Abstract
Recent cryoEM density maps of Aβ42 fibrils obtained at low pH revealed two protofilaments winding around a hollow core raising the question if such tubular structures also exist at physiological pH. Based on the cryoEM measurements and on NMR data, we probe amyloid fibril organizations corresponding to the observed cryoEM density map. Our study demonstrates that the tubular Aβ42 fibril models exist at both acidic and physiological pH; however, the relative populations of the polymorphic models shift with pH. At acidic pH, the hollow core model exhibits higher population than the other models; at physiological pH, although it is less populated compared to the other models, structurally, it is stable and represents 8% of the population. We observe that only models with C termini facing the external surface of the fibril retain the hollow core under acidic and physiological conditions with dimensions similar to those observed by cryoEM; on the other hand, the hydrophobic effect shrinks the tubular cavity in the alternative organization. The existence of the hollow core fibril at physiological pH emphasizes the need to examine toxic effects of minor oligomeric species with unique organizations.
Keywords: amyloid fibril structures, polymorphism, self-assembly
Alzheimer’s disease (AD) is widespread with large social impact. For more than a century, the accumulation of insoluble Aβ40/Aβ42 fibrils in the brain has been recognized as a pathognomic AD feature and its eventual end-stage product. To understand fibril assembly pathways, considerable research has focused on structural characterization of the oligomers and fibrils. X-ray fiber diffraction revealed that Aβ fibrils have a cross-β structure, with backbone hydrogen bonds between β-strands aligned in parallel, perpendicular to the main fibril axis (1–3). Although this arrangement is universal, Aβ fibrils are highly polymorphic due to different packing in the three-dimensional structures. Experimentally, different approaches under different environmental conditions obtained distinct Aβ40/Aβ42 fibril morphologies (4–12). Among the fibril forms, a recent cryoEM study revealed a unique structure of Aβ42 fibrils with a tubular shape and a hollow core (8). The tubular structure was incubated under acidic conditions (pH ∼ 2.0), and the question arises whether such structure is relevant at physiological pH. If so, it illustrates the range of oligomeric polymorphic forms that have to be considered when addressing Aβ toxicity.
To probe molecular interactions that could lead to an amyloid fibril that would correspond to the cryoEM density map (8), we constructed various tubular Aβ42 fibril-like models based on the cryoEM measurements (8) and on NMR data (4). Fitting structures of Aβ42 based on NMR (4) to cryoEM density maps is a reasonable strategy: Both are experimental data with well-correlated dimensions (SI Text). The constructed models are subjected to explicit solvent molecular dynamics simulations to test their structural stability, and their relative thermodynamic energies are subsequently evaluated. Our simulations cover the pH range from neutral to acidic (pH ∼ 3–7). Through extensive search, we found that if the C-terminal faces the inner fibril surface, hydrophobic interactions lead to collapsed stable structures without a hollow core. On the other hand, a stable structure with a hollow core can be obtained only when the C-terminal faces outside and forms hydrophobic interactions between the N termini and the loop regions that stabilize the hollow core structure. Fig. 1 demonstrates the fitting of our constructed model to the cryoEM density map. The relative stability and population of the hollow core structure increase at low pH, providing an explanation for the cryoEM measurements at low pH (8). Nonetheless, at neutral pH, although the hollow core structure is less populated, structurally it is stable, indicating its existence also at physiological pH. Because the hollow core structure is less populated at physiological pH, it is expected that it would be more difficult to detect in experiments under these conditions.
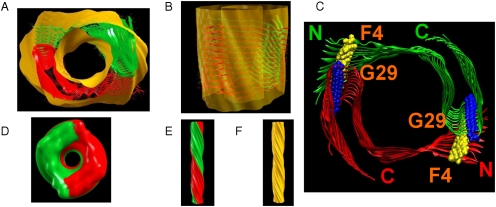
Fig. 1.
The fitting of model M1 to the cryoEM density map (ref. 8) is demonstrated. The experimental cryoEM density map (yellow) was generated from the PNAS program (SI Text). Top view (A) and side view (B): Coordinates for Aβ17–42 were extracted from ref. 4 (PDB ID code 2BEG), and fragments 1–16 were linked to the L17 of each monomer by forming intermolecular hydrophobic interactions between F4 (yellow) in the N termini of one 12-mer and G29 (blue) in the loop regions of second 12-mer (C). Top view (D) and side view (E) of the twofold axial symmetry of the simulated cryoEM map of model M1 at resolution of 10 Å, generated by the PNAS program. The continuous helix of the simulated cryoEM map of model M1 (E) is in agreement with the experimental cryoEM map (F).
Results
Experiment-Based Aβ42 Fibril Models Construction.
Recently, based on cryoEM measurements Zhang et al. (8) suggested that Aβ42 fibrils can have a tubular-like shape with a hollow core. They further proposed that Aβ42 modeled based on the Aβ17–42 coordinates (PDB ID code 2BEG) (4), can be arranged to fit the tubular density obtained from the cryoEM density map. Considering these cryoEM (8) density maps and using these NMR-based coordinates of Aβ17–42 (4) (SI Text) we constructed ten 24-mer tubular polymorphic models (Table S1, Fig. 1, and Figs. S1–S3) with two 12-mer oligomers in a parallel arrangement for each model.
The number of possibilities to join the N termini of one oligomer to the loop regions in the second is very large. For tubular models, two possibilities exist for the C termini: one with the C-terminal of each Aβ monomer facing the external surface of the fibril and the second when it faces the internal surface of the fibril cavity. To examine both possibilities, we constructed four tubular models, M1, M7, M8, and M10 with the C termini of the Aβ monomers facing the external surface of the fibril, and six tubular models M2, M3–M6, and M9 arranged with the C termini facing the internal surface of the fibril. Within each category the differences relate to different interactions between the N termini and the loops.
M1–M6 models demonstrate hydrophobic interactions between the N termini and the loops. Model M1 (Fig. 1C and Fig. S1) presents contacts between F4 in the flexible N termini of one 12-mer and G29 in the loop regions of the second 12-mer. Each monomer was rotated 5° along its fibril growth axis to obtain the tubular 24-mer. Models M2 and M3 (Fig. S1) were constructed by forming interactions between F4 in the N termini of one 12-mer and A30 in the loop region of the second 12-mer. Models M2 and M3 differ in the distances between the C termini of the two 12-mers. M4 has similar structure and interactions between the N termini and the loop region as M2 and M3, with additional F4-I31 hydrophobic interactions in this domain (Fig. S1). Models M5 and M6 present hydrophobic interactions between the N termini and the loops: F4 and I31 for M5, and A2 and I31 for model M6 (Fig. S2).
Considering possible nonhydrophobic interactions between the flexible N termini and the loop region, we constructed four models to test various interactions: M7 (Fig. S2) and M8 (Fig. S3) with D1 and N27 interactions and M9 and M10 (Fig. S3) to test side-chain interactions (D1 and I31 and D1 and G29 interactions, respectively). Each monomer in M8 was rotated 10° along its fibril growth axis, whereas in M7 the monomers were not rotated. Monomers in M9 and M10 were also rotated 10° along the fibril axis.
Tubular Structural Stabilities of Aβ42 Models at Physiological and Acidic pH.
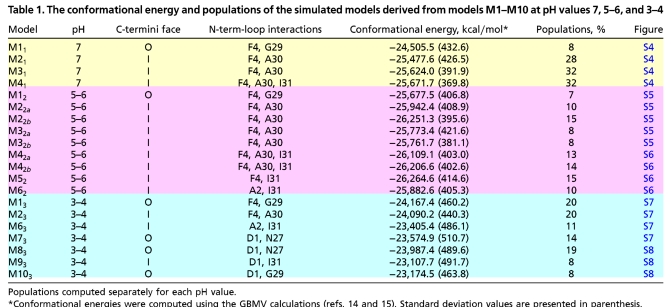
We simulated the tubular models M1–M10 at different pH values to analyze their stability and fit to the cryoEM density map (8). Table 1 and Table S2 summarize variants derived from models M1–M10. Four variants were examined at pH = 7: M11, M21, M31, and M41 (derived from models M1, M2, M3, and M4, respectively) (Fig. S4). All four reveal that intermolecular hydrophobic interactions between the N termini of one 12-mer and the loop regions of the second 12-mer exist along the simulations. Variants M21, M31, and M41 collapsed during the simulations, forming strong hydrophobic interactions in the internal surface of the fibril, between V39 of one 12-mer and A42 of the second 12-mer. In contrast, M11 demonstrates a stable hollow core, fitting the cryoEM density map (8). Apart from the differences in the hydrophobic interactions between the N termini and the loops, the main difference between M11 and M21, M31 and M41 is in the orientation of the ends of the C termini. In M21, M31, and M41, the ends of the C termini are protected in the internal core of the fibril, whereas in M11 the ends of the C termini face the external fibril surface. Previously (8), it was suggested that the C termini face the internal surface of the fibril; however, our models M21, M31, and M41 revealed that this morphology results in strong intermolecular hydrophobic interactions between the two ends of the C termini of the two 12-mers and the hollow core does not appear to be sustainable.
Table 1.
The conformational energy and populations of the simulated models derived from models M1–M10 at pH values 7, 5–6, and 3–4
To examine the effect of acidic pH on the models, we simulated nine models at pH = 5–6: M12 (derived from M1), M22a and M22b (derived from M2), M32a and M32b (derived from M3) (Fig. S5), M42a and M42b (derived from M4), M52 (derived from M5), and M62 (derived from M6) (Fig. S6). M2a and M2b, M3a and M3b, and M4a and M4b differ in the initial simulation times with the geometrical constraints (Table S2). We further simulated seven variants at pH = 3–4: M13 (derived from M1), M23 (derived from M2), M63 (derived from M6), M73 (derived from M7) (Fig. S7), M83 (derived from M8), M93 (derived from M9), and M103 (derived from M10) (Fig. S8). The hydrophobic interactions between the N-termini-loop regions in both M12 and M13 remain stable, and intermolecular hydrophobic interactions in the internal surface of the fibril are not observed along the simulations. Overall, the structural features of variants derived from model M1 that fit well the cryoEM density map are not affected by pH. However, as in physiological pH, variants derived from M2–M10 collapsed and/or exhibit less N-termini-loop hydrophobic interactions at lower pH, e.g., M22b and M62 at pH = 5–6, and M23 and M63 at pH = 3–4. Furthermore, whereas at pH = 5–6 some variants demonstrate intermolecular hydrophobic interactions in the internal surface of the fibril, at pH = 3–4 only M23 illustrates such interactions. Consequently, acidic pH decreases both the N-termini-loop regions’ hydrophobic interactions and intermolecular hydrophobic interactions in the internal surface of the fibril. Typical examples of such a case are models M62 and M63, both derived from M6.
We further investigated the effect of pH on the tubular model M1 that fits the cryoEM density map. We compared the F4-G29 distances of the three simulated variants derived from M1 at different pH values: M11 at 7, M12 at 5–6, and M13 at 3–4. Fig. S9 illustrates three different F4-G29 distances for each model along the simulations: The first is between the N-termini and loop regions in the same plane of the initial model. The second distance is between the N termini of one plane and the loop regions, in a cross-first sequential plane. The third is between the N termini of one plane and the loop regions in a cross-second layer plane. Because the β-sheets and the N-termini fragments are initially twisted and continue to twist along the simulations, the F4-G29 distance at the same plane is relatively large for all three models. Whereas in acidic pH, this distance does not vary, at physiological pH this distance increases. The second distance is similar in all models (∼8 Å). Finally, the third distance F4-G29 slightly increases in acidic pH: for M11 the distance is ∼8 Å, for M12 it is ∼8.5 Å, and for M13 it is ∼9 Å. Interestingly, though the N termini fluctuate extensively during the simulations, the F4-G29 interactions hold the hollow core both at physiological and acidic pH. To further examine the structural stability of model M1 under different pH conditions, we followed the change in the number of hydrogen bonds between the β-sheets along the simulations for models M11, M12, and M13. Fig. S10 shows that all variants derived from M1 demonstrate stable backbone hydrogen bond interactions (SI Text).
Remarks on EM Reconstruction and Segmentation.
The atomic model of Aβ42 established in this study leads to two suggestions regarding helical EM reconstruction and the subsequent image analysis of a β-sheet filament. The construction of a 2-start helical Aβ42 filament from a single monomer structure requires a combination of two symmetry operations: the first is based on given helical parameters (azimuthal angle and axial rise) to specify how the monomers arrange in the protofilament, and the second applies a twofold axial symmetry to relate one protofilament to the other. Two possible axial symmetries can construct a seamless helical tube, i.e., where each monomer in the constructed tube has the exact same environment (interactions with neighboring monomers). In the earlier Aβ42 model (8) only an axial symmetry (C2 symmetry along the fibril axis) was applied in the helical EM reconstruction. Such C2 axial symmetry corresponds to the (2,0) symmetry notation specifying a helical tube (13). This suggests that the other possible (2,1) symmetry was not applied in the EM reconstruction process (8), although it is unclear if it would have led to a better fitting.
In order to align an atomic model into an EM map, the first step is to identify the regions in the density map that correspond to distinct components. This process is usually called EM map segmentation. Isosurfaces in different colors usually reflect the result of segmentation. Separation between two protofilaments, which are shown in two different colors, appears reasonable; on the other hand, an inherent continuous helical symmetry in the cryoEM map implies that the boundary between the Aβ42 monomers cannot be identified. The fitting of our model into cryoEM map further suggests that segmentation either through an interactive, manual method or via an automatic segmentation algorithm for a β-sheet filament might not give a correct separation.
Interpretation of the Density Map.
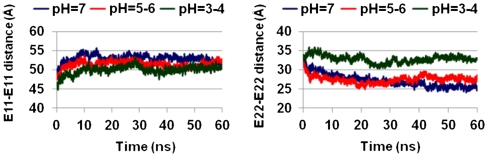
Model M1 appears compatible with the experimental density map (8). Fig. 1 demonstrates the fitting of model M1 to the cryoEM density map. The cross-section of the density region gives the dimensions of ∼40 Å × 20 Å. This size is in agreement with the cryoEM image (8). Fig. 1 D and E illustrate the top and side views of the simulated density map of M1 at 10-Å resolution, generated by our newly developed Protein Nanoscale Architecture by Symmetry (PNAS) program using only one monomer. Each protofibril was constructed with two helical parameters: unit twist of -4.0° and unit pitch of 4.9 Å, consistent with the experimental cryoEM density map. A twofold axial symmetry was then applied to generate the fibril. Fig. 1E presents a left-handed twist, which is consistent with the experimental density map (Fig. 1F). Our model M1 demonstrates two parallel oligomers winding together around a hollow core with a diameter of ∼50 Å between E11 of one oligomer and E11 of the second oligomer and with a diameter of ∼30 Å between E22 of one oligomer and E22 of second oligomer. Fig. 2 exhibits the simulated variants that derived from M1 at different protonation states: M11, at pH = 7, M12, at pH = 5–6, and M13 at pH = 3–4. To investigate the diameters in these variants, we computed these two distances along the simulations (Fig. 3). The E11-E11 distance is around 50 Å for all variants. The E22-E22 distance decreases by ∼5 Å for models M11 and M12; however, in M13 this distance increases by ∼5 Å. Consequently, the diameters hold for all variants and in particular in M13. The experimental tubular fibril was incubated under extreme conditions; however, the hollow core dimensions were not reported.
Fig. 2.
Top view of model M1 of the Aβ42 fibril at different pH values: (A) M11, at pH = 7, (B) M12, at pH = 5–6, and (C) M13, at pH = 3–4. First, the simulated models were averaged for the last 5 ns of the simulations, and then the average structures were generated from the PNAS program. The hollow core holds for all three models: Fig. 3 demonstrates the estimated dimensions of the hollow core.
Fig. 3.
The hollow core of models derived from model M1 holds both at neutral and low pH. The structural stability examined by following the averaged E11-E11 and E22-E22 distances along the simulations for models derived from model M1. M11 (pH = 7), M12 (pH = 5–6), and M13 (pH = 3–4) show slight changes.
Populations and Relative Stabilities for Aβ42 Tubular Models Are Affected by pH.
For the complex kinetics of amyloid formation, the 20 tubular models are likely to represent only a very small percentage among the ensemble. Nevertheless, the carefully selected models cover the most likely organizations. Based on Monte Carlo simulations with the energy landscape computed with the generalized Born method with molecular volume (GBMV) (14, 15) (SI Text), we estimated the overall populations and stability for each tubular conformer for each pH value (Table 1).
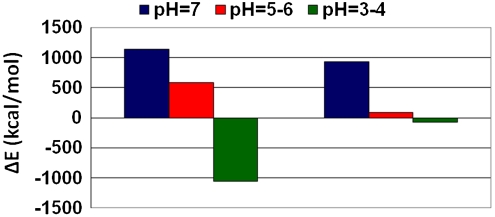
Based on the populations and the relative stabilities, we divide the 20 simulated models into two categories: the first includes models with the C termini facing outside that retain the organization and the dimensions in agreement with the experimental cryoEM density maps (8), among them M11, M12, and M13 (all derived from M1). The second includes the other 17 that do not fit the density map. At physiological pH, i.e., pH = 7, M11 is relatively less stable than other models with a similar protonation state (M21, M31, and M41) that demonstrate fairly high populations compared to M11. At acidic pH, i.e., pH = 5–6, M12 is relatively less stable compared to the other models with a similar protonation state, but the energy differences between M12 and the other models are smaller than at physiological pH, the population distribution is wider, and the differences decrease. At more acidic pH, i.e., at pH = 3–4, the “story” switches: M13 is relatively more stable than the other models with a similar protonation state, and it has the highest and similar populations as M23. Interestingly, at neutral pH, the three variants (M21, M31, and M41) have a significant preference over M11 that fits the cryoEM measurements. However, at low pH (pH = 5–6) M12 and other variants with a similar protonation state have similar preferences, and at acidic pH (pH = 3–4) M13 is preferred over the others (Fig. 4). The cryoEM density map (8) is observed at pH ∼ 2, and therefore our study provides a model that fits the cryoEM observations and insight as to why this model is observed at acidic pH and not at physiological pH. However, at physiological pH, although this model is not highly populated, it still represents ∼8% of the ensemble. Polymorphic behavior may also be seen by comparing the intermolecular interactions between the two 12-mers in the fibril at different pH values (SI Text).
Fig. 4.
Population of polymorphic states of Aβ42 is shifted by pH. The relative energy differences between models derived from M1 [M11 (simulated at pH = 7, blue), M12 (simulated at pH = 5–6, red), and M13 (simulated at pH = 3–4, green)] and the most and least stable models at different pH values: At pH = 7: E(M11) - E(M41) and E(M11) - E(M21). At pH = 5–6: E(M12) - E(M52) and E(M12) - E(M32b). At pH = 3–4: E(M13) - E(M23) and E(M13) - E(M93).
The stabilization of the hollow core structure at acidic pH could derive from the E22 neutralization. Water molecules interacting with backbone carbonyl groups may be trapped by β-sheets (16). Our calculations show that at pH = 7 and pH = 5–6, the average backbone atom solvation of A21 and E22 is higher than that of the nearby hydrophobic residues L17-F20. However, at lower pH, the protonation of E22 decreases the A21 and E22 backbone solvation, thus stabilizing the β-sheet interactions (Fig. S10).
Discussion
It is well documented that fibril morphology depends on environmental conditions, e.g., pH, temperature, and agitation (9, 17–21). However, even under the same conditions and within the same sample, variation of the fibril morphology may exist. The same Aβ segment may interact in various ways to form the basic cross-β structure leading to variations in the three-dimensional fibril organization. Experimentally, Petkova et al. (9) illustrated that distinct fibril morphologies at different pH reflect the variation in the structure of Aβ1–40 at the protofilament level. However, the effect of the pH on structural morphology at the detailed molecular level has not yet been investigated. Here, using all-atom molecular dynamics (MD) simulations in explicit solvent, we investigate polymorphic architectures of Aβ42 models with a tubular-like shape at physiological and acidic pH. All constructed models were based on the cryoEM density map (8) and the NMR-based coordinates (4). Overall, our study provides an insight into the molecular morphology of a recent cryoEM density map of a tubular-like shape of Aβ42 fibrils that form a hollow core (8). We further present other polymorphic tubular-like shape states of Aβ42, which do not form a hollow core and are the prevailing species at physiological pH.
To interpret the recent cryoEM density map of Aβ42, we examined two main possibilities of the arrangement of the C termini along the main fibril axis: C termini facing the internal fibril cavity and C termini facing the external surface of the fibril. Our simulations reveal that when the C termini face the external surface of the fibril, the hollow core exists along the simulations, whereas when the C termini face the internal surface of the fibril, the hydrophobic effect involving interactions between residues at the C termini leads to fibril collapse. Zhang et al. (8) performed a partial proteolysis assay to determine which of the residues are protected in the fibrillar structure. The observed cleavage sites for the Aβ monomer include L17, F19, F20, and A30 in the C-terminal segments and residues in the N-terminal segments. The pH of the buffer used in the assay (pH = 6.5 to pH = 7.5) differs from that of the fibril incubation (pH = 2.0), suggesting an altered amyloid landscape, with a different distribution of the polymorphic states. Cleavage at positions 20 and 30 was observed for nonhollow Aβ40 fibril (22). Correlation of polymorphic fibrils and proteolysis products is best illustrated by the observation that the N terminus in ∼20% of the peptides could be protected, and buried C terminus can be cleaved around Met35 by chymotrypsin (23). Thus, the cleavage data may be for the ensemble, rather than corresponding to the specific low pH state of fibril growth.
Two important observations emerge from our study. First, we demonstrate how pH values shift the population of the structural morphologies. At physiological pH, the model that fits the cryoEM density map is less populated, whereas other polymorphic states are more populated. At acidic pH, this model is more populated than the other polymorphic states. Our study offers an explanation why this model is observed in acidic pH and provides an interpretation to the density map. Moreover, our simulations further reveal that at physiological pH, the tubular hollow structure is also stable and exhibits nonnegligible population and, therefore, may be present also at physiological pH. Obviously, its population is low compared to other models, including those not tested here; nonetheless, it does exist in physiological pH. Second, the polymorphic tubular-like shape Aβ42 fibril models studied here reveal that the N-terminal of the Aβ peptide plays a crucial role in the protofilament assembly. So far, it was believed that the disordered N-terminal segment is not involved in fibril formation because of its high flexibility, although earlier studies suggested that the N-terminal segments may form β-sheet in parallel orientation (24, 25), which is also observed here (SI Text and Fig. S10). Our results show that the intermolecular interactions between the flexible N-terminal of one Aβ peptide and the loop region of a second stabilize the models; these interactions may also present polymorphic forms. We also note that in the “standard” fibril form, the charged states and N-terminal association could relate to fibril formation. Hori et al. (26) reported that the D7N and H6R N-terminal mutations enhance fibril elongation, but do not change protofibril formation.
To conclude, the amyloid landscape is well documented to present a broad range of aggregated conformational states (27), and the relative populations of the species are the outcome of conditions (4, 28): Different conditions will shift the landscape (4, 28, 29). Conformational ensembles can present monomer states with different turns (30) and parallel/antiparallel organizations (30). The availability of the cryoEM map of the tubular organization at low pH (8) allows a clear demonstration of the population shift of preexisting states as a function of conditions (4, 28) and emphasizes the role of the N-terminal in amyloid aggregation.
From the standpoint of toxicity, our conclusion of the existence of the hollow core fibril at physiological pH emphasizes that nonconventional polymorphic forms should also be considered in AD drug regimes. Currently, overwhelming data point to oligomers as the toxic species in AD, rather than fibrils. Our study highlighted the stability of the hollow core seed, which is expected to be the toxic species rather than the mature hollow core fibril that is the end product. This suggests that it is necessary to examine also the toxic effects of the minor oligomer forms with unique arrangements that differ from those seeding the dominant fibril forms. We expect that the polymorphic range is broad; the discovered species simply reflect higher population states under the experimental conditions (27, 29, 30).
Material and Methods
Determination/Estimation of pH Conditions.
To study the effect of the pH on the structural stability and the populations, we used different protonation states of titratable side chains to simulate constructed models at different pH values. For pH = 7, only the positive charged residues (Lys and Arg) were protonated. For pH = 5–6, all histidines were also protonated, whereas for pH = 3–4, all Lys, Arg, His, Glu, and Asp residues were protonated.
Protein Nanoscale Architecture by Symmetry Program.
We generated the cryoEM image of our constructed model M1 using the PNAS program (SI Text). The helical cryoEM reconstruction of our model was based on the C2 axial symmetry (13).
MD Simulations Procedure.
MD simulations of solvated Aβ42 oligomers were performed in NPT (N, number of particles; P, pressure; and T, temperature) ensemble using the NAMD program (31) with the CHARMM27 force field (32, 33) for 60 ns. The oligomers were explicitly solvated with TIP3P water box. The Langevin piston method with a decay period of 100 fs and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. The temperature (300 K) was controlled by Langevin thermostat with a damping coefficient of 10 ps-1 (31). The short-range van der Waals interactions were calculated using the switching function, with a twin range cutoff of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 12.0 Å for all simulations. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm (SI Text).
Analysis Details.
The fitting to the cryoEM density map and the simulated properties in comparison with the experimental measurements were performed by the PNAS program (SI Text).
To obtain the relative stability and populations of the tubular Aβ42 models, the Aβ trajectories of the last 5 ns were first extracted from the explicit MD simulation excluding water molecules. The solvation energies of all systems were calculated using the GBMV (14, 15) (SI Text).
Finally, for the structural stability analysis details of the Aβ42 oligomeric tubular M11, M12, and M13 variants, see SI Text.
Supplementary Material
Acknowledgments.
All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health (NIH), Bethesda, MD (http://biowulf.nih.gov). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004704107/-/DCSupplemental.
References
- 1.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 2.Kirschner DA, Abraham C, Selkoe DJ. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc Natl Acad Sci USA. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 4.Luhrs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachse C, Fandrich M, Grigorieff N. Paired beta-sheet structure of an Abeta(1-40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci USA. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Abeta(1-40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol. 2009;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt M, et al. Comparison of Alzheimer Abeta(1-40) and Abeta(1-42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci USA. 2009;106:19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R, et al. Interprotofilament interactions between Alzheimer’s Abeta1-42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci USA. 2009;106:4653–4658. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 10.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai CJ, Zheng J, Nussinov R. Designing a nanotube using naturally occurring protein building blocks. PLoS Comput Biol. 2006;2:e42. doi: 10.1371/journal.pcbi.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MS, Salsbury FR, Brooks CL. Novel generalized Born methods. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 15.Lee MS, Feig M, Salsbury FR, Brooks CL. New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J Comput Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 16.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in beta-amyloid fibrils. Proc Natl Acad Sci USA. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen JS, et al. The changing face of glucagon fibrillation: Structural polymorphism and conformational imprinting. J Mol Biol. 2006;355:501–523. doi: 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 18.Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–238. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 19.Makarava N, Baskakov IV. The same primary structure of the prion protein yields two distinct self-propagating states. J Biol Chem. 2008;283:15988–15996. doi: 10.1074/jbc.M800562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verel R, et al. Polymorphism in an amyloid-like fibril-forming model peptide. Angew Chem Int Edit. 2008;47:5842–5845. doi: 10.1002/anie.200800021. [DOI] [PubMed] [Google Scholar]
- 21.Klement K, et al. Effect of different salt ions on the propensity of aggregation and on the structure of Alzheimer’s A beta(1-40) amyloid fibrils. J Mol Biol. 2007;373:1321–1333. doi: 10.1016/j.jmb.2007.08.068. [DOI] [PubMed] [Google Scholar]
- 22.Yan P, et al. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J Biol Chem. 2006;281:24566–24574. doi: 10.1074/jbc.M602440200. [DOI] [PubMed] [Google Scholar]
- 23.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Abeta amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 24.Urbanc B, et al. In silico study of amyloid beta-protein folding and oligomerization. Proc Natl Acad Sci USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melquiond A, Dong X, Mousseau N, Derreumaux P. Role of the region 23-28 in Abeta fibril formation: insights from simulations of the monomers and dimers of Alzheimer’s peptides Abeta40 and Abeta42. Curr Alzheimer Res. 2008;5:244–250. doi: 10.2174/156720508784533330. [DOI] [PubMed] [Google Scholar]
- 26.Hori Y, et al. The Tottori (D7N) and English (H6R) familial Alzheimer disease mutations accelerate Abeta fibril formation without increasing protofibril formation. J Biol Chem. 2007;282:4916–4923. doi: 10.1074/jbc.M608220200. [DOI] [PubMed] [Google Scholar]
- 27.Miller Y, Ma B, Nussinov R. Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape. Chem Rev. 2010 doi: 10.1021/cr900377t. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: Dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller Y, Ma B, Nussinov R. Zinc ions promote Alzheimer Aβ aggregation via population shift of polymorphic states. Proc Natl Acad Sci USA. 2010;107:9490–9495. doi: 10.1073/pnas.0913114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller Y, Ma B, Nussinov R. Polymorphism of Alzheimer’s Abeta17-42 (p3) oligomers: The importance of the turn location and its conformation. Biophys J. 2009;97:1168–1177. doi: 10.1016/j.bpj.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kale L, et al. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- 32.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Brooks BR, et al. Charmm—a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.