Abstract
We show herein that the Salmonella pathogenicity island 2 (SPI2) response regulator SsrB undergoes S-nitrosylation upon exposure of Salmonella to acidified nitrite, a signal encountered by this enteropathogen in phagosomes of macrophages. Mutational analysis has identified Cys203 in the C-terminal dimerization domain of SsrB as the redox-active residue responding to nitric oxide (NO) congeners generated in the acidification of nitrite. Peroxynitrite and products of the autooxidation of NO in the presence of oxygen, but not hydrogen peroxide, inhibit the DNA-binding capacity of SsrB, demonstrating the selectivity of the reaction of Cys203 with reactive nitrogen species (RNS). These findings identify the two-component response regulator SsrB Cys203 as a thiol-based redox sensor. A C203S substitution protects SsrB against the attack of RNS while preserving its DNA-binding capacity. When exposed to SPI2-inducing conditions, Salmonella expressing the wild-type ssrB allele or the ssrB C203S variant sustain transcription of the sifA, sspH2, and srfJ effector genes. Nonetheless, compared with the strain expressing a redox-resistant SsrB C203S variant, wild-type Salmonella bearing the NO-responsive allele exhibit increased fitness when exposed to RNS in an NRAMPR, C3H/HeN murine model of acute oral infection. Given the widespread occurrence of the wild-type allele in Salmonella enterica, these findings indicate that SsrB Cys203 increases Salmonella virulence by serving as a redox sensor of NO resulting from the host immune response to oral infection.
Keywords: macrophages, pathogenesis, two-component regulatory system
Nitric oxide (NO) and its congeners react with metal prosthetic groups, organic and inorganic radicals, lipids, and DNA molecules. This rich biochemistry probably mediates the broad-spectrum antimicrobial activity of reactive nitrogen species (RNS) against phylogenetically diverse microorganisms (1). Diarrheagenic Salmonella elicit a burst of NO in rectal content in infected humans (2) and stimulate the accumulation of iron-nitrosyl adducts in affected viscera of experimental animals (3). Mounting evidence indicates that these RNS are not mere signatures of the infection but are active components of the anti-Salmonella host arsenal. The gross susceptibility of mice unable to synthesize NO in response to experimentally induced salmonellosis is arguably the most dramatic proof of the role for RNS in the host response to Salmonella (3, 4).
Despite being a radical, NO exhibits remarkable specificity in its reactions with organic and inorganic molecules. Most direct effects of NO on biological systems stem from its interaction with transition metals in terminal cytochromes of the electron transport chain and [4Fe-4S] prosthetic groups in dehydratases (5, 6). The selectivity of NO for metalloproteins also applies to Salmonella, as shown by the nitrosylation of the bd quinol cytochrome oxidase (7); in that case, NO is a positive signal that stimulates the expression of antioxidant defenses. The biochemistry of this diatomic radical is enriched through its interactions with other radicals. The reaction of NO and superoxide (O2•-), species, which are concomitantly produced by professional phagocytes (8), yields peroxynitrite (ONOO−). ONOO− is a potent oxidizing species with high affinity for tyrosine residues and the solvent-exposed Feα of [4Fe-4S] clusters (9, 10). The autooxidation products nitrogen dioxide (NO2•) and dinitrogen trioxide (N2O3) further enrich the biological chemistry of NO (11). N2O3, which can arise independently from the condensation of acidified nitrite (NO2−), oxidizes and nitrosylates redox-active thiol groups. Salmonella are likely to be exposed to N2O3 in the context of IFNγ-primed macrophages or during transit through the stomach (12–14). In turn, microorganisms sense NO through the nitrosylation of either iron or sulfhydryl metal groups in response regulators, thereby engaging signaling pathways that promote the enzymatic and nonenzymatic detoxification of RNS. By avoiding contact with inducible NOS (iNOS)-containing vesicles, the Salmonella pathogenicity island 2 (SPI2) type III secretion system has been shown to contribute to the antinitrosative defenses of Salmonella (15).
Both positive and negative regulation of SPI2 are critical for Salmonella pathogenesis (16). The capacity of Salmonella to survive within host phagocytes and proliferate within epithelial cells relies on SPI2-dependent remodeling of the Salmonella-containing vesicle. Salmonella reside intracellularly within vacuoles that establish contact with the secretory pathway but minimize fusion with late endosomes and lysosomes (17, 18). By doing so, SPI2 decreases contact of Salmonella with the hydrolytic enzymes of lysosomes as well as reactive oxygen species (ROS) and RNS generated by NADPH oxidase and iNOS (17, 19). Nonetheless, the overexpression of SPI2 can have adverse effects on Salmonella pathogenicity in vivo. In fact, negative regulation of SPI2 by YdgT is needed for Salmonella virulence at late stages of the infection (20). RNS also appear to exert negative control of SPI2 transcription (13, 21). RNS repress genes within and outside the SPI2, including those encoding for chaperones, the type III secretion apparatus, translocon, and effectors. Collectively, the widespread repression of SPI2 loci suggests that RNS inhibit a critical signaling pathway upstream of or involving the SsrAB two-component regulatory system. The goals of the work presented here are to test whether the SsrB response regulator is a target of RNS and to establish the extent to which the interaction of SsrB and RNS contributes to Salmonella pathogenesis.
Results
S-Nitrosylation of the Redox-Active Cys203 Thiol in the SsrB Response Regulator.
We first sought to determine possible targets of NO-mediated inhibition of SPI2. In silico analysis revealed the presence of three cysteines in SsrB, raising the possibility that these residues could be targets of RNS. To determine whether SsrB contains redox-active cysteines, strain AV07104 expressing a C-terminal 3xFLAG-tagged SsrB variant was constructed using the λ Red system. This strain proved to be as virulent as its parent control when tested in an i.p. model of acute infection in C57BL/6 mice (Fig. S1). Having established its full virulence potential, we exposed strain AV07104 to nitrosative stress generated by acidified NO2−, which at low pH is protonated to its acid conjugate nitrous acid (HNO2). Two molecules of HNO2 in turn condensate to form the strong oxidative and nitrosative species N2O3. Acidified NO2− is a relevant source of nitrosative stress in our system because it adds to the RNS of Salmonella-infected macrophages (14). Strain AV07104 expressing an SsrB::3xFLAG was cultured for 6 h in EG medium, pH 5.5, containing 750 μM NO2− or NO3−. The amount of SsrB protein was similar in NO2−- and NO3−-treated Salmonella (Fig. 1A, Upper). The cytoplasmic extracts were tested for the presence of S-nitrosylated SsrB after derivatization of S-nitrosylated proteins in a biotin switch assay. Immunoblots of NeutrAvidin-purified, biotinylated proteins demonstrated the S-nitrosylation of SsrB in Salmonella cultures exposed to acidified NO2− but not in controls grown in NO3− (Fig. 1A). To test the hypothesis that Cys203 is a redox-active cysteine modifiable by RNS, we took advantage of the C-terminal position of this residue to engineer a serine substitution by the λ Red system. The C203S substitution is a conservative mutation, because the redox thiol is exchanged for a nonreactive hydroxyl group. Wild-type and SsrB C203S variants expressed by AV07104 and AV08171 strains, respectively, were grown for 6 h in EG medium, pH 5.5, in the presence of 750 μM NO2−. Fig. 1B shows that Salmonella strains AV07104 and AV08171 harbor similar concentrations of the SsrB protein. However, SsrB is solely S-nitrosylated in the wild-type Salmonella strain AV07104 but not in its C203S-expressing isogenic control. The lack of S-nitrosylated SsrB in the ΔssrB::FRT strain AV0321 attests to the specificity of the assay. Collectively, these data demonstrate that Cys203 not only is redox active but also can be S-nitrosylated by the RNS generated by NO2− at an acid pH normally found in Salmonella-containing phagosomes.
Fig. 1.
SsrB Cys203 is S-nitrosylated in Salmonella cultures grown in acidified NO2−. (A) Salmonella were grown in EG medium, pH 5.5, in the presence of 750 μM NO3− or NO2−. S-nitrosothiols were derivatized in the biotin switch assay. SsrB::3xFLAG was detected in unfractionated bacterial cytoplasmic extracts (Upper) or in affinity-purified, biotinylated fractions (Lower). (B) S-nitrosylated SsrB was determined in Salmonella strains AV0321 (ΔssrB), AV07194 (WT; ssrB::3xFLAG), or AV08171 (SNO; C203S; ssrB C203S::3xFLAG) grown for 6 h in 750 μM NO2− EG medium, pH 5.5. Representative data from two or three independent experiments are shown.
DNA-Binding Activity of SsrB Is Selectively Inactivated by NO Congeners.
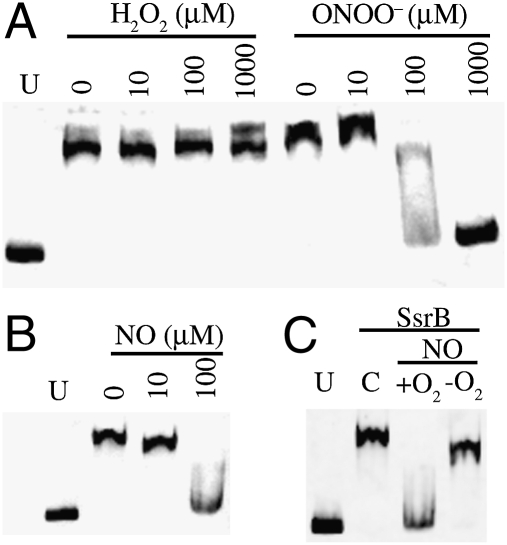
The effects that RNS have on the DNA-binding capacity of SsrB were used to gain insights into the biological relevance of the redox-active Cys203. We directed our efforts to the study of the DNA-binding, SsrB C-terminal domain (SsrBc) that encompasses amino acids 137–212, including Cys203. SsrBc was found to bind in a concentration-dependent manner to the ssrA promoter (Fig. S2A). Binding was abrogated with 100- to 200-fold excess of unlabeled ssrA or srfH SPI2 promoters but not by a similar-sized DNA fragment from the rpoD gene (Fig. S2B). The specificity in the interaction between SsrBc and the ssrA DNA was demonstrated further by the supershift noted when antibodies to the His-tag were added to the reaction (Fig. S2C). Because the affinity of SsrBc was higher for ssrA than for srfH, the DNA fragment containing the ssrA promoter was used subsequently as a readout to measure the sensitivity of SsrB to oxidative and nitrosative stress. The binding activity of SsrBc to its ssrA cognate promoter was inhibited by 100 μM ONOO− but not by 1 mM H2O2 (Fig. 2A). These data suggest that SsrB is modified selectively by ONOO− but not H2O2, both of which are produced by Salmonella-infected macrophages (8). That SPI2 transcription can be inhibited in gp91phox-deficient macrophages unable to synthesize ONOO− (8) indicates that other RNS also may inhibit SsrB binding to DNA. Consequently, NO itself was tested for its ability to modify the DNA-binding capacity of SsrBc. Addition of 100 μM authentic NO prevented the formation of an SsrBc-ssrA complex (Fig. 2B). In the absence of O2, however, NO did not affect the binding of SsrBc to ssrA (Fig. 2C). These results indicate that nitrosative and/or oxidative species such as NO2• and N2O3 generated during the autooxidation of NO in the presence of O2 can inhibit the DNA-binding activity of SsrBc.
Fig. 2.
Effect of ROS and RNS on the ability of SsrBc to bind to its ssrA cognate promoter. Effect of increasing concentrations of authentic H2O2, ONOO−, (A) or NO (B) on the binding of SsrBc to ssrA DNA. (C) Effects of NO autooxidation products on the binding of SsrBc to the ssrA promoter. The blot shows the binding of SsrBc with ssrA in untreated (U), control (C), or NO-treated samples in the presence or absence of O2. Representative data from two independent experiments are shown.
Oxidation of Cys203 Inactivates the DNA-Binding Capacity of SsrB.
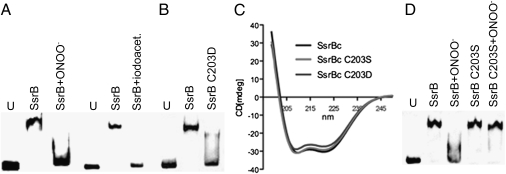
Mass spectrometry revealed that ONOO− oxidizes Cys203 to a mixture of sulfenic (−SOH), sulfinic (−SO2−), and sulfonic (−SO3−) groups (Fig. S3). It therefore is possible that the RNS-mediated oxidation of the thiol group in Cys203 inhibits binding of SsrBc to DNA. In support of the notion that oxidation of the redox-active thiol group of Cys203 can alter the function of SsrB, the thiol-specific oxidizer iodoacetate effectively inhibited SsrB DNA binding (Fig. 3A). Based on the oxidation patterns seen in the mass spectrometric analysis, genetic studies were performed to evaluate whether the oxidized, negatively charged Cys203 could compromise the DNA-binding capacity of SsrB. A C203D substitution was generated to mimic the negative charge of the SO2− and SO3− modifications of ONOO−-treated SsrBc. An SsrBc C203D variant failed to bind to the ssrA promoter (Fig. 3B). The failure of SsrBc C203D to bind to ssrA does not appear to reflect overall changes in secondary structure, because circular dichroism spectra revealed an α-helical pattern similar to the wild-type protein (Fig. 3C). Nor does an SsrBc C203S variant affect the α-helical structure of SsrBc (Fig. 3C). In contrast to the SsrB C203D allele, the C203S variant bound to the ssrA promoter with affinity apparently similar that of the wild-type protein (Fig. 3D). These findings demonstrate that the cysteine in SsrBc is not required for DNA binding. Nonetheless, Cys203 provides a mechanism for redox sensing, because the SsrBc variant bearing the C203S allele retained its ability to bind to the ssrA promoter after the protein was treated with 1 mM ONOO− (Fig. 3D). Together, these data suggest that Cys203 serves as a redox sensor of RNS, modulating the binding of SsrB to DNA.
Fig. 3.
Effects of the oxidation of Cys203 on SsrB DNA binding. (A) Binding of 300 ng SsrBc to 20 fmol of its cognate ssrA promoter after treatment with 1 mM ONOO− or 25 mM iodoacetate. The unbound, biotinylated ssrA DNA probe (U) is shown for comparison. (B and D) DNA binding of the SsrBc C203D or C203S alleles. Where indicated, selected samples in D were treated with 1 mM ONOO− before binding to the DNA. (C) Circular dichroic (CD) spectra of 0.2 mg/mL SsrBc recombinant variants. Representative data from two or three independent experiments are shown.
SsrB C203S Variant Sustains SPI2 Expression.
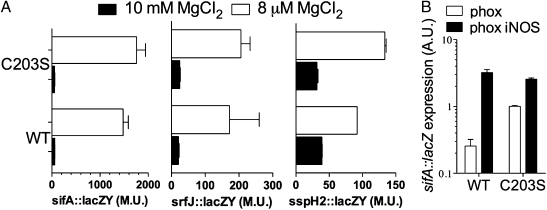
We tested whether the SsrBc C203S variant can stimulate SPI2 expression. As expected, the sifA::lacZY, srfJ::lacZY, and sspH2::lacZY transcriptional fusions were induced in wild-type Salmonella grown in 8 μM MgCl2 N salts medium, pH 6.9 (Fig. 4A). Salmonella expressing the SsrB C203S variant also supported the expression of the sifA::lacZY, srfJ::lacZY, and sspH2::lacZY transcriptional fusions. Of note, the expression of sifA::lacZY and sspH2::lacZY was slightly higher in Salmonella bearing the ssrB C203S allele. Because of their wild-type nitrosative capacity but inability to kill Salmonella (8, 14), macrophages lacking the gp91phox subunit of the NADPH oxidase were used to determine the contribution of the redox-active Cys203 to the transcriptional activity of SsrB in Salmonella exposed to host-derived RNS. Macrophages deficient in gp91phox iNOS hemoproteins unable to sustain nitrosative stress were used as controls. The intracellular expression of a sifA::lacZY transcriptional fusion was higher in the strain AV08270 expressing SsrB C203S than in the AV08267 control bearing the wild-type ssrB allele (Fig. 4B). The lack of iNOS abrogated the differences in sifA::lacZY expression in these two strains, suggesting that the presence of a redox-active Cys203 modulates the transcriptional activity of SsrB in response to RNS produced in the innate response of macrophages.
Fig. 4.
Regulatory functions of SsrB C203S. (A) SPI2 expression was induced in wild-type and ssrB C203S-expressing Salmonella by shifting the bacteria grown to an OD600 of 0.5 in 10 mM MgCl2 N9 medium into 8 μM MgCl2 N9 medium. Expression of SPI2 genes quantified as β-galactosidase of the lacZY transcriptional fusions is expressed in Miller Units (M.U.) 3 h after the shift. The data represent eight observations from three independent experiments. (B) Transcriptional sifA::lacZY activity also was measured in gp91phox (phox) and gp91phox iNOS-deficient (phox iNOS) macrophages as described in SI Materials and Methods. Data are represented as arbitrary units of light/106 bacteria (A.U.) ± SEM from four independent observations.
Redox-Active Wild-Type SsrB Allele Increases Salmonella Fitness in a Model of Acute Intestinal Infection.
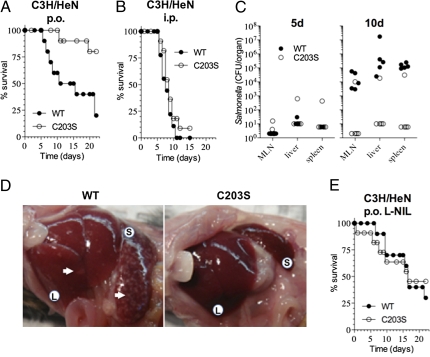
Given that the RNS-resistant SsrB C203S allele sustains SPI2 transcription (Fig. 4), it seems paradoxical that diverse populations of S. enterica adapted to a wide range of vertebrate hosts maintain a redox-sensitive wild-type ssrB allele in their genome. We reasoned that the presence of a redox-sensitive cysteine in the dimerization domain of SsrB may prove advantageous at some point during the association of this enteropathogen with its vertebrate host. Because Salmonella is transmitted primarily through contaminated food and water, we tested the virulence of wild-type and its ssrB C203S variant in a model of acute gastrointestinal infection. C3H/HeN mice were selected in these studies because the presence of the wild-type NRAMP1R locus is associated with increased NO production by macrophages (22). Strain AV07104 expressing the wild-type ssrB allele exhibited higher virulence than its isogenic ssrB C203S variant when tested in an NRAMP1R C3H/HeN murine model of acute oral infection (Fig. 5A). The Salmonella burden in mesenteric lymph nodes, liver, and spleen was similar for the wild-type and the SsrB C203S variant expressing bacteria 5 d after oral infection (Fig. 5C, Left). However, 103–105 times more wild-type bacteria were isolated from most mice 10 d after oral infection (Fig. 5C, Right). Hepatomegaly and splenomegaly were evident 10 d after C3H/HeN mice had been infected with wild-type Salmonella (Fig. 5D), probably reflecting increases in bacterial burden (Fig. 5C) and the inflammatory response. In fact, abscesses were noted on the surface of livers and spleens of C3H/HeN mice 10 d after infection with wild-type bacteria (Fig. 5D). The virulence of Salmonella bearing the wild-type ssrB allele in a C3H/HeN mouse model of acute oral infection appears to be related to the RNS-sensing capability of this response regulator, because bacteria expressing the RNS-resistant SsrB C203S allele regained full pathogenicity in C3H/HeN mice treated with 500 μg/mL of the iNOS-specific inhibitor N6-(1-iminoethyl)-l-lysine, dihydrochloride (l-NIL) administered daily in drinking water (Fig. 5E). In contrast to the differences noted when the bacteria were tested in an oral model of infection, wild-type and ssrB C203S-expressing Salmonella exhibited identical virulence (P = 1.0, log-rank Martel–Cox test) when tested in an acute i.p. C3H/HeN model of systemic infection (Fig. 5B).
Fig. 5.
The wild-type, RNS-sensitive SsrB allele provides a selective advantage in an acute model of oral salmonellosis. The virulence of wild-type (WT) Salmonella strain AV07104 or its isogenic SsrB C203S variant strain AV08171 (C203S) was investigated in acute intragastric and i.p. models of infection. (A) Survival of NRAMP1R C3H/HeN mice challenged per os (p.o.) with 107 cfu Salmonella. (B) NRAMP1R C3H/HeN mice were inoculated i.p. with 3 × 103 cfu of Salmonella. (C) The bacterial burden was recorded in mesenteric lymph nodes (MLN), liver, and spleen 5 and 10 d after C3H/HeN mice were inoculated p.o. with 107 cfu of Salmonella. (D) Gross pathological findings of C3H/HeN mice infected p.o. with Salmonella for 10 d. Liver (L) and spleen (S) are identified; white arrows indicate hepatic and splenic abscesses. (E) Effect of 500 μg/mL of the iNOS-inhibitor l-NIL administered in drinking water on survival of C3H/HeN mice infected p.o. with 107 cfu of Salmonella. Data represent two independent experiments (n =5–11 mice per group).
Discussion
Thiol groups can undergo oxidation and/or S-nitrosylation in bacteria exposed to RNS (23, 24). Our investigations have identified the two-component response regulator SsrB Cys203 as a thiol-based, redox sensor. A wild-type SsrB bearing a redox-active thiol in the Cys203 is advantageous to the interactions of Salmonella with its host, as demonstrated by the increased fitness of wild-type Salmonella over an isogenic strain expressing the RNS-resistant SsrB C203S variant in a C3H/HeN murine model of acute oral salmonellosis. The full recovery of virulence of Salmonella expressing the RNS-resistant SsrB C203S variant upon l-NIL treatment argues that Cys203 is a redox sensor of reactive species generated by the iNOS hemoprotein. The redox-sensing capabilities of SsrB appear to be dispensable for virulence of Salmonella in a model of acute systemic infection that is dominated by the antimicrobial activity of the NADPH phagocyte oxidase. This observation is in keeping with the fact that Cys203 in SsrB is relatively resistant to oxidation by H2O2.
Alignment of the ssrB allele of Salmonella isolates logged in the GenBank database shows that most typhoidal and nontyphoidal Salmonella strains encode for a conserved cysteine at position 203. The only exception is an isolate of S. Typhimurium strain St. Paul, which instead encodes for a tyrosine. Similar to cysteine, tyrosine is a common target of ONOO− (9). Also, in analogy to cysteine oxidation, the nitrotyrosine formed upon ONOO− treatment can disrupt protein function. The expression of cysteine or tyrosine residues at position 203 of SsrB raises the intriguing possibility that a redox-active moiety at this position provides Salmonella with a selective advantage. Data presented herein indicate that the fitness afforded by Cys203 is manifested in the context of RNS generated by the host response to Salmonella encountered in an oral infection. The expression of the SPI2 type III secretion system is essential for Salmonella pathogenesis. Nonetheless, its overexpression attenuates the virulence of Salmonella at later time points of the infection (20). In this context, the transcriptional control of SPI2 expression through the RNS-mediated modifications of SsrB Cys203, analogous to the importance that YdgT- and Hha-mediated silencing, is important for Salmonella virulence (20, 25).
Chemical production of RNS by the acidification of NO2− in the gastric lumen and enzymatic synthesis of NO from the oxidation of l-arginine by epithelial, inflammatory, and stromal cells are likely sources of NO in the gastrointestinal tract (2, 13). The nitrosative chemistry generated by acidified NO2− not only inhibits SPI2 expression (13) but also, as shown herein, S-nitrosylates SsrB. The advantage of Salmonella expressing the wild-type, RNS-responsive ssrB allele is evident in mice on an NRAMP1R background, indicating that sensing of RNS by Cys203 must take place within macrophages where the NRAMP1 metal transporter is expressed (26). Accordingly, the increased bacterial burden of Salmonella expressing the wild-type ssrB locus was most evident after 10 d of infection. A Cys203 residue could be advantageous for Salmonella in at least three different ways. First, the RNS-dependent repression of SPI2 is concomitant with the transcriptional up-regulation of the SPI1 type III secretion system, flagella, and fimbrial operons (13). Thus, RNS may favor the expression of Salmonella virulence factors that promote invasion, inflammation, adhesion, and motility in the gastrointestinal phase of the infection. The effects of RNS on these Salmonella virulence factors are likely to occur in the gastrointestinal lumen, before the NRAMP1-dependent, RNS-mediated effects take place. Therefore, we do not favor the idea that SsrB Cys203 helps Salmonella sense RNS in the gastrointestinal lumen. Second, RNS inhibit Salmonella-induced apoptosis (3), in which the SPI2 type III secretion system plays a critical role (27). In this context, the inhibition of phagocyte cell death by the RNS-dependent repression of SPI2 may be advantageous later in the course of infection. Third, redox control of SsrB may down-regulate intracellular SPI2 expression and thereby minimize detection by T and B lymphocytes. The second and third models are not necessarily mutually exclusive.
Biochemical analyses have revealed that Cys203 can undergo several covalent modifications after Salmonella or recombinant SsrBc protein is exposed to a variety of NO congeners. The thiol group of Cys203 is S-nitrosylated in Salmonella challenged with acidified NO2−, whereas recombinant SsrBc is oxidized upon exposure to ONOO−. The range of covalent modifications induced by NO congeners unequivocally demonstrates the redox nature of Cys203. Structural studies have shown that the Cys203 side chain contacts Leu192 in the homodimer interface (28). The negatively charged modifications of ONOO−-treated SsrB may block these interactions and thereby inhibit the formation of dimers necessary for the binding of SsrB to SPI2 promoters. Binding of SsrB to DNA also is inhibited by NO autooxidation products. N2O3-mediated oxidation of the Cys203 thiol group may have mechanistic consequences for binding of SsrB to DNA, similar to those seen in ONOO−-treated protein or an SsrB Asp203 variant. It also is possible that S-nitrosylation of Cys203 may itself interfere with the binding of SsrB with DNA or may promote formation of mixed disulfides with glutathione or other small molecular weight thiols, thereby increasing the complexity of the side chain of Cys203.
Redox-active Cys203 is not equally responsive to ROS and RNS. The DNA-binding activity of SsrB is preserved after treatment with H2O2 or NO but is inhibited after exposure to ONOO−, oxidative byproducts of NO, or the thiol oxidizer iodoacetate. The apparent susceptibility of the Cys203 thiol group to ONOO−-mediated oxidation is reminiscent of the preferential reactivity of ONOO− over H2O2 with thiols in serum albumin (k = 2,700 M−1s−1 vs. k = 2.26 M−1s−1, respectively) (29). Our observation also is consistent with the fact that most protein sulfhydryls in Escherichia coli are differentially affected by RNS and H2O2 (24). Functionally, the preservation of DNA binding in H2O2-treated SsrB may help explain why SPI2 protects Salmonella from NADPH phagocyte oxidase-derived oxyradicals (19, 30). The resistance of SsrB to biologically relevant ROS also may explain the comparable virulence of Salmonella strains expressing the wild-type or the redox-resistant SsrB C203S variant in the i.p. model of infection, because this phase is dominated by the antimicrobial activity of the NADPH phagocyte oxidase.
The number of sensors of nitrosative stress in bacteria continues to increase. The transcriptional regulators [2Fe-2S] SoxR and [4Fe-4S] Fnr serve as examples (31, 32). To the best of our knowledge, OxyR is the only identified bacterial regulator in which sulfhydryls have been co-opted as sensors of nitrosative stress (33). In this respect, Cys203 in the C-terminal H4 helix of the dimerization domain of SsrB serves a function analogous to that of OxyR Cys199. There are, nonetheless, significant differences between these two bacterial regulators. The ability of OxyR to sense nitrosative stress appears to be secondary to its recognition of endogenous H2O2. In contrast, as discussed above, SsrB appears to be more specific for RNS than ROS. Furthermore, S-nitrosylation of Cys203 is manifested readily in wild-type Salmonella, whereas the RNS-dependent modifications of OxyR are best noted in E. coli lacking the reductive power of glutathione (33). Another important difference between these two regulators lies in the activation of DNA binding by OxyR in response to nitrosative stress, whereas RNS inhibit the formation of SsrB-DNA complexes. In this sense, Cys203 of SsrB serves a function analogous to that of Cys13 of the Staphylococcus global regulator SarZ, whose DNA-binding activity is disrupted upon oxidation with H2O2 or organic hydroperoxides (34).
In conclusion, Cys203 in the C-terminal dimerization domain of the SsrB response regulator is a sensor of RNS. RNS-mediated posttranslational modifications of Cys203 repress the binding of SsrBc to DNA. This redox-active cysteine provides Salmonella with a selective advantage in the context of nitrogen oxides encountered when the infection takes place through the oral mucosa. The widespread occurrence of a cysteine at this position in SsrB of medically important Salmonella isolates indicates that RNS exert a positive selective pressure in the effector domain of SsrB in both typhoidal and nontyphoidal strains of Salmonella enterica.
Materials and Methods
Bacterial Strains.
Salmonella enterica serovar Typhimurium strain ATCC 14028s was used as wild type and as the background for the construction of ssrB mutations and lacZY-transcriptional fusions (Table S1). Bacterial strains were constructed using primers in Table S2 following the λ Red method as described in SI Materials and Methods.
EMSA.
EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific,) as described in SI Materials and Methods.
In Vivo Detection of S-Nitrosylated SsrB Protein.
S. Typhimurium strains AV07104 and AV08171 expressing 3xFLAG-tagged SsrB and 3xFLAG-tagged SsrB C203S variants were grown overnight in LB medium. Strain AV321 (ΔssrB::FRT) was used as a negative control. The bacteria were subcultured at an OD600 of 0.3 in EG medium, (0.2 g/L MgSO4, 2 g/L C6H8O7-H2O, 10 g/L K2HPO4, 3.5 g/L Na(NH4)HPO4-4 H2O, and 4 g/L d-glucose) pH 5.5, with 750 μM NaNO2− or NaNO3− at 37 °C for 6 h, with shaking. Bacterial soluble lysates were obtained by sonication in 250 mM Hepes, 1 mM EDTA, 0.1 mM neocuproine (HEN) buffer, pH 7.7, and the protein concentration in the clear lysates adjusted to 0.8 mg/mL. S-nitrosylation was detected by the biotin switch method (35) as described in SI Materials and Methods.
Circular Dichroism Spectroscopy.
Circular dichroism spectroscopy was performed on a Jasco-810 spectrometer as described in SI Materials and Methods.
Transcriptional Analysis.
Transcription of SPI2 genes was induced in vitro by culturing Salmonella in 8 μM MgCl2 N salts medium (36). SPI2 transcriptional analysis also was studied in gp91phox and gp91phox iNOS-deficient macrophages exhibiting different nitrosative capacities. The methods are described in SI Materials and Methods.
Virulence of Salmonella in Murine Models of Infection.
C3H/HeNcrl/Br mice (8–10 wk old) were used to assess oral virulence of Salmonella as described in SI Materials and Methods.
Statistical Analysis.
Data are presented as means ± SEM or SD. The statistical significance was calculated with a two-way ANOVA, followed by a Bonferroni post test. Differences in mouse survival after Salmonella infection were determined by a log-rank Martel–Cox test. Data were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Linda J. Kenney for helpful discussions, Jessica Prenni for help with the mass spectrometry, and the Biophysics Core Facility at the University of Colorado Denver for assistance with the CD spectrometry. This work was supported by National Institutes of Health Grants AI054959, AI053213, AI07447, RR16082, and T32 AI52066 and by the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005299107/-/DCSupplemental.
References
- 1.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enocksson A, Lundberg J, Weitzberg E, Norrby-Teglund A, Svenungsson B. Rectal nitric oxide gas and stool cytokine levels during the course of infectious gastroenteritis. Clin Diagn Lab Immunol. 2004;11:250–254. doi: 10.1128/CDLI.11.2.250-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam MS, et al. Role of nitric oxide in host defense in murine salmonellosis as a function of its antibacterial and antiapoptotic activities. Infect Immun. 2002;70:3130–3142. doi: 10.1128/IAI.70.6.3130-3142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastroeni P, et al. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren B, Zhang N, Yang J, Ding H. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol Microbiol. 2008;70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Husain M, et al. Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J Biol Chem. 2008;283:7682–7689. doi: 10.1074/jbc.M708845200. [DOI] [PubMed] [Google Scholar]
- 8.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ischiropoulos H. Biological tyrosine nitration: A pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 10.Castro L, Rodriguez M, Radi R. Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 11.Herold S, Röck G. Mechanistic studies of S-nitrosothiol formation by NO*/O2 and by NO*/methemoglobin. Arch Biochem Biophys. 2005;436:386–396. doi: 10.1016/j.abb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Björne H, Weitzberg E, Lundberg JO. Intragastric generation of antimicrobial nitrogen oxides from saliva—physiological and therapeutic considerations. Free Radic Biol Med. 2006;41:1404–1412. doi: 10.1016/j.freeradbiomed.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Bourret TJ, et al. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One. 2008;3:e1833. doi: 10.1371/journal.pone.0001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCollister BD, et al. N2O3 enhances the nitrosative potential of IFNγ-primed macrophages in response to Salmonella. Immunobiology. 2007;212:759–769. doi: 10.1016/j.imbio.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravortty D, Hansen-Wester I, Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med. 2002;195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiya K, et al. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 1999;18:3924–3933. doi: 10.1093/emboj/18.14.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez-Torres A, et al. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 20.Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc Natl Acad Sci USA. 2005;102:17460–17465. doi: 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCollister BD, Bourret TJ, Gill R, Jones-Carson J, Vázquez-Torres A. Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. J Exp Med. 2005;202:625–635. doi: 10.1084/jem.20050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Formica S, Roach TI, Blackwell JM. Interaction with extracellular matrix proteins influences Lsh/Ity/Bcg (candidate Nramp) gene regulation of macrophage priming/activation for tumour necrosis factor-α and nitrite release. Immunology. 1994;82:42–50. [PMC free article] [PubMed] [Google Scholar]
- 23.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci USA. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandes N, Rinck A, Leichert LI, Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol. 2007;66:901–914. doi: 10.1111/j.1365-2958.2007.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fass E, Groisman EA. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol. 2009;12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabado N, et al. Natural resistance to intracellular infections: Natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect Immun. 2000;68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll RK, et al. Structural and functional analysis of the C-terminal DNA binding domain of the Salmonella typhimurium SPI-2 response regulator SsrB. J Biol Chem. 2009;284:12008–12019. doi: 10.1074/jbc.M806261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 30.Gallois A, Klein JR, Allen LA, Jones BD, Nauseef WM. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J Immunol. 2001;166:5741–5748. doi: 10.4049/jimmunol.166.9.5741. [DOI] [PubMed] [Google Scholar]
- 31.Nunoshiba T, deRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci USA. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz-Ramos H, et al. NO sensing by FNR: Regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Nitrosative stress: Activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 34.Chen PR, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;86:1–9. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 36.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.