Abstract
MicroRNAs influence hematopoietic differentiation, but little is known about their effects on the stem cell state. Here, we report that the microRNA processing enzyme Dicer is essential for stem cell persistence in vivo and a specific microRNA, miR-125a, controls the size of the stem cell population by regulating hematopoietic stem/progenitor cell (HSPC) apoptosis. Conditional deletion of Dicer revealed an absolute dependence for the multipotent HSPC population in a cell-autonomous manner, with increased HSPC apoptosis in mutant animals. An evolutionarily conserved microRNA cluster containing miR-99b, let-7e, and miR-125a was preferentially expressed in long-term hematopoietic stem cells. MicroRNA miR-125a alone was capable of increasing the number of hematopoietic stem cells in vivo by more than 8-fold. This result was accomplished through a differentiation stage-specific reduction of apoptosis in immature hematopoietic progenitors, possibly through targeting multiple proapoptotic genes. Bak1 was directly down-regulated by miR-125a and expression of a 3′UTR-less Bak1 blocked miR-125a-induced hematopoietic expansion in vivo. These data demonstrate cell-state-specific regulation by microRNA and identify a unique microRNA functioning to regulate the stem cell pool size.
Hematopoietic stem cells (HSC) self-renew and differentiate to form all blood cells throughout animal life. The intricate balance between these two characteristic stem cell states is required for maintaining hematopoietic homeostasis and responding to tissue injury. Stem cell population size is tightly regulated and thought to be dictated by rates of proliferation, relative frequency of differentiative versus self-renewal outcomes, and apoptosis. Disruption of any of these processes could lead to stem cell exhaustion or increased risk of leukemogenesis (1–5). However, the molecular events specifying stem cell population size are still poorly understood.
MicroRNAs are emerging as a class of important cellular regulators that mediate cell state, with specific patterns of microRNA expression demarcating developmental or differentiation stages (6–8). They are transcribed as longer primary microRNAs and their maturation is dependent on the RNase III enzyme, Dicer (9–13). In the blood system, multiple microRNAs have been found to direct differentiation, such as miR-181 for T cells (14), miR-150 for B cells (15, 16), and miR-223 for granulocytes (17–19). We have shown that miR-150, shunts megakaryocyte and erythrocyte common progenitors (MEP) toward megakaryocytes (20). To date, all known microRNAs reinforce specific lineage outcome and no specific microRNAs are known to regulate the number of heatopoietic stem/progenitor cells (HPSCs) in the hematopoietic system.
Results
Hematopoietic Ablation of Dicer Impaired the Hematopoietic Stem/Progenitor Compartment.
We hypothesized that microRNAs regulate HSCs and first evaluated this using a mouse with a conditional allele of the microRNA processing-enzyme Dicer (10, 13). Dicerlox/lox mice were bred with MxCre mice, which express the Cre recombinase in response to IFNs and can be experimentally induced with high efficiency in blood cells, including HSCs, via peritoneal injection of polyI:polyC (pIpC) (4, 5, 21). Mice with the genotypes of Cre+Dicerlox/lox (mutant) and Cre+Dicerwt/wt or Cre+Dicerlox/wt littermates (control) were used as we did not observe differences between Cre+Dicerwt/wt and Cre+Dicerlox/wt mice.
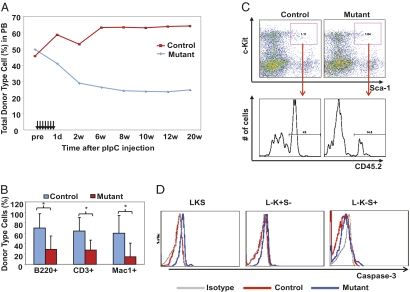
HSC alteration by Dicer loss was assessed by long-term repopulation, a definitive assay for HSCs. Whole bone marrow (BM) from control or mutant mice (CD45.2+) before pIpC treatment were mixed 1:1 with wild-type competitor BM (CD45.1+) and transplanted into lethally irradiated recipient mice (CD45.1+). Seven doses of pIpC were administered 5 wk after transplantation given every other day over a course of 13 d. The day of the last pIpC injection was counted as day 0. The contribution to T, B, and myeloid lineages in the peripheral blood was monitored over time (Fig. 1 A and B and Fig. S1). Although both mutant and control groups showed ∼50% overall donor-type (CD45.2+) reconstitution before pIpC injection, reconstitution by mutant marrow markedly declined after pIpC treatment, and remained reduced until 20 wk post-pIpC, when donor contribution was primarily stem cell-derived. The reduction in reconstitution by mutant BM could also be observed in secondary transplant recipients (Fig. S1C), underscoring the importance of Dicer in HSCs.
Fig. 1.
Dicer deletion abolishes functional and immuno-phenotypic HSPCs. (A) Peripheral blood chimerism by control and mutant BM in a 1:1 competitive transplantation assay. The seven arrows indicate pIpC injections. Each dot on the line indicates the average donor-type cell percentage (%CD45.2+) at the indicated time points (d, days; w, weeks after pIpC injection). n = 15. (B) Lineage contribution by donor-type cells 20 wk after pIpC injections. Lineages analyzed include myeloid (Mac-1+), B (B220+), and T cells (CD3+). Error bars indicate SD. *P < 0.05. (C) Representative FACS plot showing donor-type LKS cells in recipient BM 6 mo post-pIpC injections. (D) Intracellular flow cytometry for activated caspase-3 in three BM populations including the Lin-c−Kit+Sca+ (LKS), Lin-c−Kit+Sca− (L-K+S-), and Lin-c−Kit−Sca+ (L-K-S+) cells.
Two lines of evidence support the notion that HSPCs are impaired by Dicer loss, rather than the alternative possibility that the reduction in mutant-marrow repopulation capacity is completely because of impairing multiple independent committed lineages. First, mutant marrow gave reduced donor-cell contribution to the Lin−Kit+Sca+ (LKS) population (Fig. 1C), which paralleled in magnitude the reduction in committed cell types. It should be noted, however, that the HSPC immuno-phenotype was found to be modified by other manipulations that affect substrates of Dicer (Fig. S2C), and therefore LKS numbers may not accurately reflect primitive cell numbers in these experiments. On the other hand, we also observed a reduction of donor-cell contribution in the secondary transplant recipients (Fig. S1C), strongly suggesting HSPCs were reduced following Dicer deletion. Second, the decline of mutant BM reconstitution occurred progressively after pIpC treatment over a period of 8 to 10 wk (Fig. 1A). Importantly, the drop in myeloid reconstitution was evident as early as 1 d after the last dose of pIpC (13 d after initiation of pIpC), although the lymphoid lineages were still intact at this time (Fig. S1D). This finding is consistent with the difference in the turnover rate among these lineages, with the rapidly turning-over myeloid cells reflecting damage in the immature hematopoietic pools earlier than the longer-lived lymphoid populations. Although we regard these data as consistent with an effect on more primitive cells, we cannot exclude and do not assert that Dicer deletion does not have effects on lineage-committed cells.
Donor cells were present months after pIpC treatment, with largely normal lineage distribution (Fig S1A), raising the possibility that Dicer was not essential for HSPC function or that some HSCs had escaped Cre-mediated Dicer excision. To evaluate this, we sorted donor cells from the peripheral blood 6 mo posttransplant (Fig. S2A). Analyses of the genomic DNA indicate that all mice transplanted with control Cre+Dicerlox/wt cells showed complete deletion of the loxed alleles; however, in mice that received mutant Cre+Dicerlox/lox cells, the loxed allele (functionally wild-type allele) persisted (Fig. S2A, Upper). To test deletion at a clonal level, donor-type (CD45.2+) BM cells were sorted and plated into methylcellulose. No colonies could be identified with a DicerΔ/Δ genotype (0/34) (Fig. S2A, Lower). In contrast, the control Dicerlox/wt colonies all showed a DicerΔ/wt genotype (38/38). DicerΔ/Δ colonies were also completely absent from BM cells of unmanipulated mutant animals following pIpC injections, whereas control Dicerlox/wt BM colonies again displayed 100% deletion of the loxed allele (Fig. S2B). These data support that homozygous Dicer deletion is incompatible with a functional HSC state.
To evaluate the basis for cell loss, apoptosis was assayed immediately following pIpC treatment in otherwise unmanipulated mice. LKS cells, Lin−Kit+Sca− (L-K+S-) cells (containing myeloid progenitors) and another heterogeneous population Lin−Kit−Sca+ (L-K-S+) (22, 23) were examined for caspase-3 activation. Mutant LKS cells displayed a consistent and significant increase in apoptosis, whereas the L-K+S- myeloid progenitors and L-K-S+ population were less affected (Fig. 1D and Fig. S3A). Meanwhile, mutant BM demonstrated increased Ki67 staining, suggesting a compensatory response to cell loss (Fig. S3 B and C) following pIpC treatment. Similar results were observed when Dicer deletion was induced by IFN-β in FACS-purified HSPCs in vitro (Fig. S4).
The notion that Dicer loss causes apoptosis in HSPCs predicts that stem cell niches could be vacated to allow engraftment of exogenous stem cells. We tested this possibility by transplanting 5 × 106 wild-type BM (CD45.2+) mononuclear cells into control or mutant animals (CD45.1+) following seven doses of pIpC, but without irradiation (Fig. S3D). Indeed, minimal engraftment was observed in control mice, as would be expected from the few available stem cell niches at homeostasis. In contrast, robust CD45.2+ donor engraftment was observed in mutant mice, with contribution to both myeloid and lymphoid lineages 5 mo posttransplantation (Fig. S3D, red gate). These data further support HSPC impairment by Dicer loss and suggest HSC death caused by Dicer deficiency.
Taken together, the data above indicate that Dicer is necessary for HSPCs and its loss compromises HSPC function in a manner consistent with stem cell death.
A MicroRNA Cluster Preferentially Expressed in Long-Term HSCs.
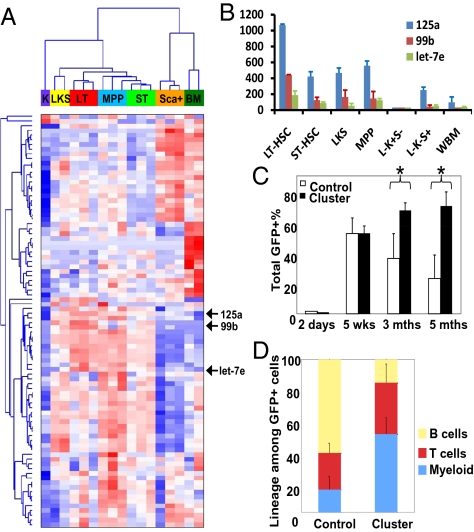
Because microRNAs are major substrates for Dicer and microRNAs specify cellular states, we hypothesized that specific microRNAs contribute to the functional maintenance of a HSC state, as defined by persistent self-renewal capability coupled with multilineage differentiation capacity. To identify such microRNAs, we performed microRNA expression-profiling in multiple stem cell and progenitor populations with well-defined markers, using a bead-based expression-analysis platform (6, 20) followed by RT-PCR validation (Fig. 2 A and B and Fig. S5A). Interestingly, hierarchical clustering indicates that microRNA expression profiles reflect self-renewal and multipotency, with the more primitive population clustered closely (Fig. 2A). Specifically, we found multiple microRNAs preferentially expressed in populations that self-renew (Table S1). Of particular interest, three microRNAs, miR-99b, let-7e, and miR-125a, were highly expressed in long-term HSCs compared with other populations (Fig. 2B and Fig. S5A). These three microRNAs display a complete evolutionary conservation among mammals and organize in a cluster spanning a ∼600-bp region on chromosome 19 in humans and 17 in mice (Fig. S5B).
Fig. 2.
MiR-99b-let-7e-miR-125a cluster expression enhances long-term multilineage reconstitution. (A) Heatmap of microRNA expression profiles of hematopoietic cells. Population designation: Sca+, Lin-c−Kit−Sca+; BM, whole BM; K, Lin-c−Kit+Sca-; LKS, Lin-c−Kit+Sca+; LT, Lin-c−Kit+Sca+CD34-Flk2-; MPP, Lin-c−Kit+Sca+CD34+Flk2+; and ST, Lin-c−Kit+Sca+CD34−Flk2+. Each column represents an independent sample. Populations were sorted from pooled BM cells from multiple animals on multiple days. Red color indicates higher expression; blue for lower. (B) Bar graph of data in A for miR-125a, miR-99b, and let-7e. (C) Primed wild-type donor marrow (5FU) was transduced with retrovirus expressing either control vector or miR-99b-let-7e-miR-125a (cluster) in addition to GFP. Contribution to peripheral blood by control or cluster-transduced BM cells (GFP+) at indicated time is shown. The first time-point indicates GFP+% in the culture 2 d posttransduction. wks: weeks; mths: months. *P < 0.05. (D) Multilineage differentiation into myeloid (Mac-1+), B (B220+), and T lineages (CD3+) among GFP+ cells 5 mo posttransplantation. C and D: n = 4–7 each. Error bars indicate SD.
We tested the function of the miR-99b-let-7e-miR-125a cluster in regulating HSCs using retroviral expression (vector also expresses GFP) followed by transplantation (20). BM cells expressing the microRNA cluster showed enhanced reconstitution in all major blood lineages after long-term transplantation (Fig. 2 C and D). In contrast, reconstitution by control vector-transduced cells declined over time (Fig. 2C). The increase in multilineage reconstitution by the miR-99b-let-7e-miR-125 cluster persisted in secondary transplantation (Fig. S6A), consistent with an effect on HSCs. This increase in reconstitution was not because of a difference in transduction efficiency of the cluster versus control vectors, as both were similar immediately posttransduction (Fig. 2C).
A Single MicroRNA, miR-125a, Augmented HSC Activity.
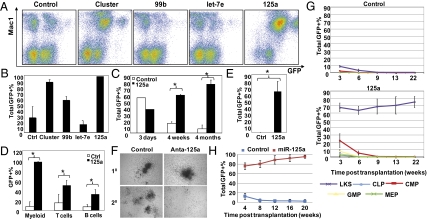
Individual microRNAs in the cluster were then analyzed. The microRNA miR-125a alone, but not miR-99b or let-7e, provided comparable increase in long-term multilineage reconstitution (Fig. 3 A–D). Remarkably, this enhancement appears to occur in the absence of a specialized BM microenvironment or niche, as 18-d ex vivo culture in the presence of miR-125a preserved robust stem cell activity, which was completely lost in the control cultures (Fig. 3E and Fig. S6B).
Fig. 3.
miR-125a enhances HSC function. The miR-99b-let-7e-miR-125a cluster or individual miRNA was transduced and transplanted as in Fig 2C. Peripheral blood contribution by transduced BM cells was analyzed 4 mo posttransplantation. (A) Representative FACS plots. (B) Quantification of data shown in A. (C) Comparison of contribution to blood formation by control or miR-125a alone (125a). The first time point (3 d) indicates GFP+% in the culture posttransduction. (D) Multilineage differentiation by transduced BM cells. For B to D, n = 1–5 per group per time point shown. (E) Contribution to blood formation by transduced BM cells that were cultured ex vivo for 18 d. n = 4 for control (Ctrl) and n = 5 for miR-125a (125a). *P < 0.05. (F) Serial methylcellulose colony formation in the presence of a control or miR-125a-specific antagomir. Representative fields from primary (10) and secondary (20) cultures. (G and H) Purified HSPCs were transduced with control or miR-125a and transplanted. Peripheral blood contribution in recipients was quantified. See SI Materials and Methods for population definition. Each animal received either (H) 100 SLAM, (G) 1,000 LKS, or 10,000 progenitors together with 2.5 × 105 supporting BM cells. n = 5 except for miR-125a–transduced CMP (n = 4). Error bars reflect SD.
We next sought to determine if endogenous miR-125a regulates primitive hematopoietic cell function. To this end, we used antagomir against miR-125a, a chemically synthesized cell-permeable microRNA inhibitor (24). This sequence-specific inhibitor of miR-125a did not affect methylcellulose colony morphology or numbers in primary cultures, but drastically reduced colony formation in subsequent serial replating cultures (Fig. 3F). Because this assay is an in vitro surrogate for self-renewal, these data are consistent with miR-125a regulation of HSPC self-renewal.
MicroRNA miR-125a Amplifies the HSC Pool Size.
To more precisely quantify the effect of miR-125a on stem cells, we performed the competitive limiting dilution assay. The need for this functional assay was substantiated by the observation that conventional stem cell surface markers were markedly changed with miR-125a ectopic expression (Fig. S2C), precluding accurate immuno-phenotypic enumeration. Limiting dilution-assay analysis was performed using total GFP+ BM cells from recipients after 4 to 5 mo in primary transplantation (Table 1). The number of myeloid cells predominated with a reduced proportion of lymphoid cells (Fig. 3D and Table S2). We scored both myeloid (Mac1+) and lymphoid lineages (B220+ or CD3+) in our limiting dilution assay with ≥1% as the cutoff. Although the apparent frequency of reconstituting HSCs varied between experiments (presumably because of different extents of HSC exhaustion under this experimental setting), we could consistently detect >8-fold expansion of the reconstituting HSC pool (Table 1).
Table 1.
Experimental scheme and positive responders in two independent limiting-dilution assay experiments
| Experiment 1 |
Experiment 2 |
||||||||||||
| Donor GFP+% | Donor BM cellularity (2 legs) | Cell doses (x105) | Positive responders*/total scored animals | GFP+ HSC frequency† (x10−6) | GFP+ HSC numbers (2 legs) | Donor GFP+%‡ | Donor BM cellularity (2 legs) | Cell doses (x105) | Positive responders*/ total scored animals | GFP+ HSC frequency† (x10−6) | GFP+ HSC numbers (2 legs) | ||
| Control | 35.6% | 119 × 106 | 20 | 4/6║ | 1 in 2.1§ | 20.2 | 16.6% | 127 × 106 | 25 | 6/10 | 1 in 2.4¶ | 8.8 | |
| 2 | 0/2║ | 17.7% | 183 × 106 | 5 | 3/10 | 13.5 | |||||||
| 0.5 | 0/10 | 8.4% | 96 × 106 | 1 | 0/10 | 3.4 | |||||||
| 0.1 | 0/10 | 0.3 | 0/9 | ||||||||||
| miR-125a | 86.5% | 94 × 106 | 20 | 9/9 | 1 in 0.5§ | 162.6 | 78.1% | 177 × 106 | 25 | 10/10 | 1 in 0.18¶ | 768 | |
| 2 | 2/10 | 5 | 9/10 | ||||||||||
| 0.5 | 2/10 | 1 | 4/10 | ||||||||||
| 0.1 | 0/10 | 0.3 | 3/10 | ||||||||||
| GFP+ HSC expansion | 8.1 X | 56.9–225.9 X | |||||||||||
Total GFP+ BM cells were FACS-sorted from primary recipients 4 to 5 mo posttransplantation and transplanted into secondary recipients together with 200,000 unfractionated wild-type BM cells at indicated cell doses. The number of GFP+ HSCs per two legs in each donor animal was calculated by multiplying the best estimate of GFP+ HSC frequency, donor GFP+% and donor BM cellularity.
*Positive responder called if ≥1% GFP+ cells present in myeloid (Mac1+) and lymphoid (B220+ or CD3+) lineages 16 wk posttransplantation.
†Calculated with L-Calc. The 95% confidence intervals of GFP+ HSC frequency for experiment 1 are 1 in 5.65 × 106 to 1 in 0.77 × 106 for control and 1 in 1.04 × 106 to 1 in 0.24 × 106 for miR-125a; for experiment 2 they are 1 in 4.77 × 106 to 1 in 1.22 × 106 for control, and 1 in 0.32 × 106 to 1 in 0.1 × 106 for miR-125a. The difference in GFP+ HSC frequencies between control and miR-125a groups is indicated by §P = 0.02 and ¶P = 0.0001.
‡BMs from three control mice were pooled to yield enough donor cells.
║Certain animals were lost as a result of accidental dehydration. Initially, 10 animals were in the group.
The miR-125a–Induced Stem Cell Amplification Is Cell Stage-Specific.
We next sought to determine if miR-125a expression confers self-renewal capacity regardless of the ground-cell state. Purified common lymphoid progenitors, common myeloid progenitors, granulocyte and macrophage progenitors, and MEP, as well as the LKS cells, were tested for their ability to gain or maintain the stem cell property of self-renewal with forced expression of miR-125a (Fig. 3G). The production of mature cells in the peripheral blood from common lymphoid progenitors, common myeloid progenitors, granulocyte and macrophage progenitors, and MEP was minimal and disappeared completely in the early weeks (3–6 wk) posttransplantation. In contrast, LKS cells transduced with miR-125a reconstituted >70% of peripheral blood starting from 3 wk posttransplantation and lasted 5 mo (Fig. 3G) and through secondary transplantation (see limiting dilution assays Table 1 and Table S2). Similar results were obtained with further purified HSCs (LKSCD48-CD150+) as the starting population (Fig. 3H). These results indicate that miR-125a was insufficient to induce self-renewal in committed progenitors; its effect depends upon the underlying cell state.
The miR-125a Protected Primitive Hematopoietic Cells from Apoptosis.
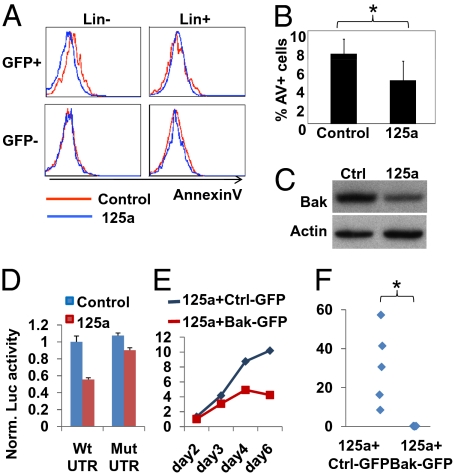
Finally, because Dicer loss induced apoptosis in HSPCs, we asked whether expression of miR-125a may protect primitive cells from apoptosis. BM from mice transplanted with control- or miR-125a–transduced cells were analyzed for apoptosis after reestablishment of homeostasis. We consistently observed decreased apoptosis in miR-125a–tranduced cells in the lineage-negative population, but not in the more mature lineage-positive cells (Fig. 4 A and B), indicating a cell-type-specific effect of miR-125a. These data are consistent with the antiapoptotic effect being at least partially responsible for the increase in the reconstituting cell pool.
Fig. 4.
MiR-125a inhibits immature hematopoietic cell apoptosis and targets Bak1. (A) Selective protection against apoptosis by miR-125a in lineage-negative cells. BM cells were analyzed for lineage markers (Lin), AnnexinV, and 7-AAD with flow cytometry. AnnexinV histograms show Lin− and Lin+ populations after gating for 7-AAD− cells. Lin− population is defined as the lowest 3% cells expressing lineage markers. n = 5. (B) Quantification of data in A. Percentage of AnnexinV+ (AV+) cells present in 7AAD−Lin−GFP+ cells are shown. n = 5. *P < 0.05. (C) HL-60 cells were transduced with miR-125a or a control vector (Ctrl). Western blot was probed for Bak1 and β-actin. (D) Luciferase reporters of WT (wild-type) or Mut (mutant for miR-125a site) Bak1 3′UTR was analyzed in the presence of miR-125a or a control vector in 293T cells. Normalized luciferase activities are shown. Error bars represent SD. (E and F) Primed wild-type donor marrow (5FU) was cotransduced with miR-125a (DsRedExpress+) and a virus for either control or Bak1 (GFP+). Cells were (E) cultured in vitro for 6 d or (F) transplanted (n = 4) and analyzed 4 wk afterward. The percentage of cotransduced cells (GFP+DsRedExpress+) in culture or in peripheral blood is shown. *P < 0.05.
The miR-125a Regulates the Proapoptotic Protein Bak1.
Given the reduced apoptosis in primitive cells and the greatly expanded HSC pool size, we reasoned that miR-125a could target proapoptotic proteins to tilt the cellular balance of pro- and antiapoptotic signals. The miR-125a is predicted to target over 500 evolutionarily conserved targets (25), a number of which have been reported with roles in apoptosis either directly or indirectly. We examined the proapoptotic protein, Bak1 (Bcl-2 antagonist/killer1). HL-60 or BaF3 cells were transduced with miR-125a or the control vector. In both cases, miR-125a expression reduced endogenous Bak1 protein by ∼40 to 50% (Fig. 4C and Fig. S7A). Scanning the 3′UTR of Bak1 revealed one conserved miR-125a–targeting site (Fig. S5C). To ascertain whether the inhibitory effect of miR-125a was mediated through the specific target site in its 3′UTR, we fused the UTR sequence to a luciferase reporter. The miR-125a caused ∼50% inhibition of the luciferase activity. In addition, mutation of the conserved targeting site alleviated most of the inhibition by miR-125a (Fig. 4D). Consistent with the observation that miR-125a is the single microRNA within the miR-99b-let-7e-miR-125a cluster that mediates HSC expansion, miR-125a inhibited the Bak1 3′UTR construct (Fig. S7B), whereas miR-99b and let-7e had minimal effect.
We next asked whether forced expression of Bak1 could block miR-125a–mediated hematopoietic expansion. We coexpressed miR-125a with either control or Bak1 in donor marrow (Fig. 4 E and F and Fig. S8). Coexpression events were marked with DsRed-Express and GFP double-positive cells as miR-125a and Bak1 are expressed from vectors that also carry these two fluorescent proteins, respectively. The percentages of all three populations increased during 6 d of in vitro culture. Although the increase for cells expressing Bak1 was always slower, Bak1-expressing cells were clearly detectable at all times and therefore the levels of Bak1 were not simply eliminating transduced cells (Fig. 4E and Fig. S8). Upon transplantation, miR-125a and control-vector cotransduced marrow expanded as expected (Fig. 4F). In contrast, donor marrow coexpressing miR-125a and Bak1 failed to contribute to peripheral blood at detectable levels, despite similar percentages of double-positive cells 2 d after infection (1.12% and 1.02% for control and Bak1, respectively) (Fig. 4E). These data demonstrate that sustained Bak1 expression blocks miR-125a–induced hematopoietic expansion. To evaluate whether blocking Bak1 mimics the effect of miR-125a, we examined HSCs in mice engineered to be deficient in Bak1 (26, 27). We did not observe significant alteration in phenotypic and functional HSCs from Bak1−/− marrows, suggesting either that miR-125a achieves hematopoietic expansion through targeting additional targets simultaneously (Discussion) or that the compensatory expression of Bak1 family members in the constitutive knock-out may obscure a more potent effect of Bak1.
Taken together, our data indicate that miR-125a protects HSPCs from apoptosis and promotes extensive expansion of the hematopoietic stem cell pool.
Discussion
We report an essential role of Dicer for HSPC maintenance and the identification of a single microRNA, miR-125a, capable of positively regulating HSC regeneration of hematopoiesis at least in part by reducing apoptosis. We provided multiple lines of evidence indicating HSPC impairment and increased apoptosis induced by Dicer deficiency, but our data do not exclude that Dicer loss may also impair more differentiated cells or the differentiation process itself (12, 15, 28, 29). Furthermore, they cannot exclude that some of the impact of Dicer deletion may be augmented by the conditions under which we deleted that gene. The use of pIpC intentionally induces IFNs and recent findings indicate that IFN signaling modulates HSCs (30, 31); it is thus possible that pIpC-activated HSPCs are more susceptible to the deleterious effect of Dicer loss.
The basis for the Dicer effect on HSPC is thought to be its role in microRNA processing. To that end, we identified miR-125a to amplify HSC number and protect primitive cells against apoptosis. Apoptosis modulation has been shown to alter HSCs in vivo with either forced Bcl-2 expression (32, 33) or MCL-1 deletion (34). We show that miR-125a has a differentiation stage-specific effect, increasing the relative abundance of cells in the stem cell state by preventing their apoptosis. The antiapoptotic effect of miR-125a is associated with its ability to down-regulate a proapoptotic protein Bak1, which is a direct target of miR-125a. We did not observe altered HSC number/activity in Bak1−/− mice. These data could be because of compensation by other Bak1 relatives in the constitutive knock-out or because additional direct or indirect proapoptotic protein targets are required for the phenotype induced by miR-125a. Given that miRNAs generally have multiple targets (25), it is certainly possible that their effect depends upon the combinatorial action of several molecules. Our data merely support that Bak1 is likely to be one of those molecules. This notion is supported by the fact that Bak1−/−Bax−/− mice displayed increases in both myeloid and lymphoid lineages (27). Indeed, we observed Puma (Bbc3) protein down-regulation by miR-125a in BaF3 cells, and others have reported that the proapoptotic gene, BMF, is a target for a miR-125a family member (35). Although it is technically difficult to mimic the down-regulation of multiple antiapoptotic proteins in HSCs simultaneously, we demonstrate that sustained Bak1 expression ablates the ability of miR-125a to induce hematopoietic expansion, supporting a role of the apoptotic pathway in mediating the effect of miR-125a. Recently, human p53 has been reported to be targeted by miR-125b (36), a homolog of miR-125a. However, the targeting site identified in this study is not conserved in mouse, consistent with our observation of a lack of significant effect of miR-125a on mouse p53 3′UTR reporter. Hence, p53 is an unlikely candidate that accounts for the superior HSC expansion seen with miR-125a.
In addition to the effect on stem cells and the amplification of all major lineages, we also noticed that sustained expression of miR-125a skewed lineage distribution, favoring the myeloid fate and compromising the B-lymphoid fate (Fig. 3D and Table S2). Although the endogenous level of miR-125a is high in long-term HSCs and much lower in progenitors, we do not exclude a role for miR-125a in committed hematopoietic progenitors during lineage commitment or in more mature blood cells. Because HSCs are heterogeneous, it is also possible that miR-125a may have selectively expanded a more myelogenic subtype [designated α-type HSC by Dykstra et al. (37)] or influenced other subtypes to be more α-like. We also note that miR-125b has been reported to be involved in leukemic translocation, suggesting the possibility that this microRNA can participate in malignant hematopoiesis (38), although the mechanism and cell of origin remain to be investigated.
In summary, we report that microRNAs are actively participating in regulating the HSC state with sensitivity of HSPCs to the loss of the microRNA processing enzyme Dicer and with the unique capability to have HSC number increased by a single microRNA, miR-125a. The ground state of the cell affects its response to microRNAs and suggest that microRNA-based cell modification may be a means to achieve stem cell-specific therapeutics.
Materials and Methods
The Subcommittee on Research Animal Care of the Massachusetts General Hospital and the Institutional Animal Care and Use Committee of Yale University approved all animal work. The Dicerlox/lox mice were described previously (28). All other mice were purchased from the Jackson Laboratory. Methylcellulose M3434 (StemCell Technologies) were used for colony forming assays. Luciferase reporter assay was preformed as described (20). MicroRNA expression constructs were cloned into pMIRWAY-GFP as described (20). Alternatively, GFP was replaced with DsRed-Express (Clontech). For protein expression, the Bak1 ORF was purchased from Invitrogen and subcloned into the pMIRWAY-GFP vector. Viral production, infection, and BM transplantation were performed as described (20). Except where specified, all data are mean ± SD. P values were calculated using two-tailed, unequal variance Student's t test. See SI Materials and Methods for more information.
Supplementary Material
Footnotes
Conflict of interest statement: A patent has been filed on data presented in this article. D.T.S. is a founder, consultant, and stockholder in a company focused on stem cell research, Fate Therapeutics.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE22450).
This article is a PNAS Direct Submission. M.J.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913574107/-/DCSupplemental.
References
- 1.Cozzio A, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 3.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 4.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Fukao T, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 19.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Fossati V, Israel M, Snoeck HW. Lin-Sca1+kit- bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol. 2008;181:7507–7513. doi: 10.4049/jimmunol.181.11.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klarmann K, Ortiz M, Davies M, Keller JR. Identification of in vitro growth conditions for c-Kit-negative hematopoietic stem cells. Blood. 2003;102:3120–3128. doi: 10.1182/blood-2003-04-1249. [DOI] [PubMed] [Google Scholar]
- 24.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsten T, et al. The proapoptotic activities of Bax and Bak limit the size of the neural stem cell pool. J Neurosci. 2003;23:11112–11119. doi: 10.1523/JNEUROSCI.23-35-11112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L, et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci USA. 2009;106:10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 31.Sato T, et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 32.Domen J, Cheshier SH, Weissman IL. The role of apoptosis in the regulation of hematopoietic stem cells: Overexpression of Bcl-2 increases both their number and repopulation potential. J Exp Med. 2000;191:253–264. doi: 10.1084/jem.191.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orelio C, et al. The role of apoptosis in the development of AGM hematopoietic stem cells revealed by Bcl-2 overexpression. Blood. 2004;103:4084–4092. doi: 10.1182/blood-2003-06-1827. [DOI] [PubMed] [Google Scholar]
- 34.Opferman JT, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–1104. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 35.Xia HF, et al. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23:347–358. doi: 10.1159/000218181. [DOI] [PubMed] [Google Scholar]
- 36.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Bousquet M, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.