of amino acid substitutions and deletions targeted to the putative top and bottom surfaces of gpFII (Fig. 3). The activity of these mutant proteins was assessed in vivo by measuring their ability to complement a nonsense mutation in the FII gene (FIIam) and in vitro by mixing purified proteins with an extract made by inducing a λ FIIam prophage. Changes in residues predicted to lie on the top or bottom surface of the gpFII ring caused large decreases in the activity of gpFII both in vitro and in vivo (Table 1). Importantly, all mutants bearing substitutions or deletions in positions at the bottom surface of gpFII also displayed a dominant-negative phenotype. For example, the plating efficiency of WT λ phage on cells expressing the R57E substitution was reduced by almost 100-fold. Even inactive mutants with most or the entire β3–β4 unstructured region deleted (Δ53–61 and Δ46–61) exhibited this strong dominant-negative effect. Deletions of the N-terminal unstructured region, which is predicted to lie on the top surface, caused no dominant-negative behavior. Furthermore, combining an N-terminal deletion with the Δ46–61 deletion resulted in no dominant-negative phenotype, implying that activity of the putative head-binding gpFII N terminus is required for imparting a dominant negative phenotype. The expression level and thermodynamic stability of all mutants was similar to WT (SI Methods).
Table 1.
Mutational analysis of the gpFII model
| Complementation | Effect on WT assembly | ||||
| Type | In vivo | In vitro | In vivo | In vitro (10:1) | Temperature melt, °C |
| WT gpFII | 1 | 1 | 1 | 1 | 52.8 |
| Top | |||||
| Δ1–9 | 6.4 × 10−3 | 1.1 × 10−2 | 1.2 | — | 61.1 |
| Δ1–24 | 1.1 × 10−3 | — | 1.3 | — | — |
| Bottom | |||||
| R57E | <10−6 | 1.2 × 10−2 | 1.1 × 10−2 | 7.8 × 10−2 | 59.0 |
| R77E | <10−6 | 1.2 × 10−5 | 1.6 × 10−1 | 5.4 × 10−3 | 58.7 |
| W89A | 2.0 × 10−5 | 6.4 × 10−2 | 3.9 × 10−1 | 3.7 × 10−2 | 56.7 |
| R92E | 2.5 × 10−4 | 4.5 × 10−2 | 1.4 × 10−2 | 1.3 × 10−1 | 59.1 |
| Δ53–61 | <10−6 | 2.5 × 10−5 | 8.3 × 10−3 | 8.9 × 10−5 | 61.4 |
| Δ46–61 | <10−6 | 1.3 × 10−5 | 9.2 × 10−3 | 3.3 × 10−3 | 54.1 |
| Δ115–117 | <10−6 | 2.1 × 10−3 | 2.4 × 10−2 | 2.2 × 10−4 | 60.3 |
| Δ109–117 | <10−6 | — | 1.0 × 10−2 | — | — |
| Δ1–9, D96R | 1.1 × 10−4 | — | 7.2 × 10−1 | — | — |
| Δ1–24, Δ46–61 | 4.9 × 10−4 | 1.1 × 10−5 | 1.1 | 1.1 | 54.5 |
| Δ1–24, Δ109–117 | 9.0 × 10−5 | — | 8.3 × 10−1 | — | — |
| (-) control | 1.0 × 10−6 | — | 1.5 | — | — |
| gpV 1–160 | 1 | — | 1 | — | — |
| gpV 1–154 | 3.0 × 10−3 | — | 3.0 × 10−3 | — | — |
All values represent the average of three experiments; the average SEs for the gpFII experiments in vivo and in vitro are 26% and 47%, respectively. Values for gpFII complementation experiments were performed with FIIam lysates and those for gpV complementation experiments with Vam lysates. Values for in vitro ratio given as test protein to WT protein.
We surmised that the dominant-negative phenotype of the bottom surface mutants could be a result of their competing with the WT protein for head-binding, and then preventing tail-binding after they have been incorporated into the head. Thus, we set out to further characterize this phenomenon. The dominant-negative effect of bottom surface mutants could be recapitulated in vitro by adding a 10-fold excess of mutant protein over WT into a gpFII activity assay. The addition of this large excess of mutant protein accentuated the dominant-negative phenotype of most mutants. In particular, the W89A and R77E mutants, which showed only mild dominant-negative phenotypes in vivo, were both approximately 10-fold more inhibitory in vitro. One of the most strongly dominant-negative mutants, Δ53–61, was titrated into a reaction containing a constant level of WT protein (Fig. S4). The inhibitory effect of this mutant was found to be strongly dose-dependent, and the curve of inhibition versus protein concentration could be fit by an exponential function assuming a simple competition between WT and mutant gpFII molecules. Fitting of these curves indicated that, if one third of the gpFII molecules incorporated into a phage were mutant, assembly would be inhibited. Further in vitro activity assays and EM experiments clearly showed that the Δ53–61 mutant inhibited phage assembly through its ability to bind heads, but not tails (SI Methods and Tables S2 and S3).
Taken together, the behavior of the gpFII mutants supports our model of the oligomeric structure of gpFII within phage particles. Mutants bearing amino acid substitutions or deletions on the putative bottom surface displayed dominant-negative phenotypes because they could still bind heads and thereby inhibit the assembly of phage even in the presence of WT gpFII. By contrast, deletion of the N-terminal region, which is expected to be involved in head-binding, caused loss of activity without a dominant-negative effect.
C-Terminal Truncations of Both gpFII and gpV Result in Dominant-Negative Phenotypes.
The phenotypes of deletions and amino acid substitutions in the β3–β4 unstructured region of gpFII parallel the dominant-negative phenotype for D61A/D62A substitution lying in the same region of gpV (3). This double mutant, when expressed concomitantly with the induction of WT λ prophage, caused the formation of truncated tails, indicating that it was able to oligomerize and incorporate into tails, but blocked the subsequent step of tail polymerization. Similarly, the gpFII β3–β4 unstructured region mutants are able to oligomerize and incorporate into heads, but are not able to bind tails.
Our previous study did not probe the function of the C-terminal unstructured region of gpV. Therefore, we investigated C-terminal truncations of both gpV and gpFII in an effort to uncover further functional congruence between these two proteins. GpV is a two-domain protein (14) and we have found that only residues 1 to 160 are necessary for tail tube formation, yet residues from position 149 onward are unstructured (3). As shown in Table 1, deletion of six unstructured residues from the N-terminal domain of gpV completely abolished its biological activity as measured by in vivo complementation of a λ Vam mutant phage. Similarly, truncations of part or all of the C-terminal unstructured residues of gpFII (Δ115–117 and Δ109–117) caused a complete loss of gpFII in vivo activity (Table 1). Interestingly, strong dominant-negative phenotypes were observed for the C-terminal truncations of both gpFII and gpV, indicating that these regions of both proteins perform similar functions. As was the case for deletions in the β3–β4 unstructured region of gpFII, further in vitro analysis and EM studies showed that the C-terminal truncation mutants of gpFII were able to bind to heads, but not to tails (SI Methods and Tables S2 and S3), implying that this region of gpFII also forms part of the tail-binding interface. Structural comparisons indicated that the unstructured C terminus of gpV may form a turn and another β-strand upon assembly into the phage tail (3). If the unstructured C-terminal region of gpFII underwent a similar rearrangement upon assembly, it would be brought to the bottom surface of the ring. This positioning would explain the dominant-negative phenotype of truncations to this region.
GpW Comprises the Middle Ring of the λ Connector.
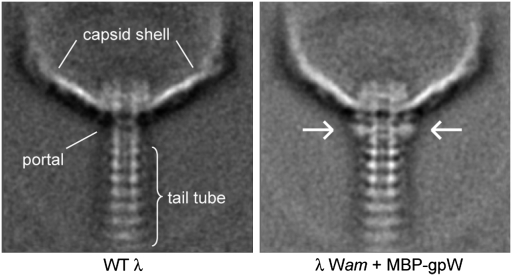
We previously showed that many phages possess an all helical protein, homologous to gp15 of SPP1 (Fig. 1 A and C), that comprises what we referred to as the “middle ring” of the connector (2). This middle ring lies between the portal protein and homologues of SPP1 gp16 that are positioned at the bottom of the connector (Fig. 1A). Although the structure and function of gpFII imply that it is a homologue of gp16, phage λ possesses no protein with any sequence or structural similarity to gp15 of SPP1. As the incorporation of λ gpW into the head is a prerequisite for gpFII addition (6), gpW has been assumed to occupy the middle position in the λ connector. However, experimental proof for this assumption has been lacking. To visualize gpW in assembled phage, we N-terminally tagged gpW, a 68-residue protein, with maltose-binding protein (MBP; 367 residues). This fusion protein was biologically active; thus, we expected it to be incorporated into phage and provide a detectable tag for the position of gpW. Comparison of electron micrographs of WT phage particles with those in which the MBP-gpW fusion protein had been incorporated showed a clear region of extra density at the position of the connector below the portal protein, gpB (Fig. 4). The symmetrical appearance of this density suggests that gpW is distributed evenly around the connector, probably in a ring-like structure. These data strongly support the conclusion that gpW forms the middle ring of the λ connector and that gpFII likely interacts with gpW in this region of the phage.
Fig. 4.
GpW is part of the connector. Averages are shown from images of negatively stained head–tail junction regions of WT λ particles (Left) and λ particles with MBP-gpW (Right) incorporated. The extra density corresponding to MBP, which shows where gpW is located, is indicated with arrows. Major structural elements are indicated on the WT image. These images were produced by averaging 280 and 300 images for the WT and MBP-gpW phage, respectively.
gpFII Unstructured N Terminus May Be an Evolutionary Adaptation for gpW Binding.
The localization of gpW within the λ connector presents a paradox because the structure of gpW is very different from gp15, the protein in SPP1 that comprises the middle ring of its connector (2, 9, 15). Thus, gpFII possesses a similar structure to gp16 of SPP1, but it must bind to a structurally divergent surface in the λ head comprised of gpW. The structure of gpFII is distinguished from the structures of its putative homologues, SPP1 gp16 and PBSX XkdH (Fig. 1C), in possessing a 24-residue unstructured region at its N terminus, whereas the commencement of defined secondary structure is preceded by only five or six residues in these other structures. As this unstructured region protrudes on the top surface of our oligomeric model of gpFII and the phenotype of its deletion was consistent with a role in head-binding (Table 1 and Tables S2 and S3), the presence of this region may partially account for the ability of gpFII to recognize the distinct structure of the λ head. Secondary structure prediction indicates a high probability of helix formation for this region, suggesting that it may become helical upon gpFII incorporation into the head. Supporting this idea, the crystal structure of STM1035, a homologue of gpFII (25% sequence identity; Figs. S5 and S6) encoded by the Gifsy-2 prophage of Salmonella, possesses a fully formed helix at its N terminus. We hypothesize that helices formed at the N-termini of gpFII and STM1035, which would be amphipathic in both proteins (Fig. S5), play a key role in the interaction with gpW incorporated into the head.
The importance of the helical N-terminal extension on gpFII is underscored by examining gpFII homologues. Through extensive iterative PSI-BLAST (16) searches, we identified greater than 150 proteins from phages and prophages that displayed significant sequence similarity to gpFII. An alignment of 38 diverse representatives of these sequences (Fig. S6) showed that each maintains an N-terminal region that displays similarity to the unstructured N-terminal region of gpFII and is predicted to be amphipathic and helical (Fig. S6). For each representative gpFII sequence (Fig. S6), it was possible to identify a protein with significant sequence similarity to gpW that was encoded in the same genomic position (i.e., between the end of the large terminase gene and the beginning of the portal gene; Fig. S7). These data indicate that the presence of a gpW homologue in a phage genome is correlated with the presence of the helical N-terminal extension on gpFII. No genome was found that contained both a homologue of gpW and a homologue of SPP1 gp15. In addition, no proteins were found with significant sequence similarity to both SPP1 gp16 and λ gpFII even though they perform the same function in their respective phages and are very likely to be homologues. Gp16 and gpFII appear to be distinct subfamilies of head–tail connector proteins evolved from the same primordial protein. The addition of the N-terminal extension to gpFII may be an evolutionary adaptation to allow for interaction with gpW (Fig. S5).
gpFII/gpV Fold Is Also Found in Baseplate Hub Proteins.
In contractile-tailed phages, the bottom of the tail is attached to a trimeric protein called the baseplate hub (Fig. 1B). The baseplate hub structures of phages T4 [gp27; Protein Data Bank (PDB) ID no. 1K28] and Mu (gp44; PDB ID 1WRU) are extremely similar even though sequence similarity cannot be detected between these proteins. The upper region of the hub structure, which interacts with the tail tube (17), forms a pseudohexameric ring because each monomer contains two copies of the same fold (Fig. 3C). It was previously observed that the hexameric structure of Hcp1, the putative type VI secretion system tube protein, is structurally similar to the pseudohexameric hub ring (18). We have found that gpFII can be well overlayed upon the structure of one domain of the hub ring (rmsds of 2.9 Å over 53 resudies and 2.9 Å over 57 residues for the N-terminal and C-terminal domains, respectively), and gpV can also be fit with similar statistics (Fig. S3C). Strikingly, superimposition of gpFII onto one subunit of the baseplate hub ring places gpFII in the same orientation as when it was superimposed into the gp16 or Hcp1 ring, generating a putative top and bottom surface that would involve the same regions as identified earlier (Fig. 3C). These data demonstrate that proteins positioned at the bottom of the tail tube, the tail tube protein itself, and the family of head–tail connector proteins lying at the top of the tail all possess both the same tertiary structure and the same quaternary structure when incorporated into phage. As the genes encoding these proteins are always positioned within the same vicinity of the phage genome, it is reasonable to hypothesize that these tail-associated structures could have arisen through the duplication of a gene encoding a primordial tail tube protein. It should be noted that there are currently no structures of the proteins lying at the base of the λ tail tube, so it is not known whether these proteins will adopt the tail tube fold. However, a recent publication has shown the presence of the tail tube fold in two different proteins present in the tip of a noncontractile tailed phage that infects the Gram-positive bacterium Lactococcus lactis (19).
gpFII/gpV Fold to the gpU Fold: Possible Case of Fold Evolution.
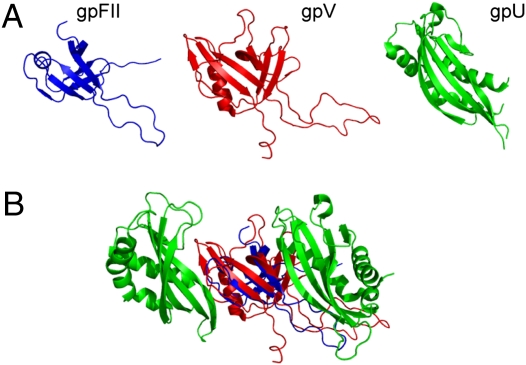
The λ tail is capped by a hexameric ring of gpU, which comprises the interface for binding to gpFII in the head. Although homologues of gpU are widely spread among contractile and noncontractile tailed phages, and its structure is conserved (13), the structure of gpU is different from that of gpFII and gpV, comprising a β-sheet packed against two large helical regions. Despite clear differences in the gpU structure, a structural similarity search using the structure of gpV as a query detected the structure of gpU with a significant score. An overlay of the structures of gpV and gpU shows that one β-sheet and part of one helix overlay very well, whereas the other β-sheet of gpV is mostly replaced in gpU by a long helix and unusual loop structure that is appended N-terminally to where the region of structural similarity commences (Fig. 5). Strikingly, the portion of these structures that is most similar is the sheet that forms the inside of the putative hexameric ring structure of gpV. As the structure of the biologically relevant hexameric form of gpU has been solved, it can be seen that this sheet also forms the inner surface of the gpU ring when it is assembled into phage particles (13).
Fig. 5.
Protein structure comparisons with the λ tail terminator protein, gpU. (A) gpFII and gpU were structurally aligned to gpV (rmsds of 2.4 Å over 46 residues and 4.5 Å over 72 residues, respectively) and the structures are displayed side by side in their aligned orientations. (B) gpFII (blue) and gpV (red) are positioned within the gpU hexameric ring structure (half the ring is shown) based on their structural superimposition upon the monomeric structure of gpU. The flanking subunits of gpU are shown (green) with gpFII and gpV in the center. This view shows the inside of the gpU ring from with the same orientation as the rings shown in Fig. 3.
Although it cannot be superimposed over as many amino acid positions, gpFII possesses the same regions of structural similarity to gpU as does gpV. When gpFII is overlaid onto one monomer of the gpU hexameric ring (rmsd of 3.1 Å over 45 residues), the gpFII orientation is the same as was seen in the overlays with the gp16 and Hcp1 rings. Once again, the 46–62 loop, Arg77, Trp89, and Arg92 are on the bottom surface and the N-terminal extension is on the top surface. This bottom surface of gpFII is in the same position as the surface of the gpU ring that was shown to interact with the top of the tail tube (13). Thus, the “bottom” of the gpU ring is the same surface as would form the bottom surface of our proposed gpFII oligomer. Remarkably, the solution structure of the monomeric form of gpU determined by NMR spectroscopy indicated that the 17-residue loop between strands 2 and 3, which protrudes from the bottom of the gpU ring structure, is disordered (20). This loop changes structure dramatically when gpU hexamerizes and a single amino acid substitution in this loop abrogates the tail-binding activity of gpU (13). Thus, just as in gpFII, a large disordered loop in the same topological position (Fig. 5A) plays a crucial functional role on the bottom surface of gpU.
Although the structural and functional similarities between gpU and gpFII/gpV could be coincidental, it is also possible that the gpU fold evolved from the primordial tail tube fold. Most observed additions and removals of protein domains following gene duplication occur at protein termini (21). Thus, the addition of an N-terminal helix to an ancestral tail tube protein to make a “gpFII” protein, or addition of an N-terminal helix and loop structure to make a “gpU” protein are both feasible mechanisms by which these proteins may have evolved new functions. There are several cases in which an evolutionary link has been implied between proteins with different folds (22, 23), and fold evolution has clearly occurred in the case of the phage Cro proteins, some of which adopt an all-helical fold, whereas others adopt a mixed helical and β-sheet fold (4, 24). As in the case of gpU, the divergent Cro structures overlay very well in one region, but the secondary structures diverge in another.
Conclusions
GpFII, the head–tail connector protein of phage λ, and its tail tube protein, gpV, possess the same tertiary fold and display functionally important unstructured regions in the same positions. Structural modeling combined with analysis of the dominant-negative behaviors of gpFII and gpV mutants provide strong evidence that gpFII and gpV adopt the same quaternary structure when they are incorporated into phage particles. Furthermore, the tertiary structures of both gpFII and gpV match the subunit fold of the pseudohexameric ring of baseplate hub proteins, which are found at the bottom of the tail tube in contractile-tailed phages. Given the proximity of the genes encoding these proteins in typical phage genomes, we propose that all of these proteins evolved from a single ancestral tail tube protein, and that gene duplication followed by differentiation led to the specialized roles of these proteins seen in phages today. This evolutionary mechanism has been proposed to occur in many systems and is believed to be a common means by which proteins evolve (22). The example of this mechanism presented by our work is striking because we have been able to present an evolutionary mechanism for a group of proteins with different functions that bear no sequence similarity. It could be argued that the similarities observed among these proteins are a result of convergent evolution. However, there are many different proteins with diverse structures that form rings of similar proportion to the tail; thus, it seems unlikely that three different proteins involved in forming the phage tail tube would have converged on the same structure by coincidence.
A further important conclusion of our work is that SPP1 gp16 and λ gpFII, although possessing the same fold and performing the same function, use their common structures in different fashions. A cryoEM study on the SPP1 connector suggested that the large unstructured β2–β2′ loop of gp16 (analogous to β3–β4 of gpFII) is involved in plugging the hole in the connector and preventing premature DNA egress from the head, whereas a long unstructured β1–β2 loop extends downward and may interact with the tail. Conversely, the β3–β4 unstructured loop of gpFII is used for tail binding and only a short loop is present between strands 1 and 2. As gpW of λ is known to fulfill the function of stabilizing packaged DNA within the head (6), gpFII need not perform this role, which may explain its alternate use of the β3–β4 loop. GpFII also differs from gp16 and its homologues by the addition of a long unstructured N-terminal region. We propose that this region becomes helical upon gpFII assembly into the head and that it forms part of the surface that interacts with gpW. The conserved nature of the gpFII N-terminal extension among proteins related by sequence and its correlated occurrence with gpW homologues in phage and prophage genomes suggests that the addition of this region was a key evolutionary step required for gpFII to gain the ability to bind gpW.
Our work proposes that gene duplication and addition of extra structural elements onto a fold can provide a mechanism for the evolution of a complex structure like the phage tail. As examples of protein fold evolution are very difficult to identify and prove (25), further studies using the same approaches as we have used could lead to important progress in this field. We are confident that mechanisms similar to those described here may account for the evolution of many other large multiprotein complexes.
Methods
Protein Expression, Mutagenesis, and Functional Assays.
N-terminally 6-His–tagged gpFII, expressed from a pET15 (Novagen)–based vector, was purified by Ni-affinity chromatography and in vitro activity assays were performed as previously described (26). Site-directed mutations were created by the QuikChange (Stratagene) approach, and deletions were made by the PCR-based SOEing method (27). In vivo complementation assays using λFIIam or λVam phages were also performed as previously described (3, 26). Complementation was scored by counting plaques and comparing to cells carrying WT plasmids or empty vector controls. Dominant-negative phenotypes were observed by plating WT λ phage with cells expressing mutant proteins of interest.
Database Searches and Structural Comparisons.
Searches of the structural database and pairwise structural comparisons were performed with DaliLite (28) or FATCAT (29). Sequence database searches were performed using PSI-BLAST (16).
Supplementary Material
Acknowledgments
We thank Paul Sadowski for critical reading of the manuscript. This work was supported by Operating Grant MOP-77680 from the Canadian Institutes of Health Research (to A.R.D.). L.C. was supported by a Canada Graduate Scholarships (Masters) and Postgraduate Scholarship (Doctoral) from the Natural Sciences and Engineering Research Council of Canada, and an Ontario Graduate Scholarship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KX4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005822107/-/DCSupplemental.
†We noticed that one region of our NMR structure of gpFII (PDB accession no. 1K0H) was not well defined when originally solved. A more thorough analysis of the data has allowed us to refine this region, and this new structure has been deposited in the PDB with accession no. 2KX4. For a description of the new structure, see Fig. S1 and Table S1.
References
- 1.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardarelli L, et al. The crystal structure of bacteriophage HK97 gp6: Defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 3.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roessler CG, et al. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci USA. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casjens S. Bacteriophage lambda FII gene protein: Role in head assembly. J Mol Biol. 1974;90:1–20. doi: 10.1016/0022-2836(74)90252-6. [DOI] [PubMed] [Google Scholar]
- 6.Perucchetti R, Parris W, Becker A, Gold M. Late stages in bacteriophage lambda head morphogenesis: In vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology. 1988;165:103–114. doi: 10.1016/0042-6822(88)90663-0. [DOI] [PubMed] [Google Scholar]
- 7.Casjens S, Horn T, Kaiser AD. Head assembly steps controlled by genes F and W in bacteriophage lambda. J Mol Biol. 1972;64:551–563. doi: 10.1016/0022-2836(72)90082-4. [DOI] [PubMed] [Google Scholar]
- 8.Orlova EV, et al. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lhuillier S, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsura I. Morphogenesis of bacteriophage lambda tail. Polymorphism in the assembly of the major tail protein. J Mol Biol. 1976;107:307–326. doi: 10.1016/s0022-2836(76)80007-1. [DOI] [PubMed] [Google Scholar]
- 11.Katsura I, Tsugita A. Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail. Virology. 1977;76:129–145. doi: 10.1016/0042-6822(77)90290-2. [DOI] [PubMed] [Google Scholar]
- 12.Katsura I, Kühl PW. Morphogenesis of the tail of bacteriophage lambda. II. In vitro formation and properties of phage particles with extra long tails. Virology. 1975;63:238–251. doi: 10.1016/0042-6822(75)90388-8. [DOI] [PubMed] [Google Scholar]
- 13.Pell LG, et al. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol. 2009;389:938–951. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 14.Katsura I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J Mol Biol. 1981;146:493–512. doi: 10.1016/0022-2836(81)90044-9. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell KL, et al. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 18.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sciara G, et al. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci USA. 2010;107:6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmonds L, et al. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: Identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J Mol Biol. 2007;365:175–186. doi: 10.1016/j.jmb.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 21.Buljan M, Bateman A. The evolution of protein domain families. Biochem Soc Trans. 2009;37:751–755. doi: 10.1042/BST0370751. [DOI] [PubMed] [Google Scholar]
- 22.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Belogurov GA, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newlove T, Konieczka JH, Cordes MH. Secondary structure switching in Cro protein evolution. Structure. 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Davidson AR. A folding space odyssey. Proc Natl Acad Sci USA. 2008;105:2759–2760. doi: 10.1073/pnas.0800030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–1404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 27.Rapley R. The Nucleic Acid Protocols Handbook. Totowa, NJ: Humana Press; 2000. [Google Scholar]
- 28.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 29.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.