Abstract
The spindle assembly checkpoint (SAC) is essential for proper sister chromatid segregation. Defects in this checkpoint can lead to chromosome missegregation and aneuploidy. An increasing body of evidence suggests that aneuploidy can play a causal role in tumorigenesis. However, mutant mice that are prone to aneuploidy have only mild tumor phenotypes, suggesting that there are limiting factors in the aneuploidy-induced tumorigenesis. Here we provide evidence that p53 is such a limiting factor. We show that aneuploidy activates p53 and that loss of p53 drastically accelerates tumor development in two independent aneuploidy models. The p53 activation depends on the ataxia-telangiectasia mutated (ATM) gene product and increased levels of reactive oxygen species. Thus, the ATM-p53 pathway safeguards not only DNA damage but also aneuploidy.
Faithful transmission of genetic materials is of fundamental importance to the survival of all organisms. In eukaryotes, replicated chromosomes are held together as sister chromatids by the cohesin complexes established during the replication and are segregated to daughter cells in mitosis. The timing of the sister chromatid separation is controlled by the spindle assembly checkpoint (SAC), which monitors the status of microtubule attachment at kinetochores. The SAC is activated when kinetochores are not attached (i.e., occupied) by microtubules and/or when there is a lack of tension at sister kinetochores (1, 2), under both of which situations separation of sister chromatids needs to be actively prevented or missegregation of chromosomes would ensue. The activation of SAC leads to the inhibition of the anaphase-promoting complex or cyclosome (APC/C), a multisubunit E3 ubiquitin ligase that targets securin and cyclin B1 for destruction (3–6). Both securin and cyclin B1 are recognized and brought to APC/C by the adaptor protein Cdc20. Not surprisingly, APCCdc20 is inhibited by the SAC. The inhibition is carried out by two proteins, Mad2 and BubR1 (for more detailed and recent reviews, see refs. 7 and 8). Genetic analyses in budding yeasts unequivocally demonstrated that the spindle assembly checkpoint was essential in maintaining chromosomal stability (9, 10). Studies of engineered mouse strains carrying mutations in SAC components also indicated the importance of the checkpoint in maintaining chromosome stability (11–19), and BUBR1 was found mutated in a rare human disorder, mosaic variegated aneuploidy (20).
A hallmark of human cancers is genomic instability including chromosomal instability (CIN). CIN can be numerical changes in whole chromosomes (aneuploidy) or structural alterations such as translocations. Aneuploidy is found in nearly all of the major human tumor types (21), and it was the abnormal chromosome numbers in cancerous cells that prompted Boveri to propose nearly a century ago that cancer was caused by aneuploidy (22). Nearly all SAC-compromised mouse strains develop spontaneous tumors, although the rates vary substantially (for a summary see ref. 23). Together with the finding of BUBR1 mutation in mosaic variegated aneuploidy, a condition that predisposes patients to childhood cancers (20), the tumor results in SAC mutants strongly argue that aneuploidy can induce tumorigenesis. However, the spontaneous tumor development in SAC mutant mice is usually late onset and at relatively low rates, indicating that aneuploidy does not present a serious risk of tumor development. The low risk of aneuploidy-induced tumorigenesis suggests that there are limiting factors. One such factor could be the general unfitness of aneuploid cells when compared with euploid cells, which is true from budding yeasts to mammals (24, 25). This unfitness likely stems from the imbalances in gene dosage that lead to changes in a score of physiological parameters including energy metabolism (24, 25). Mouse embryonic fibroblasts (MEFs) trisomy for chromosome 1, 13, 16, or 19 grew less robustly than the diploid MEFs and showed resistance to transformation (24). These findings support the notion that aneuploidy can be tumor suppressing under certain circumstances (15, 26). However, the fact that most human cancer cells are aneuploid (21) indicates that there must be ways to overcome the unfitness barrier and that once this barrier is overcome, aneuploidy is beneficial to the tumor development.
Here we report that p53 is another limiting factor in aneuploidy-induced tumorigenesis. We provide evidence that p53 is activated by aneuploidy and the activation depends on ataxia-telangiectasia mutated (ATM). We further show that aneuploidy resulted in heightened energy metabolism and increased levels of intracellular reactive oxygen species, which caused oxidative DNA damage and ATM activation.
Results
p53 Is Activated in SAC-Deficit Cells.
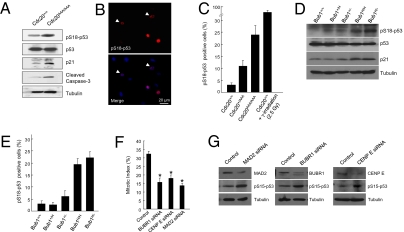
We previously reported the generation of a Cdc20 allele (Cdc20AAA) that harbors three point mutations in the Mad2-interacting motif (19). Cdc20AAA can no longer be inhibited by Mad2 and its association with BubR1 also is substantially impaired, resulting in a dysfunctional SAC. Cdc20AAA/AAA mice are embryonic lethal, whereas Cdc20+/AAA mice develop normally but succumb to spontaneous tumors late in life. MEFs derived from Cdc20AAA/AAA mice are highly aneuploid and grow poorly (19). In an effort to elucidate the mechanism(s) behind the poor growth, we discovered that p53 is activated in these cells (Fig. 1A). As expected, p21Cip1 (Cdkn1a) was elevated, as was active caspase 3 (Fig. 1A). Caspase 3 activation might explain the embryonic lethality of Cdc20AAA/AAA mice and the high apoptosis rates in MEFs derived from these mice (19). Interestingly, despite the increase in the level of phosphorylated p53, we did not observe a substantial increase in the total amount of p53 protein in the mutant MEFs. This could be explained if only a fraction of the mutant cell population contained the activated p53. To test this idea, we immunostained Cdc20AAA/AAA MEFs for Ser18-phosphorylated p53. Indeed, a broad range of labeling intensities was observed, from strong to weak (Fig. 1B). Overall, the percentages of Ser18-phosphorylated p53-positive Cdc20AAA/AAA, Cdc20+/AAA, and wild-type MEFs were 25, 10, and 3%, respectively (Fig. 1C). In contrast, >80% of wild-type MEFs were Ser18-phosphorylated p53-positive following irradiation (Fig. 1C).
Fig. 1.
p53 is activated in SAC-deficient cells. (A) Western blot analysis of wild-type and Cdc20 mutant MEFs. (B) Immunofluorescent staining of Ser18-phosphorylated p53. Arrowheads indicate more weakly stained cells. (C) Quantitation of the results of B. (D) Western blot analysis of wild-type and Bub1 mutant MEFs. (E) Quantitation of Bub1 mutant MEFs immunostained for phospho-Ser18 p53. (F) Mitotic index analysis of HCT116 cells depleted of BUBR1, CENP E, or MAD2. * indicates statistical significance between control and knockdown cells. (G) Western blot analysis of the cells in D.
Next, we asked whether the activation of p53 is unique to the effect of the point mutations in Cdc20 or a more common response to SAC deficiencies. We first examined MEFs with graded reduction in Bub1 expression (14). Western blot analysis demonstrated that levels of Ser18-phosphorylated p53 and p21 were indeed elevated in Bub1H/H and Bub1H/− MEFs, but not in wild-type or Bub1+/H or Bub1+/− cells (Fig. 1D). Overall, Bub1H/− (lowest Bub1 expression) MEFs had 22% pS18-p53–positive cells by immunostaining, and Bub1H/H MEFs had 20% (Fig. 1E). Similar to the result of Western analysis (Fig. 1D), Bub1+/H cells contained essentially the same percentage of p53-positive cells as the wild-type control, whereas the percentage in Bub1+/− MEFs was slightly above background (Fig. 1E). Thus, there is a correlation between p53 activation and the level of aneuploidy in Bub1-insufficient cells.
We then depleted the expression of three other mitotic regulators, MAD2, BUBR1, and CENP-E in HCT116 cells via siRNA. HCT116 is a human colon cancer cell line that maintains a near diploid karyotype and is chromosomal stable (27). Knocking down these three genes impaired the spindle checkpoint function (Fig. 1F) and caused activation of p53 (Fig. 1G). Together, these results suggest that loss of spindle assembly checkpoint function can lead to p53 activation.
Activation of p53 Cannot Be Suppressed by Restoring SAC Function.
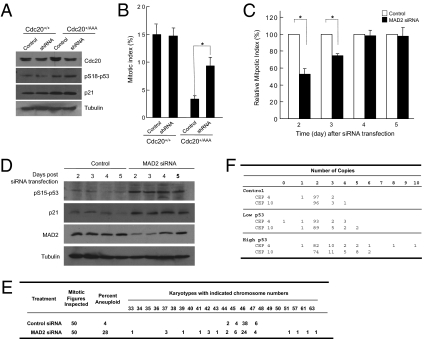
To determine whether the defect in SAC function itself is the cause of p53 activation, we restored SAC function in the mutant cells. First, we depleted the expression of mutant Cdc20 in Cdc20+/AAA MEFs via shRNA designed specifically against Cdc20AAA (by taking advantage of the unique sequence introduced by the mutations). The shRNA reduced the expression of total Cdc20 protein by ∼50% in Cdc20+/AAA MEFs (Fig. 2A). The cells could respond to nocodazole treatment after the depletion, although the response was not as robust as that of the wild-type MEFs (Fig. 2B). This is most likely due to residual AAA-Cdc20 protein. Nonetheless, there was not any decrease in the phospho-p53 level (Fig. 2A). Second, we sampled MAD2 knockdown HCT116 cells at various times after siRNA transfection. Four days after the transfection, both the expression of MAD2 and the SAC function were restored to the pretransfection level (Fig. 2 C and D). However, still ∼28% of the cells remained aneuploid at day 5 post-MAD2 siRNA transfection (Fig. 2E). These aneuploid cells must have resulted from previous mitoses when Mad2 function was impaired. Importantly, the level of p53 activation as well as the level of p21 expression remained the same even at day 5 after the MAD2 siRNA transfection. Thus, SAC dysfunction per se is unlikely the trigger of p53 activation. Rather, the consequence of SAC deficiencies, aneuploidy, is the most likely activator of p53.
Fig. 2.
The activation of p53 correlates with aneuploidy. (A) Western blot analysis of wild-type and Cdc20+/AAA MEFs treated with shRNA against Cdc20AAA. (B) Mitotic index analysis of the cells in A. (C) Mitotic index analysis of MAD2-depleted HCT116 cells. (D) Western blot analysis of MAD2-depleted HCT116 cells. (E) Karyotyping of analysis of MAD2-depleted HCT116 cells at 5 d after siRNA transfection. (F) FISH karyotyping with centromeric probes against chromosomes 4 and 10 (CEP 4 and CEP 10) of FACS-sorted, MAD2-depleted HCT116 cells. One hundred nuclei were scored. * Statistical significance (P < 0.05).
p53 Activation Correlates with Aneuploidy.
If it is aneuploidy that activates p53, there should be a correlation between p53 activation and aneuploidy. To demonstrate that, we FACS sorted Ser15-phospho p53-positive HCT116 cells after MAD2 knockdown (Fig. S1A). On the FACS profile, 5.6% of MAD2 knockdown cells were pS15-p53 positive, whereas 0.28% of control knockdown cells were (Fig. S1B). We divided the pS15-p53–positive MAD2 siRNA-transfected cells into two halves, one with high and one with low staining intensity. These two subpopulations plus the control cells were subjected to fluorescence in situ hybridization (FISH) analysis using centromeric probes for human chromosomes 4 and 10. Chromosome copies normally vary depending on cell cycle stage, with G1 and G2 cells having two and four copies, respectively, whereas S-phase cells can have two to four copies. Thus we counted the cells with copy numbers of two to four as normal. Above or below that range was considered as aneuploid. As shown in Fig. 2F, the cells with high levels of phospho-p53 are much more aneuploid (5% for chromosome 4 and 10% for chromosome 10) than those with low levels (2% for chromosome 4 and 3% for chromosome 10), and the aneuploidy was more severe (in terms of the number of chromosomes gained or lost) in the p53-high population than that in the p53-low population. The control cells showed a low background rate of aneuploidy (1% for chromosome 4 and 0% for chromosome 10). Given that the cells with high levels of active p53 were most likely in G1, those with three copies probably were aneuploid as well. Thus the percentage of aneuploidy in p53-positive cells was likely underestimated. Nonetheless, these data strongly correlate aneuploidy with p53 activation. It is unclear at present where the threshold for p53 activation lies, nor is it clear if certain combinations of chromosomes activate p53 stronger than others.
p53 Induces Apoptosis of Aneuploid Cells and Prevents Aneuploidy-Induced Transformation.
We showed previously that Cdc20AAA/AAA MEFs ceased proliferation by passage 8 due to continuous apoptotic cell death (19), which may be mediated by p53. If so, inactivation of p53 should rescue these cells. To that end, we bred the Cdc20AAA allele onto a p53 null background. Although p53 loss did not produce live-born Cdc20AAA/AAA mice, it did postpone embryonic death from embryonic day (E)12.5 to >E17.5. Importantly, Cdc20AAA/AAA/p53−/− MEFs did not show high rates of apoptosis as Cdc20AAA/AAA MEFs did and quickly became immortalized. Moreover, when thymocytes from adult mice were analyzed for apoptosis, we found that AAA-Cdc20 heterozygosity caused significant increases in the apoptotic death of both CD4+ and CD8+ thymocytes (Fig. S2). Those killed cells were most likely aneuploid and were eliminated by p53. Indeed, the increase in the number of cell deaths caused by AAA-Cdc20 was completely suppressed in Cdc20+/AAA/p53−/− mice (Fig. S2).
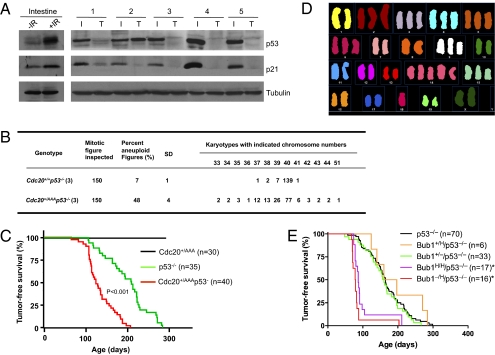
The above results suggest that p53 clears the body of aneuploid cells (at least those with high degree of aneuploidy). These results also predict that the p53 pathway would be disrupted in tumors that arise in Cdc20+/AAA mice. To examine this, we subjected tumor-bearing Cdc20+/AAA mice to 15 Gy γ-irradiation and harvested the small intestines and the tumors 4 h later. The radiation strongly induced p53 and p21 in the intestine, but in five of five tumors there was little response (Fig. 3A). Tumor 2 did show an accumulation of p53 (however, we do not know if the accumulation was induced by the irradiation or was there before the irradiation), but there was no p21 induction. The p53 protein in this tumor might be a mutant form.
Fig. 3.
Loss of p53 enhances tumorigenesis in SAC-deficient mice. (A) Western blot analysis of DNA damage-induced p53 response. Intestines from unirradiated and irradiated (15 Gy) wild-type mice (first two lanes) were included to show the normal p53 response. Intestines (I) and tumors (T) isolated from irradiated Cdc20+/AAA mice were analyzed. (B) Karyotyping of splenocytes. (C) Tumor-free survival analysis (Kaplan–Meier method) of Cdc20 mutant mice. (D) Spectrum karyotyping of one thymic tumor developed in a Cdc20+/AAAp53−/− mouse. (E) Tumor-free survival analysis (Kaplan–Meier method) of Bub1 mutant mice.
Loss of p53 Accelerates Tumor Development in Cdc20+/AAA Mice.
The results so far suggest that p53 should act as a barrier in aneuploidy-induced tumor development. To test this further, we monitored Cdc20+/AAA/p53−/− mice for tumor formation. We expected that, by removing p53, highly aneuploid cells that otherwise would have been eliminated by p53 could now survive (for example, see Fig. S2), leading to an increase in the rate of aneuploidy. Indeed, karyotyping of splenocytes in 1-mo-old mice indicated that the frequency of aneuploid cells was much higher in Cdc20+/AAA/p53−/− mice than that in p53 null mice (48% vs. 7%, Fig. 3B). It is also higher than the 35% observed in Cdc20+/AAA mice (19). More importantly, Cdc20+/AAAp53−/− splenocytes displayed more severe aneuploidy than Cdc20+/AAA splenocytes as indicated by the appearance of cells with karyotypes deviating farther from diploidy.
Although Cdc20+/AAA/p53−/− mice appeared normal, they quickly developed thymic lymphoma (Fig. 3C). The tumors were often so large (Fig. S4A) that they blocked the airway and caused the animals to die of suffocation. Within 6 mo, all Cdc20+/AAA/p53−/− mice had died. The median was ∼4 mo. It took >8 mo for all of the p53 null control mice to die of tumors and the median was >6 mo. The development of thymic lymphoma is in agreement with the finding that a large number of thymocytes were eliminated by p53 in Cdc20+/AAA mice (Fig. S2). The cells that were eliminated likely possessed high transforming potentials. Spectrum karyotyping (SKY) analysis showed the highly aneuploid nature of the Cdc20+/AAAp53−/− tumor cells (Fig. 3D, one representative example is shown). The aneuploidy is much worse than that of Cdc20+/AAA tumor cells (19).
To test if p53 plays a protective role in another aneuploidy model, we produced Bub1 deficiency mice under a p53 null background. Because the mice heterozygous for a Bub1 deletion or hypomorphic allele do not display a large increase in aneuploidy and are not very tumor prone (14), we did not observe an increase in the rates of tumorigenesis in Bub1+/Hp53−/− or Bub1+/−p53−/− mice when compared with p53 null mice (Fig. 3E). However, Bub1H/Hp53−/− and Bub1-/Hp53−/− mice rapidly developed thymic lymphomas in a manner similar to Cdc20+/AAAp53−/− mice (Fig. 3E). Thus, loss of p53 substantially accelerated aneuploidy-induced tumorigenesis in these aneuploidy models.
ATM Is Required for p53 Activation in Aneuploid Cells.
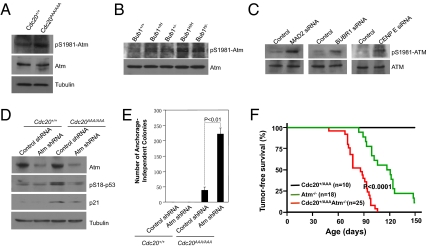
In a search for the kinase that phosphorylates p53 in SAC-deficient cells, we took a candidate approach. A number of known kinases capable of phosphorylating p53 were tested and ATM was found to be the kinase. As shown in Fig. 4 A–C, the activation of ATM (Ser1981 phosphorylation) was detected in Cdc20 mutant and Bub1-deficient MEFs as well as in HCT116 cells depleted of MAD2, BUBR1, or CENP-E. To determine if ATM is responsible for p53 activation in SAC-defective cells, we first inhibited ATM kinase with two chemical inhibitors. Indeed, both caffeine and wortmannin blocked p53 activation (Fig. S3). We then depleted Atm expression via shRNA in MEFs. Similarly, p53 activation was diminished in Atm-depleted cells (Fig. 4D). Furthermore, blocking Atm function dramatically increased the ability of Cdc20AAA/AAA MEFs to transform (Fig. 4E). These results indicate that ATM is responsible for the phosphorylation of p53 in SAC-deficient cells and prohibits the transformation of these cells.
Fig. 4.
ATM is required for p53 activation in SAC-deficient cells. (A) Western blot analysis of phosphorylated Atm in wild-type and Cdc20 mutant MEFs. (B) Western blot analysis of phosphorylated Atm in Bub1 mutant MEFs. For loading control, see Fig. 1D. (C) Western blot analysis of phosphorylated ATM in HCT116 cells. For loading control, see Fig. 1F. (D) Western blot analysis of wild-type and Cdc20 mutant MEFs depleted of Atm. (E) Colony formation assay. (F) Tumor-free survival analysis (Kaplan–Meier method).
These results also predict that the deletion of Atm should have a similar effect to the loss of p53 on aneuploidy-induced tumorigenesis. To test that, we bred the Cdc20AAA allele onto an Atm null background. As predicted, Atm−/−/Cdc20+/AAA mice very quickly developed similar gigantic thymic lymphoma as did Cdc20+/AAAp53−/− mice (Fig. 4F and Fig. S4B). In a little over 100 d, all of the double-mutant mice had had tumors and died, whereas it took >150 d for Atm null mice to do that. The Cdc20+/AAA control mice did not show any signs of tumors yet. These results indicate that Atm is a strong suppressor of aneuploidy-induced tumorigenesis.
Elevated Levels of Reactive Oxygen Species Are Responsible for the Activation of the ATM-p53 Pathway in Aneuploid Cells.
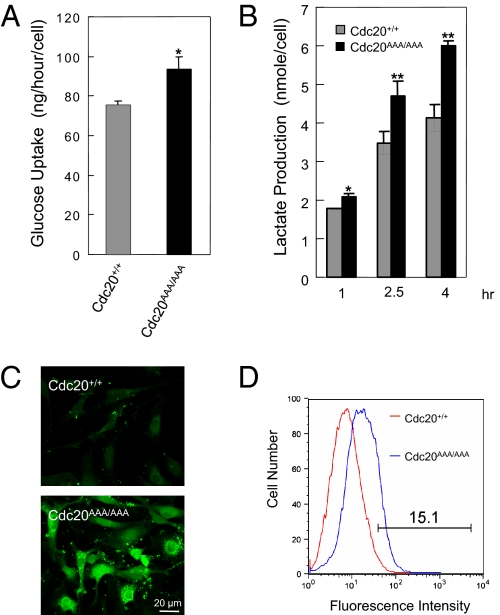
The activation of ATM and p53 in aneuploid cells suggests DNA damage might be involved. However, what could be the source of damage? Torres et al. (25) reported that aneuploid budding yeast cells display some common phenotypic characteristics regardless of which chromosome was in excess. These characteristics include increased energy metabolism. We suspected that aneuploid cells might consume more energy as the budding yeasts, resulting in an increased production of reactive oxygen species (ROS) that subsequently caused oxidative DNA damage. Indeed, alterations in energy metabolism were noted in MEFs engineered to be trisomic for chromosome 1, 13, 16, or 19 (24). To determine if Cdc20AAA/AAA MEFs (which should be more grossly aneuploid than those trisomic MEFs) consume more energy, we measured glucose consumption. Cdc20AAA/AAA MEFs indeed consumed significantly more glucose than the wild-type control (Fig. 5A). At the same time, the production of lactate was significantly increased by the mutant MEFs (Fig. 5B). We then measured ROS levels in control and mutant MEFs using the fluorescent dye CM-H2DCFDA. As shown in Fig. 5 C and D, >15% of Cdc20AAA/AAA MEFs contained much higher levels of ROS than the control cells.
Fig. 5.
Increased levels of reactive oxygen species in mutant MEFs. (A) Measurement of glucose consumption in MEFs. *P < 0.05. (B) Lactate production in MEFs. *P < 0.05; **P < 0.01. (C) Visualization of ROS with the fluorescent dye CM-H2DCFDA. (D) FACS analysis of the cells in C.
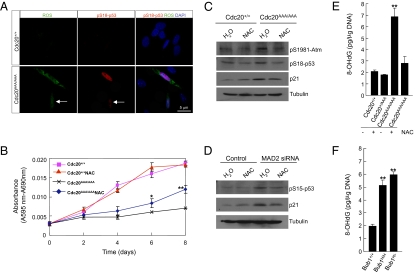
We then asked whether the elevated levels of ROS could be the signal that activated ATM-p53 in SAC-deficient cells. We first determined if the cells that contained high levels of ROS were also positive for phospho-p53. Live control and mutant MEFs were stained for ROS with CM-H2DCFDA and then fixed in 2% paraformaldehyde (PFA) and stained for Ser18-phospho p53. As shown in Fig. 6A, ROS-positive cells were also positive for phospho-p53 and there was a correlation between the level of ROS and the intensity of p53 staining. Next, we treated the MEFs with ROS scavenger (N-acetyl cysteine, NAC) to determine if reducing intracellular ROS levels would have an impact on proliferation and p53 activation. NAC greatly improved the growth of Cdc20AAA/AAA MEFs, whereas the scavenger had little effect on the growth of wild-type cells (Fig. 6B). More importantly, after only 4 h treatment, phosphorylation of both Atm and p53 was significantly reduced, and so was that of the target of p53, p21 (Fig. 6C). Finally, treating MAD2 knockdown HCT116 cells with NAC also reduced p53 phosphorylation (Fig. 6D).
Fig. 6.
ROS is responsible for ATM and p53 activation in SAC-deficient cells. (A) Visualization of ROS and phospho-p53. The cells were incubated with CM-H2DCFDA first and then processed for immunostaining of Ser18-phosphorylated p53. The arrows point to a cell with weak ROS signal and weak p53 staining. (B) Growth curve analysis of MEFs treated with NAC. *P < 0.05; **P < 0.01. (C) Western blot analysis of MEFs treated with N-acetyl cysteine (NAC). (D) Western blot analysis of HCT116 cells transfected with MAD2 siRNA and treated with NAC. (E) Analysis of the level of 8-OHdG in the genomic DNA in wild-type and Cdc20 mutant MEFs. **P < 0.01. (F) Same analysis as in E done in Bub1-deficient MEFs.
Having established that the increased ROS levels activate ATM and p53 in aneuploid cells, we asked if there were increases in the level of oxidative DNA damage in these cells. We therefore measured 8-hydroxy-2′-deoxyguanosine (8-OHdG) present in genomic DNA. As shown in Fig. 6 E and F, the amount of 8-OHdG per unit genomic DNA increased dramatically in both Cdc20AAA/AAA and Bub1-deficient MEFs. Treating Cdc20AAA/AAA MEFs with NAC effectively reduced the level of this by-product of oxidative DNA damage.
Despite the induction of oxidative DNA damage in aneuploid cells, we could not detect any γ-H2AX foci (Fig. S5A) or an increase in its levels (Fig. S5B). In addition, we failed to detect the activation of CHK1 and CHK2 in MAD2- or BUBR1-depleted HCT116 cells (Fig. S5C). These results suggest that the oxidative DNA damage activates ATM and p53 in a noncanonical way. Alternatively, the increased levels of ROS may directly cause ATM activation (28). Nonetheless, these data strongly suggest that ROS is the signal generated by aneuploidy that activates ATM and p53 to limit the proliferation of the aneuploid cells.
Discussion
Our results presented here demonstrate that aneuploidy can activate p53. A similar finding was reported recently through a completely different approach than the one used here (29). Together with the suggestion that aneuploid cells are genetically unfit for proliferation (24, 25), we propose that the cellular response to aneuploidy ranges from passive (unfitness) to active (activation of p53), depending on the severity of the aneuploidy (Fig. S6A). Highly aneuploid cells that are likely to have high potentials to be transformed are nonviable. They are eliminated by p53-mediated apoptosis. Mildly aneuploid cells may enter p53-mediated cell cycle arrest/senescence. Slightly aneuploid cells are genetically unfit and are hard to transform (24). Thus, slightly to mildly aneuploid cells do not present significant tumorigenic risks to the organisms. These cells can become more aneuploid if they divide again with mistakes, but the progeny will be eliminated by the further activated p53. In mice with weakened SAC function, cells with aneuploidy across the severity spectrum (Fig. S6A) are generated during embryogenesis and in tissues where there is a rapid turnover. Because the highly aneuploid cells are nonviable, the animals are left with only slightly to mildly aneuploid cells. Tumors can develop only when some of the highly aneuploid cells lose p53 by chance. Once the p53 pathway is inactivated, these cells can now survive and divide, acquire additional mutations, and start cancerous expansion. Thus, it takes a long time for tumors to develop in the mice with weakened SAC function and the penetrance usually does not reach 100%. However, when p53 is removed from the beginning, the latency of tumor development is drastically shortened and the penetrance becomes 100% (Fig. 3 C and E).
We demonstrate here that ATM is responsible for p53 activation in aneuploid cells. This conclusion is further supported by the finding that Atm−/−Cdc20+/AAA mice developed strikingly similar gigantic thymic lymphomas to Cdc20+/AAAp53−/− mice, strongly arguing that ATM and p53 function in the same pathway to suppress aneuploidy-induced tumorigenesis. Because ATM lies upstream of p53 and may target effectors other than p53, the tumor latency in Atm−/−Cdc20+/AAA was expectedly shorter than that in Cdc20+/AAAp53−/− mice. We showed that the ROS was responsible for the activation of ATM (Fig. 6B). ROS levels are increased in a subpopulation of SAC-deficient cells (Fig. 5A) and are correlated with p53 activation (Fig. 6A). Heightened energy metabolism in aneuploid cells (24, 25) is likely behind the surge in ROS levels. The increased ROS levels cause oxidative DNA damage (Fig. 6 E and F) that likely contributes to ATM activation, although it is also possible that ROS can directly activate ATM in aneuploidy cells. On the basis of these observations, we propose the existence of an aneuploidy checkpoint (Fig. S6B). The strength of this checkpoint response (i.e., the strength of p53 activation) depends on the severity of aneuploidy. The more severe the aneuploidy is, the higher the ROS levels and the stronger the activation of this checkpoint. In other words, the level of ROS serves as a gauge on the severity of aneuploidy.
Materials and Methods
Transgenic Mice.
Cdc20AAA mice were described previously (19) and were crossed with p53 knockout mice (a generous gift from L. Donehower at Baylor College of Medicine, Houston) to produce double-mutant mice. Bub1-deficient mice were generated previously (14). Atm-deficient mice were originally generated by Barlow et al. (30) and were obtained from C. Zhu at MD Anderson Cancer Center, Houston. All of the mouse strains were maintained on a C57BL/6 background.
Karyotyping and FISH Analysis.
Isolating and karyotyping splenocytes were described previously (19). FISH analysis was performed by the cytogenetic core laboratory in Texas Children's Hospital.
Statistic Analysis.
Statistic significance was analyzed by Student's t test or one-way ANOVA. The survival analysis of mice was carried out with the Kaplan–Meier method. More details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Cytogenetic Core Laboratory of Texas Children's Hospital for FISH analysis. M.L. is supported by a postdoctoral training grant from National Institutes of Health. This work was funded by research grants from the National Cancer Institute (CA122623 and CA116097) (to P.Z.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005960107/-/DCSupplemental.
References
- 1.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 2.Pinsky BA, Biggins S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 3.King RW, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 5.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 6.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 7.Baker DJ, Dawlaty MM, Galardy P, van Deursen JM. Mitotic regulation of the anaphase-promoting complex. Cell Mol Life Sci. 2007;64:589–600. doi: 10.1007/s00018-007-6443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Natl Rev. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 9.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 13.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 17.Iwanaga Y, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 18.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 21.Mertens F, Johansson B, Mitelman F. Isochromosomes in neoplasia. Genes Chromosomes Cancer. 1994;10:221–230. doi: 10.1002/gcc.2870100402. [DOI] [PubMed] [Google Scholar]
- 22.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci. 2008;121(Suppl 1):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 23.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Natl Rev. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 26.Weaver BA, Cleveland DW. Aneuploidy: Instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 28.Alexander A, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow C, et al. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 31.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.