Abstract
The CurA halogenase (Hal) catalyzes a cryptic chlorination leading to cyclopropane ring formation in the synthesis of the natural product curacin A. Hal belongs to a family of enzymes that use Fe2+, O2 and α-ketoglutarate (αKG) to perform a variety of halogenation reactions in natural product biosynthesis. Crystal structures of the enzyme in five ligand states reveal strikingly different open and closed conformations dependent on αKG binding. The open form represents ligand-free enzyme, preventing substrate from entering the active site until both αKG and chloride are bound, while the closed form represents the holoenzyme with αKG and chloride coordinated to iron. Candidate amino acid residues involved in substrate recognition were identified by site-directed mutagenesis. These new structures provide direct evidence of a conformational switch driven by αKG leading to chlorination of an early pathway intermediate.
Keywords: cryptic chlorination, natural products, polyketide synthases
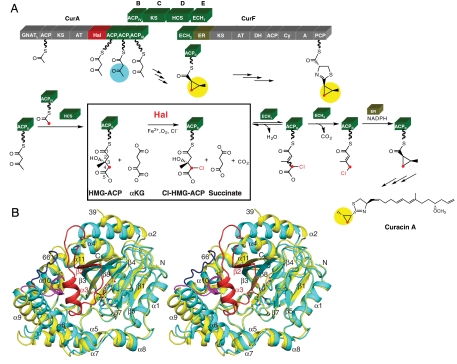
The nonheme Fe2+- and α-ketoglutarate (αKG)-dependent enzymes couple the oxidative decarboxylation of αKG to formation of an activated iron-oxygen species that catalyzes a variety of transformations including hydroxylation, desaturation, ring-closure and ring-expansion. Recently a subfamily of Fe2+/αKG-dependent oxygenases has been characterized that catalyze the chemically challenging halogenation of aliphatic C atoms (1, 2). These Fe2+/αKG-dependent halogenases function within assembly lines of two types of biosynthetic pathway. The polyketide synthases (PKS) and the nonribosomal peptide synthetases (NRPS) produce natural products of astounding chemical diversity. While a number of natural products bear halogen atoms, others use cryptic halogenation en route to other chemical end points. The multicomponent, modular PKSs and NRPSs function by sequentially modifying a growing polyketide/peptide chain as it passes from one module to another. Intermediates are tethered via thioester linkage to the phosphopantetheine arms of acyl carrier protein (ACP) domains in PKSs and peptidyl carrier protein (PCP) domains in NRPSs. Curacin A, from the marine cyanobacterium Lyngbya majuscula, is an unusual natural product with a cyclopropane ring, a thiazoline ring, an internal cis double bond, and a terminal alkene (3). The natural product exhibits antimitotic activity and inhibits the binding of colchicine to tubulin (4, 5). The 65-kbp cur gene cluster encodes a hybrid PKS-NRPS (6). An 8-kbp region within cur has strikingly high sequence identity to a region of the jamaicamaide (jam) gene cluster from a distinct strain of L. majuscula (7). The amino acid sequence identity spanning across CurA to CurF compared end-to-end with JamE to JamJ is 57%–93%. Embedded within the CurA and JamE PKS modules are catalytic domains with 92% sequence identity that were predicted to be Fe2+/αKG-dependent halogenases (2).
Recently we established the role and timing of the curacin halogenase (Cur Hal) in the biosynthesis of curacin A. Hal was shown to be an Fe2+/αKG-dependent halogenase that acted specifically on 3-S-hydroxy-3-methylglutaryl-ACP ((S)-HMG-ACP) (Fig. 1A) (8). The chlorinated product reacted sequentially with the CurE ECH1 dehydratase, the CurF ECH2 decarboxylase, and the CurF enoylreductase (ER). Following ECH1/ECH2-coupled dehydration and decarboxylation of 4-Cl-(S)-HMG-ACP, CurF ER catalyzed formation of the cyclopropane ring by nucleophilic displacement of chloride (8) on the ACP-bound substrate. As opposed to cyclopropane ring formation, the homologous JamE Hal, JamI ECH1, and JamJ ECH2 enzymes generated a vinyl chloride functional group as the endpoint of the reaction series (8).
Fig. 1.
Structure and function of Cur Hal. (A) On-assembly-line chlorination in cyclopropane ring formation. The Hal domain within CurA specifically chlorinates (S)-HMG-ACP at C4. The chlorinated product subsequently undergoes dehydration (CurE ECH1), decarboxylation (CurF ECH2), and cyclopropane ring formation (CurF ER). Catalytic and carrier domains within Cur polypeptides are rendered as blocks (A, adenylation domain; AT, acyltransferase; ACP, acyl carrier protein; Cy, condensation/cyclization domain; DH, dehydratase; GNATL, GCN5-related N-acetyltransferase-like domain with bifunctional decarboxylase/S-acetyltransferase activity; HCS, HMG-ACP synthase; KS, ketosynthase; PCP, peptidyl carrier protein). (B) Open and closed forms of Cur Hal. In this stereo diagram the open and the closed subunits of Hal are superposed. In the closed-form subunit (cyan), an ordered 27-residue lid (red) covers the active site and the α9–α10 loop (dark blue) is at the periphery. In the open form (yellow), the lid (residues 40–65) is missing, and the α9–α10 loop (magenta) is adjacent to the active site and to a 310-like helix preceding α4. Structural changes include shifts of β3 and helices α2, α4, α5, α6, α9, and α10, which were excluded from the superposition calculation (rmsd = 0.9 Å for 217 Cα atoms). In the metal center (shown in sticks), iron (orange) coordinates two histidine side chains (cyan C), αKG (gray C), chloride (green) and formate (white C). Ligands are shown with atomic colors (red O, blue N) and coordination bonds in black. Secondary structures are labeled.
Some insights into the catalytic mechanism of Fe2+/αKG-dependent halogenases came from crystal structures of the halogenases SyrB2, which converts threonine to 4-chlorothreonine in the biosynthesis of syringomycin E (9, 10) and CytC3, which doubly chlorinates 2-aminobutyric acid (11, 12). In the iron coordination sphere of these halogenases, a chloride ligand replaced the Glu or Asp ligand found in the Fe2+/αKG-dependent oxygenases (9, 11). Halogenation was shown to follow a similar mechanism to the Fe2+/αKG-dependent oxygenases (13). Following dioxygen-catalyzed decarboxylation of αKG, a reactive O = Fe4+ intermediate abstracts a hydrogen atom from the C-H bond of the substrate, yielding a Fe3+-OH and a substrate radical (13, 14). The substrate radical is halogenated by “rebound” of the chlorine atom, similar to rebound of hydroxyl in the Fe2+/αKG-dependent hydroxylases. Recently, it was shown for the halogenase SyrB2 that substrate binding triggers formation of the reactive O = Fe4+ intermediate and that hydroxylated products can be formed from alternative substrates (15, 16).
Here we describe the structure of Cur Hal in five distinct ligand states. The four crystal structures demonstrate a remarkable conformational transition between open and closed states that is triggered by αKG binding. A large “lid” covers the active site in closed-form Hal. Other concerted changes in the structure appear designed to exclude (S)-HMG-ACP from the active site of open-form Hal and to admit it to the closed form. Site-directed mutagenesis provides new insights about the basis for (S)-HMG specificity.
Results
Overall Structure.
The recombinant CurA Hal domain crystallized in four forms (Table S1), which provide a total of seven independent views of the polypeptide. The structures capture the protein in two distinct conformations, which we designate “open” and “closed,” and in several ligand states (Fig. 1B and 2). The crystallized protein included residues 1600–1919 of CurA, here designated 1–320. The CurA Hal domain possesses the cupin fold (17), a barrel-like structure consisting of a major antiparallel β-sheet (β1, β6, β4, β8, β3, and β2) and a minor β-sheet (β5 and β7) (Fig. 1B). The cupin barrel houses the active site and is surrounded by helices. Like other Fe2+/αKG-dependent enzymes, Hal possesses a large flexible active-site lid (residues 40–66), which varies from fully ordered to fully disordered among the four crystal structures. Hal is a dimer in solution and in all of the four crystal structures, consistent with the dimeric architecture of PKS modules (Fig. S1). Helices α5, α7, and α8 form the hydrophobic dimer interface, which is relatively small (940 Å2 buried surface area per monomer). Each active site is self-contained within a monomer. The two iron centers are 37 Å apart in the Hal dimer.
Fig. 2.
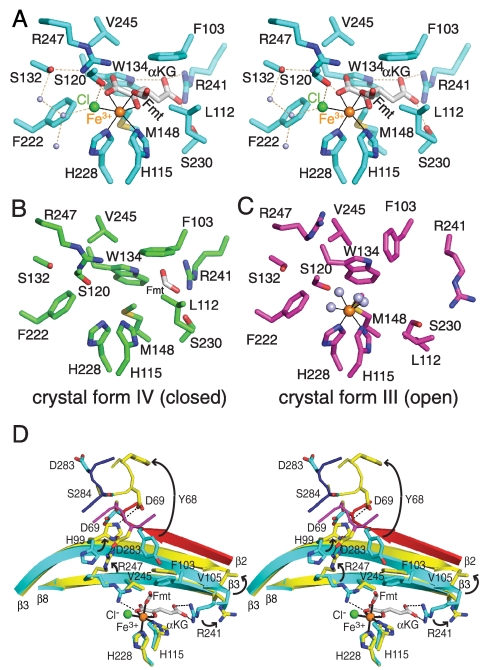
Cur Hal active site. (A) Closed-form Hal with Fe3+/αKG/Cl-/formate. The octahedral iron coordination sphere is complete in crystal form II (cyan C) with bound αKG (white C) and formate (Fmt), shown here in stereo. (B) Closed-form Hal with formate. Formate occupies the back of the αKG site and no iron is present in crystal form IV (green C). (C) Open-form Hal with Fe3+. Iron has only water (gray) in the ligand sphere of Subunit B (magenta C) of the mixed-form dimer in crystal form III. (D) αKG trigger for the conformational switch between closed and open forms. Upon loss of αKG, the Arg241 side chain (strand β8) swings out of the hydrophobic binding pocket, initiating a series of changes in strand β3, Arg247 (strand β8), Tyr68 (α4), and Asp283 (α9–α10 loop). In addition the (red) active-site lid is disordered. Open and closed forms are colored as in Fig. 1B in this stereo view. In all views, atomic coloring is used with orange iron, green chloride, blue N, red O and yellow S; coordination bonds are thin black lines and hydrogen bonds are dashed lines.
The Hal domain was originally annotated as a phytanoyl-CoA dioxygenase (PhyH) based on sequence similarity to the oxygenases (6). Indeed, Hal is most similar in structure to human PhyH (18), although only about two-thirds of Hal residues have structural equivalents in PhyH (PDB 2A1X, rmsd of 3.1 Å for the Cα atoms of 205 residues with 16% sequence identity). Hal is also similar to the two characterized nonheme Fe2+/αKG-dependent halogenases, which are closely related to each other (58% identity): SyrB2 in the syringomycin biosynthetic pathway (9) (2FCT; 3.3-Å rmsd, 216 residues, 13% identity) and CytC3 in the armentomycin pathway (11) (3GJB; 3.6-Å rmsd, 198 residues, 16% identity). Hal, PhyH, SyrB2, and CytC3 share a common cupin core with nearly identical active sites. In contrast, the outer helices, connecting loops, and active-site lid of Hal differ considerably from those of the other structures, in accord with the low 13%–16% sequence identities.
Active Site.
The Hal active site is characteristic of Fe2+/αKG-dependent oxygenase/halogenases, and is captured in five distinct ligand states in the four crystal structures (Table S1 and Fig. 2, Fig. S2). The nonheme mononuclear iron center has an octahedral coordination shell with only two protein ligands, His115 and His228 (Fig. 2A and Fig. S2). Hal was crystallized under aerobic conditions, so the iron is assumed to be in the ferric state. The cosubstrate, αKG, is a bidentate Fe3+ ligand via the C2 ketone and C1 carboxylate. Halogenase structures containing αKG also have a chloride iron ligand (Fig. 2A, Fig. S2B), as in the SyrB2 and CytC3 enzymes (9, 11). In crystal form II, a formate ion from the crystallization solution occupies the sixth coordination position (Fig. 2A), where water is thought to bind in the resting enzyme and dioxygen during catalysis. Water occupies this position in subunits containing Fe3+ and no formate (Fig. 2C, Fig. S2B). Two crystal forms lack iron (Fig. 2B, Fig. S2C). In one of these, a formate ion occupies part of the αKG site (Fig. 2B).
Both αKG and chloride form specific interactions with Cur Hal (Fig. 2A). The chloride ion is hydrogen bonded to the Ser120 hydroxyl and to water. αKG is fully buried in the enzyme in a largely hydrophobic pocket. At the bottom of the pocket, the αKG carboxylate distal to the Fe center (C5 carboxylate) is anchored by four hydrogen bonds from the side chains of Trp134, Ser230, and Arg241. The key ionic contact of both C5-carboxylate oxygens with Arg241 is conserved in the Fe2+/αKG-dependent halogenases/oxygenases. At the Fe-proximal end of αKG, the Arg247 side chain forms a hydrogen bond with the C1 carboxylate. This ionic contact is also conserved, but the position of the basic residue (Arg247 on strand β8 in Hal) is not. A stacking interaction with Phe103 stabilizes an extended conformation of αKG. Several other residues (Phe222, Leu112, Met148, and Val245) contact αKG and contribute to the hydrophobic character of the binding site (Fig. 2A).
αKG Binding Triggers a Conformational Switch.
The structures provide four views of the Hal subunit in a closed conformation and three in an open conformation (Table S1). The open and closed forms differ by a remarkable set of concerted changes in several parts of the structure (Fig. 1B). The most dramatic difference is an order-disorder transition of a 27-residue lid (40–66, red in Fig. 1B), which covers the active site in the closed form and is completely disordered in the open form. In addition, radically different structures also exist for the loop connecting helices α9 and α10 (residues 274–284) (magenta and dark blue in Fig. 1B). Finally, several secondary structures are in slightly different positions, including strand β3 and helices α2, α4, α5, α6, α9, and α10. These structural differences clearly distinguish two states of the enzyme. The individual subunits of each form are identical within experimental error (0.2 Å–0.5 Å rmsd for Cα atom superposition) whereas the open and closed forms differ (1.5 Å rmsd). It is evident that the open and closed forms are not peculiarities of crystallization conditions or iron binding status, as each form occurred both with and without bound iron and in two unrelated crystals grown in different conditions. The open and closed forms occur together in the mixed-form dimer in crystal form III, attesting to the independence of the active sites.
The structures reveal that the physiological switch between open and closed Cur Hal is αKG binding. All Hal subunits with a ligand in the αKG site are in the closed form and all subunits lacking a ligand in this site are in the open form. The key interaction in the switch is the salt bridge between αKG and Arg241 (Fig. 2D). Movement of Arg241 appears to trigger the overall conformational change between closed and open Hal. Upon departure of the αKG C5-carboxylate from the bottom of the binding pocket, the Arg241 side chain (strand β8) swings out of the binding pocket into space occupied by Val105 on the adjacent strand β3. Strand β3 shifts 3 Å toward helices α2 and α4. Strand β2 becomes disordered. Helices α2 and α4 shift slightly, repacking the hydrophobic core between α2/α4 and β3/β8. The shift of helix α4 is incompatible with the position of the α9–α10 loop, which folds into the active site. The restructured α9–α10 loop is incompatible with helix α3, which becomes disordered as the active-site lid (residues 40–66) opens. The flexible active-site lid comprises all residues between α2 and α4, including helix α3 (residues 48–55) and strand β2 (residues 64–68). Strand β2 and residues 61–63 of the α3–β2 loop cover the hydrophobic αKG binding pocket, and the pocket is solvent exposed when the lid opens.
Both forms of the Cur Hal subunit are stabilized by specific interactions (Fig. 2D). In addition to the steric effects at Arg241 that drive the switch, four parts of the protein structure converge to form a set of specific interactions in open-form Hal, including formation of a 310-like helix (residues 68–77) following helix α4, formation of a 310 helix (residues 278–282) in the α9–α10 loop, and ionic interactions of the Asp69, His99, Arg247, and Asp283 side chains. In the closed-to-open transition, Tyr68 in the 310-like helix moves 17 Å out of the active site (Fig. 2D). Asp283 in the α9–α10 loop moves 12 Å toward the active site where it forms a salt bridge with the Arg247 side chain, which has moved 5 Å out of the active site. His99 (strand β3) swings 7 Å away from the active site to hydrogen bond with Asp69 in the 310-like helix.
Substrate Binding and Catalysis.
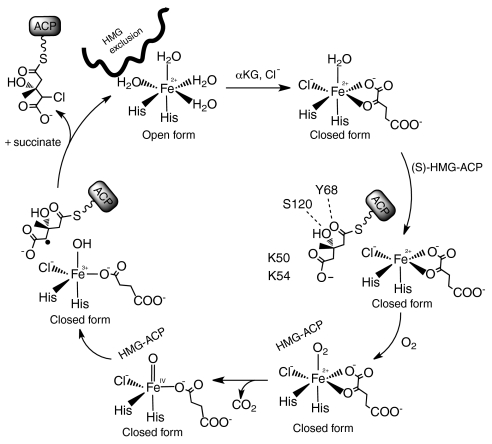
The five ligand states of the Cur Hal active site (Fig. 2 and Fig. S2) are consistent with the mechanism in which oxidative decarboxylation of αKG by iron-bound dioxygen creates a ferryl-oxo (Fe4+-oxo) intermediate (Fig. 3) (2, 13). The dioxygen position is occupied by a formate ion in the form II crystal structure (Fig. 2A). In this structure, the active-site lid (residues 40–66) is fully ordered throughout its length and the active site is inaccessible to (S)-HMG-ACP. However, in the closed subunit of the form III crystal structure, residues 48–59 including lid helix α3 are disordered while residues 60–66 are secured over bound αKG. Substrate access to the active site is presumably through the opening created by disorder of lid helix α3 in closed-form Hal.
Fig. 3.
Hal mechanism and role for conformational change. The resting enzyme (upper left) is in the open form and (S)-HMG-ACP is excluded. αKG and Cl- binding trigger the switch to the closed form, to which (S)-HMG-ACP can bind. Substrate binding permits dioxygen to coordinate the Fe2+, and triggers oxidative decarboxylation of αKG and formation of the chloro-oxo-ferryl intermediate. The intermediate abstracts a hydrogen atom from the substrate, and the substrate radical is resolved by chlorination via a “rebound” mechanism (13).
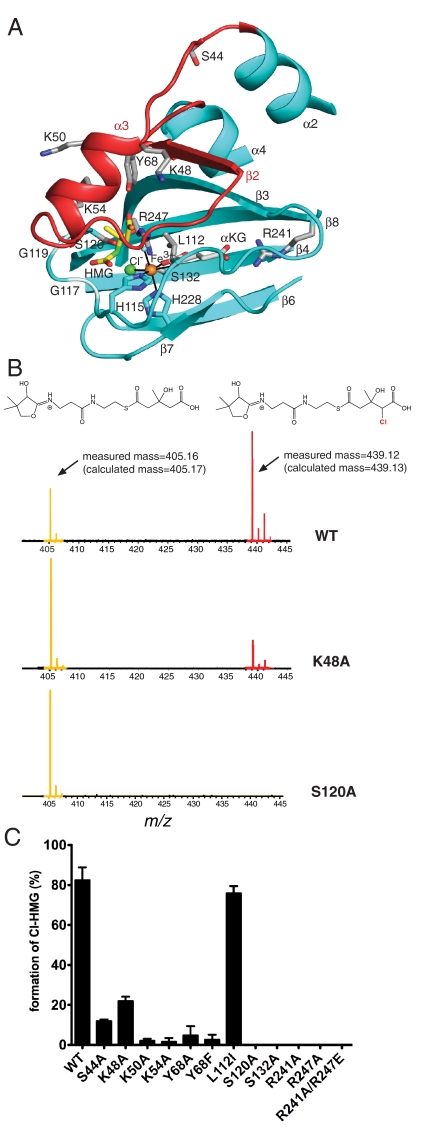
We modeled the (S)-HMG-thioester into the active site of closed-form Cur Hal (excluding lid residues 48–59) so that the HMG C4 atom was near both the Cl- ligand and the ferryl-oxo O atom (Fig. 4A). Hal is highly specific for its substrate, requiring both the S-hydroxyl group at C3 and the C5-carboxylate (8). In the model, the S-hydroxyl group is hydrogen bonded to the Ser120 side chain, which forms a hydrogen bond with Cl- in structures containing αKG (Fig. 2A and Fig. S2B). The Tyr68 side chain is hydrogen bonded to the substrate thioester carbonyl. No candidate residues for recognition of the HMG carboxylate are obvious in the core of the active site. However, lid helix α3 includes several basic side chains, which may recognize the (S)-HMG carboxylate.
Fig. 4.
HMG model and site-directed mutagenesis of Cur Hal. (A) Model of HMG binding in the active site. Residues in contact with modeled HMG (yellow C) include Ser120 to the 3-S-hydroxyl, Tyr68 to the thioester carbonyl and the backbone at residues 117–119 to a carboxylate oxygen. The C4 atom of HMG to be chlorinated is green. Side chains of residues tested by mutagenesis are shown with gray C. (B) PPant-ejection assay of Hal activity. Substrate and product were ejected from ACP by IRMPD and detected by FTICR-MS. HMG-PPant is at m/z 405.16, Cl-HMG-PPant is at m/z 439.12. (C) Activity of wild-type and Hal mutants. Activity is represented as the percent of chlorinated product compared to total species ejected. Each reaction was performed at least in triplicate. Error bars represent the standard error of measurement.
Residues that may be important for substrate recognition were probed by alanine replacement or conservative substitution (Fig. 4 and Table S2). These residues include three basic and two polar residues that may recognize (S)-HMG-ACP (Leu112, Lys48, Lys50, Lys54, Ser44, and Tyr68), and four active-site residues that directly or indirectly contact αKG or chloride (Ser120, Ser132, Arg241, and Arg247). Production of 4-Cl-HMG-ACP from (S)-HMG-ACP was monitored by Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS), including a phosphopantetheine (PPant)-ejection assay (19), which releases Cl-HMG-PPant from Cl-HMG-ACP (8). Catalytic efficiency was monitored by the relative abundance of Cl-HMG-PPant and HMG-PPant, which differ in m/z by 34 units (Fig. 4B and Figs. S3, S4, and S5).
Under our assay conditions, wild-type Hal converted about 80% of HMG-ACP to the chlorinated product (Fig. 4 B, C). As expected, substitutions for residues that anchor αKG in the active site (Arg241, Arg247) resulted in a Hal variant with no detectable activity (Fig. 4C). Ala replacement of Ser120, which is hydrogen bonded to Cl-, also abolished activity, as did the Ala substitution at Ser132, which is connected to the active site indirectly through hydrogen bonds (Fig. 2A, Fig. 4C). A conservative substitution at Leu112 did not alter activity, whereas substitutions in the active-site lid substantially diminished activity. The largest effects (greater than tenfold) were observed for the basic residues Lys50 and Lys54, and for Tyr68. The mutagenesis data are consistent with a role in (S)-HMG (or PPant) recognition for Lys50, Lsy54, and Tyr68, although no single residue was essential for catalysis. The greater than tenfold effect of conservative Phe substitution for Tyr68 suggests a critical hydrogen-bonding interaction for this side chain. The effects of the Lys48 and Ser44 substitutions were five- and sixfold, respectively, consistent with an indirect effect on catalysis, for example by altering the dynamics of the active-site lid.
Discussion
The structure of CurA Hal is the third for a halogenase from the Fe2+/αKG-dependent oxygenase/halogenase family. The structures of Hal in five ligand states (apo, Fe3+/αKG/Cl-/formate, Fe3+/αKG/Cl-, formate, Fig. 2 and Fig. S2) provide new pictures of the active site. Together with site-directed mutagenesis results (Fig. 4) the structures are relevant to the catalytic cycle and mechanism of Hal (Fig. 3) (13, 15) and two distantly related halogenases, SyrB2 and CytC3, which have active sites nearly identical to the Cur Hal active site (9, 11).
Site-directed mutagenesis revealed a critical role for Ser132. This result was unexpected because Ser132 is in the center of strand β4 where the Ala substitution should not alter the backbone conformation (Fig. 4A). However, in closed-form Hal, a hydrogen bond of the Ser132 side chain helps position the Arg247 guanidinium plane to interact with αKG in the active site (Fig. 2A). The loss of activity in Ser132Ala implies that Arg247 guanidinium positioning is critical to catalysis. We suggest that the Ser-Arg pair may assist in the departure of CO2 during formation of succinate and the chloro-oxo-ferryl intermediate (Fig. 3). This hydrogen bond may be a general feature of the Fe2+/αKG-dependent halogenases as the hydrogen-bonding network to αKG is conserved in SyrB2 and CytC3, but with a Thr/Arg pair in place of Ser/Arg.
Remarkably, the new structures reveal that Cur Hal exists in two conformations and that αKG triggers a switch between these forms (Fig. 2D). We designated the forms “open” and “closed” because the active-site lid (residues 40–65) is completely disordered in the open form. An open active site is essential to substrate access, however open-form Hal is not simply a relaxed version of the closed form nor is it equivalent to the closed form with a disordered active-site lid. Rather, the open and closed forms are characterized by distinct positions for several helices and loops, stabilized by different sets of hydrogen bonds. Open-form Hal has a total of 10 more hydrogen bonds than does the closed form (excluding the 40–65 lid), suggesting that the open form has an important function beyond exposing the active site. The additional hydrogen bonds in open-form Hal are in the α9–α10 loop and the adjacent α4 helix N terminus. Arg247 is a key element of the conformational transition because it is hydrogen bonded to αKG in the closed form and in the open form has two hydrogen bonds with the carboxylate of Asp283 at the N terminus of helix α10, as well as a hydrogen bond with the Ser120 carbonyl oxygen.
Large conformational changes triggered by αKG have not been observed for other members of the Fe2+/αKG-dependent oxygenase/halogenase family, although there are few reports of companion structures with both occupied and empty αKG sites. Examples include the structures of a cephalosporin synthase (20) and an alkylsulfatase (21), in which αKG binding is accommodated by strictly local conformational changes relative to the free enzymes.
The chemical structure of HMG may explain the need for two conformations of Hal. HMG is unusual and perhaps unique among substrates of Fe2+/αKG-dependent oxygenase/halogenases in that the reactive carbon (C4, Fig. 1A) has a carboxylate substituent (C5). The proximal carboxylate is an enormous complication for an Fe-dependent enzyme because only two of six coordination positions are occupied by protein ligands, the four nonprotein ligands turn over in each catalytic cycle, and the substrate carboxylate is not a ligand. Thus, it is critical for Hal to keep the HMG carboxylate out of the Fe2+ coordination sphere, perhaps by excluding (S)-HMG-ACP altogether until the halogenase active site is assembled by Fe2+ coordination of αKG and Cl-. In open-form Hal, the active site appears able to accommodate a carboxylate at any unoccupied position in the Fe ligand sphere. Indeed, in family members that are not halogenases, a carboxylate side chain coordinates Fe at the position of Cl- in Hal and the other halogenases (17). We propose that the purpose of a specific open form of Hal is to prevent (S)-HMG-ACP from entering the active site until both αKG and Cl- are bound (Fig. 4). In this manner, the interactions of the 310-like helix preceding α4, the α9–α10 loop, and Arg247 create a barrier to (S)-HMG-ACP in open-form Hal.
Hal has exquisite specificity for its substrate, (S)-HMG-ACP, requiring both the C3 S-hydroxyl group and the C5 carboxylate (8). Specificity for the C5 carboxylate is consistent with the need to engage the carboxylate with the protein and not with the Fe2+ center. We assume that Hal has little specificity for the C3 R-methyl group because the 92%-identical halogenase from the jamaicamide PKS (7) accommodates a C3 R-substituent with a five-carbon chain. The exquisite substrate specificity is consistent with our finding that several residues on the active-site lid are critical to activity (Figs. 3 and 4A. Ala substitution for Lys50, Lys54, and Tyr68 resulted in more than tenfold reduction in activity in our assay conditions. Lys50 and Lys54 are thus candidates to recognize the HMG carboxyl group. The Tyr68 hydroxyl may hydrogen bond with the substrate thioester carbonyl oxygen, consistent with the finding that even a conservative substitution of Tyr with Phe resulted in more than tenfold loss of activity. In the core of Hal, Ala substitution for Ser120, which is hydrogen bonded to Cl-, abolished activity. The chloride hydrogen bond is not required for halogenation because neither SyrB2 nor CytC3 has a direct hydrogen bond from protein to Cl- or a hydrogen-bonding side chain at the analogous position to Ser120. We suggest that the Ser120 side chain recognizes the C3 S-hydroxyl of HMG. In open-form Hal, the proposed substrate-recognition residues are otherwise occupied: Tyr68 is displaced 17 Å away from the active site; access to the Ser120 side chain is blocked by a hydrogen bond from the Arg247 side chain to the Ser carbonyl oxygen; and Lys50 and Lys54 are disordered.
A highly reactive oxo-ferryl (O = Fe4+) intermediate forms when dioxygen binds to the iron center and decarboxylates αKG (Fig. 3). An event of “substrate triggering” was proposed to explain how the Fe2+/αKG-dependent oxygenase/halogenases prevent formation of this reactive intermediate until the substrate is positioned (22). Substrate triggering has been observed in the SyrB2 halogenase (15), and is likely to occur in Hal as well. Thus, the substitutions at Lys50, Lys54, Tyr68, and Ser120 may prevent substrate triggering of chloro-oxo-ferryl formation in addition to any direct effects on HMG binding.
The exquisite substrate selectivity of Hal is strikingly different from the situation of the SyrB2 halogenase, which formed halogenated, hydroxylated, or mixed products when presented with molecules similar to its natural substrate (16). In contrast to these results, we observed no hydroxylation products in FTICR-MS nor in the PPant-ejection assay of Hal activity (Fig. 4 and Fig. S3). A conundrum of Fe2+/αKG-dependent halogenases is how the enzymes resolve the substrate free radical exclusively by halogenation because both chloride and hydroxyl ligands are available at the iron center (Fig. 3). The results with SyrB2 were interpreted as evidence that the halogenation vs. hydroxylation outcome depends on substrate positioning, which was thought to vary with the chemical structure of the SyrB2 substrate (16). The possibility of a hydroxylated product from a differently positioned substrate is relevant to Hal function because the enzyme may encounter molecules that are chemically similar to (S)-HMG-ACP (8). Hal is followed in the curacin pathway by a dehydratase and a decarboxylase. We showed previously that these enzymes can act sequentially on HMG-ACP, i.e., preceding the Hal reaction (23). If the HMG-ACP dehydration product (C2-C3 alkene) or the subsequent decarboxylation product were able to bind in the Hal active site, incorrect hydroxylated products may result. Because the ACP that carries these substrates is fused to Hal (Fig. 1A), it is critical that any dehydrated and/or decarboxylated products be unable to access the Hal active site. Thus, the exquisite substrate selectivity serves as a barrier to the competing hydroxylation reaction.
The combined requirements that Hal prevent Fe coordination of the HMG C5 carboxylate, allow dioxygen reaction at the appropriate moment, and exclude inappropriate substrates from the active site may have led to both the αKG trigger for transition to a substrate-accessible state, the substrate trigger to admit dioxygen, and the exquisite substrate selectivity of Cur Hal.
Methods
CurA Hal and the site-directed mutants were expressed in Escherichia coli and purified by affinity and gel filtration chromatography. Diffraction data were collected at beamline 23ID at the Advanced Photon Source (APS). The structure of crystal form I was solved by a 2.6 Å single wavelength anomalous diffraction dataset. Crystal form II, III, and IV were solved by molecular replacement using the model from crystal form I. Conversion of (S)-HMG-ACP substrate to Cl-(S)-HMG-ACP was analyzed by FTICR-MS and an infrared multiphoton dissociation (IRMPD)-based phosphopantatheine (PPant) ejection assay (8, 19). Detailed methods are available in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. D. L. Akey for helpful discussions, Dr. T. C. Terwilliger for suggestions on processing data in the cubic crystal form I, the staff of APS 23ID-D, and the Caribbean Marine Biological Institute research station and the government of Curaçao for assistance and permission to make collections of the original Curacin A-producing cyanobacterial strains. The research was supported by National Institutes of Health (NIH) Grants DK-42303 (to J.L.S.) and CA-108874 (to D.H.S. and W.H.G.), and by National Science Foundation Career award CHE-05-47699 (to K.H.). Beamline 23ID-D is supported by the NIH National Institute of General Medical Sciences (GM) and National Cancer Institute (CA) through the GM/CA Collaborative Access Team at the APS, which is supported by the Department of Energy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3NNJ, 3NNF, 3NNL, and 3NNM for structures in crystal forms I–IV, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006738107/-/DCSupplemental.
References
- 1.Fujimori DG, Walsh CT. What’s new in enzymatic halogenations. Curr Opin Chem Biol. 2007;11:553–560. doi: 10.1016/j.cbpa.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 3.Gerwick WH, et al. Structure of Curacin A a novel antimitotic, antiproliferative and brine shrimp toxic natural product from the marine cyanobacterium Lyngbya majuscula. J Org Chem. 1994;59:1243–1245. [Google Scholar]
- 4.Blokhin AV, et al. Characterization of the interaction of the marine cyanobacterial natural product Curacin A with the colchicine site of tubulin and initial structure-activity studies with analogues. Mol Pharmacol. 1995;48:523–531. [PubMed] [Google Scholar]
- 5.Verdier-Pinard P, et al. Biosynthesis of radiolabeled Curacin A and its rapid and apparently irreversible binding to the colchicine site of tubulin. Arch Biochem Biophys. 1999;370:51–58. doi: 10.1006/abbi.1999.1363. [DOI] [PubMed] [Google Scholar]
- 6.Chang Z, et al. Biosynthetic pathway and gene cluster analysis of Curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 7.Edwards DJ, et al. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Gu L, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 10.Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc Natl Acad Sci USA. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural analysis of an open active site conformation of nonheme iron halogenase CytC3. J Am Chem Soc. 2009;131:4872–4879. doi: 10.1021/ja8097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueki M, et al. Enzymatic generation of the antimetabolite gamma,gamma-dichloroaminobutyrate by NRPS and mononuclear iron halogenase action in a streptomycete. Chem Biol. 2006;13:1183–1191. doi: 10.1016/j.chembiol.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Galonic DP, Barr EW, Walsh CT, Bollinger JM, Jr, Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 14.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews ML, et al. Substrate-triggered formation and remarkable stability of the C-H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry. 2009;48:4331–4343. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews ML, et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc Natl Acad Sci USA. 2009;106:17723–17728. doi: 10.1073/pnas.0909649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clifton IJ, et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 18.McDonough MA, et al. Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease. J Biol Chem. 2005;280:41101–41110. doi: 10.1074/jbc.M507528200. [DOI] [PubMed] [Google Scholar]
- 19.Dorrestein PC, et al. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valegard K, et al. Structure of a cephalosporin synthase. Nature. 1998;394:805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 21.Muller I, et al. Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II) alpha-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43:3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- 22.Solomon EI, et al. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 23.Gu L, et al. Metabolic coupling of dehydration and decarboxylation in the curacin A pathway: functional identification of a mechanistically diverse enzyme pair. J Am Chem Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.