Abstract
Lignin is a major component of plant secondary cell walls and is derived from p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monolignols. Among higher plants, S lignin is generally considered to be restricted to angiosperms, which contain the S lignin-specific cytochrome P450-dependent monooxygenase, ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydoxylase (F5H). The transcription factor MYB58 directly regulates expression of monolignol pathway genes except for F5H. Here we show that F5H expression is directly regulated by the secondary cell wall master switch NST1/SND1, which is known to regulate expression of MYB58. Deletion of NST1 expression in Medicago truncatula leads to a loss of S lignin associated with a more than 25-fold reduction of F5H expression but only around a 2-fold reduction in expression of other lignin pathway genes. A detailed phylogenetic analysis showed that gymnosperms lack both F5H and orthologs of NST1/SND1. We propose that both F5H and NST1 appeared at a similar time after the divergence of angiosperms and gymnosperms, with F5H possibly originating as a component of a defense mechanism that was recruited to cell wall biosynthesis through the evolution of NST1-binding elements in its promoter.
Keywords: ferulate 5-hydroxylase, NAC transcription factor, pathway evolution
Lignin is an aromatic heteropolymer derived primarily from the hydroxycinnamyl alcohols p-coumaryl, coniferyl, and sinapyl alcohol, which give rise to p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) subunits, respectively (1). Lignin is one of the three main components of plant secondary cell walls and plays important roles in vascular plant growth by contributing to structural rigidity, facilitating water transport, and providing a defensive barrier against pathogens.
Lignin monomer composition varies among plant species. Although S lignin has arisen more than once during evolution (2), it does not appear to exist in the majority of gymnosperms or in ferns, whereas angiosperms deposit S units in lignin polymers in secondary cell walls. A cytochrome P450 (CYP)-dependent monooxygenase, ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydoxylase (F5H, CYP84A1) is specifically required for S lignin biosynthesis (3, 4) and diverts G monolignol intermediates into the S lignin branch pathway.
The Arabidopsis lignin-specific transcription factor MYB58 can directly activate most lignin biosynthesis genes except F5H (5). This activation is through binding to AC elements, which are present in most of the promoters of lignin biosynthetic genes (6–9), although no AC element has been detected in the F5H gene (9). Two NAC transcription factors, NST1 and SND1 (NST3), act together as key regulators of the entire secondary cell wall biosynthesis program in Arabidopsis (10, 11) and activate a cascade of downstream transcription factors, which in turn activates the biosynthetic pathways for the individual secondary cell wall components cellulose, xylan, and lignin (12). MYB46 is a direct target of SND1 in Arabidopsis. Although downstream of SND1, it is able to activate the complete secondary cell wall biosynthesis program (13).
We have recently identified a homolog of Arabidopsis NST1 and SND1 in the model legume Medicago truncatula (14). Loss of function of the single MtNST1 gene results in lack of lignification in interfascicular fibers, loss of anther dehiscence, and stomatal phenotypes associated with loss of ferulic acid in guard cell walls (14). We now report that expression of F5H is much more severely reduced than that of all other lignin biosynthesis genes in the Medicago nst1 mutant. Using an in vitro promoter assay, we demonstrate that Arabidopsis SND1 directly binds to the promoter of Medicago truncatula F5H and activates its expression, whereas activation of the expression of other lignin biosynthetic genes requires additional downstream protein factors. Conversely, Arabidopsis MYB58 activates many lignin biosynthesis genes from M. truncatula more strongly than F5H, suggesting that SND1 regulates the lignin biosynthesis pathway by directly activating F5H and indirectly activating the other lignin biosynthesis genes via MYB58. Phylogenetic analysis of CYP84, NAC, and MYB genes indicates that the evolution of the basic S lignin synthesis pathway occurred after the emergence of land plants, and the expansion and maturation of the regulatory machinery began no earlier than the appearance of angiosperms.
Results
F5H Is Regulated Differently from Other Lignin Biosynthesis Genes in the M. truncatula nst1 Mutant.
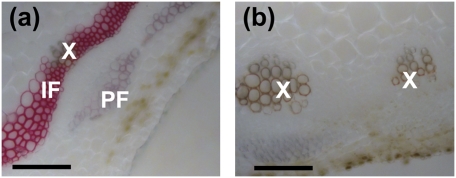
M. truncatula NST1 was previously identified from a Tnt1 retrotransposon insertion mutant population screened by analysis of stem cross sections for lignin autofluorescence. The nst1 mutants (five independent alleles) show a dramatic loss of lignin autofluorescence in the interfascicular region, and thioacidolysis analysis indicated reductions in the levels of H, G, and S monomers (14). Remarkably, S units, which account for ≈30% of the total lignin content in mature Medicago stems, were totally undetectable by thiacidolysis. To confirm this finding by an alternative approach, Mäule staining of stem cross sections was performed to selectively locate S lignin in the nst1-1 line (Fig. 1). The red coloration indicates the presence of S lignin in the vascular elements and interfascicular fibers in wild-type plants, but this was almost undetectable in the nst1 mutant, such that the fibers were not visible in the stained cross sections.
Fig. 1.
Syringyl lignin is undetectable in the M. truncatula nst mutant. Mäule staining of syringyl lignin in cross sections from the fifth internodes of nst1-1 (A) and wild type (B). X, xylem; PF, phloem fiber; IF, interfascicular fiber. (Scale bar, 100 μm.)
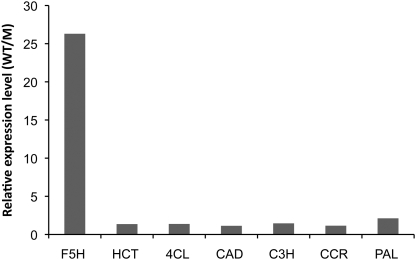
Based on microarray analysis, expression of most verified lignin biosynthetic genes, including cinnamate 4-hydroxylase (C4H), caffeic acid 3-O-methyltransferase (COMT), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), and 4-coumarate: CoA ligase (4CL), was reduced by between 1- and 2-fold in stems of the Medicago nst1 mutant (Fig. 2). However, F5H gene expression was reduced by more than 25-fold, suggesting that F5H is regulated differently from other lignin biosynthesis genes.
Fig. 2.
Expression of lignin biosynthesis genes in the Medicago nst mutant, as determined by microarray analysis. WT, wild type; M, mutant.
Arabidopsis SND1, MYB46, and MYB58 Differentially Activate M. truncatula Lignin Biosynthesis Genes.
To investigate how regulation of F5H might differ from that of other lignin biosynthesis genes, we used an in vitro Medicago leaf protoplast transcription assay to test the effects of coexpression of cell wall-related transcription factors on trans-activation of a reporter gene, firefly luciferase, driven by the promoters of various M. truncatula lignin biosynthesis genes. Because MYB58 can directly activate lignin biosynthesis genes, and MYB46 can regulate the entire secondary cell wall formation program (5, 13), these two transcription factors were chosen for the assay. However, because the Medicago orthologs of MYB58 and MYB46 have yet to be identified, Arabidopsis MYB58 and MYB46 were used. To be consistent with the use of Arabidopsis MYB58 and MYB46, Arabidopsis SND1 was used in place of Medicago NST1, because the two genes are orthologous (14).
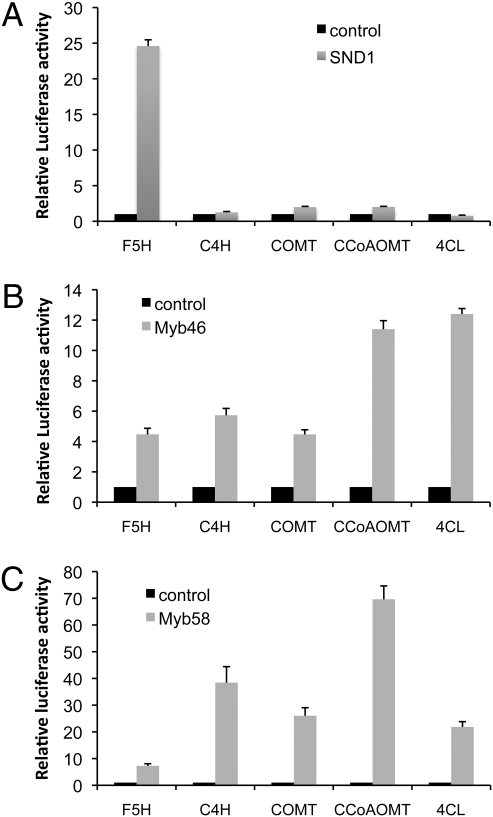
We first tested the effect of SND1 expression on monolignol pathway gene activation. Firefly luciferase reporter gene constructs were made to include around 1 kb of DNA upstream of the start codon for each of the lignin pathway genes tested. Coexpression of cauliflower mosaic virus (CaMV) 35S promoter driven SND1 activated the expression of the firefly luciferase reporter gene driven by the F5H promoter by around 25-fold (Fig. 3A). However, SND1 could not activate the promoters of the other lignin pathway genes tested (C4H, COMT, CCoAOMT, and 4CL), all of which are involved in both G and S lignin biosynthesis (1) (Fig. 3A). The same assay was then performed with the 35S promoter driving expression of MYB46 in place of SND1. The F5H, C4H, and COMT promoters were activated by around 4-fivefold, and CCoAOMT and 4CL were activated by more than 10-fold (Fig. 3B). The fact that all promoters were able to be activated rules out the possibility that the promoter regions of C4H, COMT, CCoAOMT, and 4CL selected for the assays were not functional. Using the same approach, the ability of MYB58 to activate the same set of promoters was also tested. The C4H, COMT, CCoAOMT, and 4CL promoters were strongly activated, by 20- to 70-fold, whereas the F5H promoter was only activated around 4-fold (Fig. 3C).
Fig. 3.
Lignin biosynthesis genes are induced by SND1, Myb46, and Myb58. Each transcription factor was coexpressed in M. truncatula leaf protoplasts with the firefly luciferase reporter gene driven by the lignin pathway gene promoters. The induction of the reporter gene was measured by assaying firefly luciferase activity. The control, which has no transcription factor coexpressed, is set to 1.0. A 35S promoter-driven Renilla luciferase was coexpressed to standardize the transfection efficiency. Bar charts show transactivation of lignin pathway gene promoters by SND1 (A), Myb46 (B), and MYB58 (C). Error bars represent SE of three biological replicates.
SND1 Directly Activates Expression of F5H by Binding to Its Promoter.
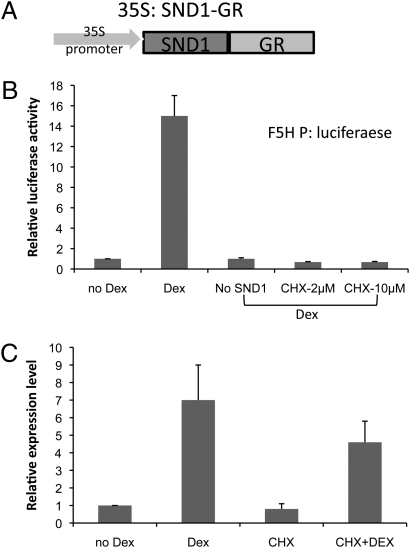
To further investigate the hypothesis that F5H is, unlike other monolignol pathway genes, directly regulated by SND1, a steroid receptor-based inducible system was used in the Medicago protoplast assay. The regulatory region of the human estrogen receptor (HER) (15) was first tested. Cotransfection of Medicago leaf protoplasts was performed with 35S promoter-driven SND1 fused with HER and the F5H promoter-driven firefly luciferase reporter construct. However, the F5H promoter was highly activated even without addition of the inducer estradiol. This could possibly be due to the high level of phytoestrogens in legume cells, which are able to mimic estradiol (16). To obviate this problem, we used a glucocorticoid receptor (GR)-based inducible system (17). The 35S promoter-driven SND1 was fused with the GR domain, and this construct was cotransfected into Medicago leaf protoplasts with the F5H promoter-driven firefly luciferase reporter gene (Fig. 4A). Luciferase activity was highly induced by the addition of dexamethasone (Dex), whereas Dex alone could not activate expression of the reporter gene (Fig. 4B).
Fig. 4.
Direct activation of F5H by SND1. M. truncatula leaf protoplasts were first transfected with 35S:SND1 fused with GR. The protoplasts were then treated with dexamethasone (Dex) or cycloheximide (CHX) plus Dex. Luciferase activity or F5H expression (luciferase transcript level) in control samples (no Dex) is set to 1.0. (A) The 35S:SND1-GR construct. (B) Luciferase activities in protoplasts cotransfected with SND1-GR and F5H promoter-luciferase. Dex alone cannot induce F5H promoter activity, and 2 μM CHX can completely abolish luciferase activity, indicating that de novo protein synthesis is inhibited. (C) Induction of F5H gene expression. M. truncatula Actin was used as internal control. Error bars represent SE of three biological replicates.
To test whether SND1 is directly activating the F5H promoter-driven reporter gene in these assays, the protein synthesis inhibitor cycloheximide was used to block de novo protein synthesis. A total of 2 μM cycloheximide was sufficient to completely abolish F5H promoter-driven firefly luciferase activity induced by Dex (Fig. 4B), confirming that this concentration effectively inhibits protein synthesis. Under these conditions, genes that are directly activated by SND1 should still be induced by addition of Dex, because SND1 is under control of the 35S promoter and has been produced before cycloheximide treatment. However, indirect activation of genes by SND1 via other proteins would be inhibited because of inhibition of new protein synthesis. Dex-induced appearance of F5H transcripts via SND1 under cycloheximide treatment clearly indicates that the activation of F5H expression does not require new protein synthesis and is therefore direct (Fig. 4C).
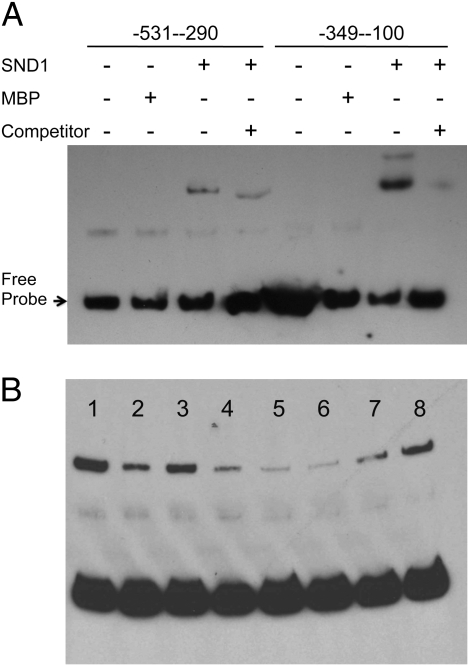
To further confirm direct activation of F5H by SND1, we investigated whether SND1 can bind to the F5H promoter using an electrophoretic mobility shift assay (EMSA). A 531-bp F5H promoter fragment, which was shown to be activated by AtSND1, was further split into two overlapping fragments. Recombinant maltose binding protein (MBP)-AtSND1 fusion protein was expressed in Escherichia coli and purified for use in the EMSA. The recombinant AtSND1 protein was able to strongly bind to the −100–349 fragment and cause a mobility shift (Fig. 5A). Addition of unlabeled fragment competed with the binding, and the mobility shift was not seen when MBP alone was used (Fig. 5A), indicating that the binding of SND1 to the F5H promoter is specific. There was also weak binding to the −290–531 fragment of the F5H promoter.
Fig. 5.
SND1 binds directly to the F5H promoter. (A) EMSA showing direct binding between SND1 and the F5H promoter. The NAC domain of SND1 fused with MBP was expressed in E. coli, and the purified recombinant protein incubated with biotin-labeled F5H promoter. (B) Competition analysis of SND1 binding to the F5H promoter shows that P2, P3, and P4 fragments of the F5H promoter contain the binding site. Lanes are as follows: 1 ,no competitor; 2, wild type 249-bp probe; 3-8, fragments P1-P6, respectively. MBP was used as a control protein. Unlabeled competitor DNA fragments are in 200-fold molar excess relative to the labeled probes.
To determine the minimal sequence required for SND1 binding, we further divided the −100–349 fragment into six overlapping fragments, each of 50 bp in length and named P1–P6, for EMSA analysis. Three fragments, P2, P3, and P4, could compete with biotin-labeled probe as effectively as the full −100–349 fragment, and P5 also somewhat weakly competed with the labeled probe (Fig. 5B). There was no conservation between sequences in the binding fragments and the recently discovered SND1 binding motif in the MYB46 promoter (13), nor any obvious sequence motif that was repeated within the −100 to −349 region of the promoter.
Phylogenetic Analysis of NST, F5H, MYB46, and MYB58.
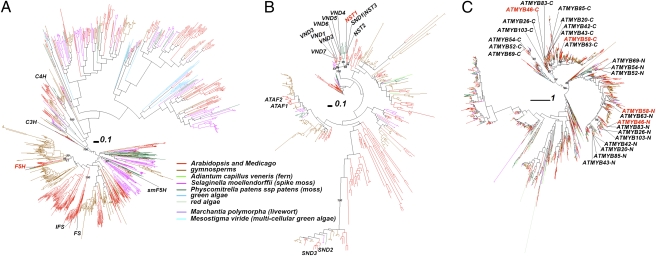
Since we have shown that F5H is regulated directly by NST1/SND1, and S lignin is often considered to be restricted to angiosperms, we next considered the evolutionary origins of F5H, NST, and MYB genes, by conducting phylogenetic analysis of plant species including algae, ferns, moss, two angiosperms, and 12 gymnosperm species (Table S1). The large phylogeny of F5H-related proteins (Fig. 6A) included the other monolignol pathway P450s coumaroyl shikimate 3′-hydroxylase (C3′H) and C4H, and other cytochrome P450 proteins. Three Medicago proteins are closest to the two Arabidopsis F5H proteins in the phylogeny (Fig. S1). Interestingly, the five proteins were clustered with one protein translated from an assembled EST UniGene from the fern species Adiantum capillus veneris. The six proteins together were further clustered with 13 Medicago proteins and then with many gymnosperm proteins. Therefore, the single fern protein may represent the ortholog of F5H, whereas true gymonosperm F5H orthologs may be missing. The many gymonosperm P450 proteins await functional characterization. Interestingly, no moss or spike moss proteins were found in the higher plant F5H clade. The previously described spike moss (Selaginella moellenodorffii) smF5H is located in a distant clade, consistent with the independent evolution of F5H activity in spike moss (18).
Fig. 6.
Phylogenies of F5H, NST, and MYB homologous proteins from 21 plant species. The multiple sequence alignments were of (A) 1,331 proteins homologous to Arabidopsis F5H, (B) 500 proteins homologous to Arabidopsis NST proteins, and (C) 2,209 proteins homologous to Arabidopsis MYB proteins, respectively. Selected supporting values >70% are shown. Plant proteins of interest are identified and highlighted by names.
In the large phylogeny of NAC transcription factors (Fig. 6B), those related to secondary cell wall synthesis (12, 19, 20) form one clade, except for SND2 and SND3. Medicago NST1 is orthologous to Arabidopsis NST1, -2, and -3 (SND1) in this clade (Fig. S2). No gymnosperm proteins are found to be orthologous to Arabidopsis and Medicago NST proteins. The closest gymnosperm proteins to MtNST1 (plantGDB ID: PUT-163a-Picea_glauca-31332_2 and PUT-162b-Picea_allspecies-73182_2) form a separate clade with AT4G10350.1 (ANAC070), AT1G33280.1 (ANAC015), and Medtr4g041220.1, all of which are not yet functionally characterized. All of the remaining Arabidopsis NAC proteins form a separate subclade, consisting of AtVND1-7, a few Medicago proteins, a few gymnosperm proteins, and two spike moss proteins basal to the other proteins.
MYB transcription factors usually contain at least two Pfam Myb_DNA-binding domains (21). In our large phylogeny (Fig. 6C), 13 MYB proteins related to secondary cell wall biosynthesis (12, 19, 20) are located in two major clades each consisting of four subclades. One of the major clades contains the N-terminal Myb_DNA-binding domain, whereas the other contains the C-terminal Myb_DNA-binding domain. ATMYB58 and -63 form a subclade, ATMYB20, -42, -43, and -85 form a subclade, ATMYB46, MYB83, MYB26, and MYB103 form a subclade, and ATMYB52, -54, and -69 form a subclade. In the subclade containing ATMYB58 (Figs. S3–S5), two Medicago EST UniGenes are orthologous to ATMYB58, ATMYB63, and the other two Arabidopsis proteins (AT1G56160.1 and AT3G12820.1); in the subclade containing MYB46 (Figs. S6 and S7), Medtr2g117450.1 is the Medicago ortholog.
We did not find algal genes homologous to NST transcription factors, but did find algal homologs of F5H enzymes and MYB transcription factors (Fig. 6). Recently it was reported that one red alga, Calliarthron cheilosporioides, makes lignins (22). However, none of the algal homologs is close to the known lignin synthesis-related F5H, C3H, and C4H genes (Fig. 6A). This indicates that either the green algae included in this study do not have lignins or that algal lignin synthesis genes evolved independently compared with their higher plant equivalents, as seen for smF5H (18). In the MYB phylogeny (Fig. 6C), a total of 32 green algal and 37 red algal homologs are found, many in the two major clades containing the N and C termini of the 13 secondary cell wall-related MYB proteins, which further locate in four subclades of each of the two major clades. Clearly three of the four subclades, excepting the one containing MYB52/54/69 homologs, are closer to each other and one single green algal protein is found basal to them, suggesting they have diverged after the green algae appeared. On the other hand, all of the remaining algal proteins of the two major clades are basal to the subclade containing MYB52/54/69 homologs, suggesting that these MYB subclades diverged from the other three before the appearance of the green algae.
Discussion
Syringyl units are the second most abundant monolignol units in the lignin of angiosperms, and, apart from a report of a sinapyl alcohol-specific cinnamyl alcohol dehydrogenase from aspen (23), F5H appears to be the only truly S-lignin–specific monolignol biosynthetic enzyme. S lignin accumulation can be decreased or increased simply by down-regulation or up-regulation of F5H (24, 25). Given the special role of F5H in the lignin biosynthetic pathway, its regulation is particularly critical for the determination of lignin composition. Early reports of transcript accumulation in plants grown under different light/dark periods first hinted that F5H may be differently regulated than other lignin pathway genes (26).
Recently, several transcription factors involved in lignin biosynthesis have been characterized (5, 10, 11, 13). NST1 and SND1 can activate the entire secondary cell wall formation program and regulate the expression of MYB46 and MYB58. Although MYB46 is a downstream target of NST1 and SND1, it alone can also activate the whole secondary cell wall biosynthesis program. MYB58 directly activates lignin pathway genes, except for F5H, by binding to the AC elements in their promoter regions (5), and there is no AC element in the F5H promoter (9). We now show that F5H is directly regulated by SND1, which in parallel regulates the secondary cell wall biosynthesis program, in part by activation of the lignin-specific MYB58.
A series of in vivo and in vitro data support the concept that F5H is in a different regulatory network from the other lignin pathway genes. First, F5H is down-regulated to a much greater degree than all of the other lignin biosynthesis genes in the Medicago nst1 mutant, which appears to essentially lack S lignin (14); it should be noted that the genes analyzed in Fig. 2 have been functionally proven to be involved in lignification in Medicago (27), and some are present in the M. truncatula genome as single genes (C3H, HCT, and F5H). Second, in a transient expression system, the F5H promoter is activated by coexpression of SND1, the Arabidopsis ortholog of Medicago NST1, but other lignin pathway promoters are not. Third, SND1 can bind the promoter region of F5H and activate F5H expression without de novo protein synthesis, indicating that the regulation of F5H by SND1 is through direct binding to the F5H promoter rather than through downstream transcription factors such as MYB46 and MYB58.
Most gymnosperms and ferns do not synthesize S lignin. Based on our phylogentic analysis, angiosperms including Arabidopsis and Medicago possess F5H, NST, MYB46, and MYB58. However, gymnosperms do not appear to have orthologs of F5H and NST, but do have orthologs of MYB46 and the lignin-specific transcription factor MYB58. Ferns, moss, and algae do not appear to have NST, MYB46, or MYB58, although there is currently only one fern species with EST data (16,944 UniGenes), compared with the EST data from 12 gymnosperm species. Ferns do have an F5H ortholog, the biochemical function of which remains to experimentally determined whereas the mosses and algae analyzed do not appear to have F5H homologs, with the exception of Selaginella moellenodorffii, which has an independently evolved F5H (18).
Although all 12 gymnosperms in our analysis lack complete genome information (relying on EST analysis), it seems unlikely that the datasets for all 12 species would contain MYB46 and MYB58 but lack F5H and SND1. The apparent absence of F5H genes in lower plants, the absence of green algal homologs of NAC genes, and the limited distribution of algal MYB genes therefore strongly support the evolution of S lignin synthesis as starting after the emergence of land plants, and the expansion and maturation of the regulatory machinery has taken a very long time, starting no earlier than the appearance of angiosperms. However, as indicated previously, the presence of S lignin in spike moss (18) and lower algal plants (22) suggests that there may have been independent, alternative evolutionary routes for the generation of S lignin synthesis and its regulation in lower plants.
The current model for secondary cell wall biosynthesis suggests that both NST1/SND1 and MYB46 can activate the entire cell wall formation program including cellulose, hemicellulose, and lignin biosynthesis. Although MYB46 is a downstream target of NST1/SND1, these two different master switches at first sight seem to be redundant. However, the fact that the NST1/SND1 deletion mutant grows quite well, whereas the growth of the MYB46:MYB83 double deletion mutant is arrested immediately after germination (28) suggests that the more recently evolved NST1/SND1 control is more dispensable. Our model suggests that NST1/SND1 directly activates both F5H and MYB46 (Fig S8). The two different master switches may be necessary to guarantee that F5H is activated coordinately with the secondary cell wall program that was originally under the master control of MYB46 alone.
Monolignols have strong antimicrobial activity, and are produced as soluble glycosides in many plant species, including Arabidopsis roots (29, 30). Among the different lignin monomers, S lignin seems to be particularly involved in plant defense, and some plants selectively accumulate S lignin in response to pathogen attack (31). Arabidopsis plants overexpressing F5H under the C4H promoter accumulate high levels of S lignin and show increased resistance to nematode infection (29). It is possible that S monolignol biosynthesis initially evolved as a defense response following the appearance of F5H, and that the new pathway was subsequently recruited for cell wall biosynthesis once its regulatory transcription factor, NST1, gained the ability to bind the MYB46 promoter. This model is consistent with the involvement of many NAC transcription factors in biotic stress tolerance (32, 33).
Materials and Methods
Mäule Staining and Microscopy.
The fifth internodes from the top of M. truncatula stems were cross-sectioned to 100 μm. To visualize S lignin, Mäule staining procedures were followed as described (27).
Microarray Analysis.
This was performed as described previously (14).
Transfection of Leaf Protoplasts for Transactivation Analysis.
Leaves from 6- to 8-wk-old greenhouse-grown M. truncatula were used as a source of protoplasts. Protoplast isolation was performed as previously described (34). Cloning of transcription factor and promoter-reporter constructs in the vector p2GW7 (35) is described in SI Materials and Methods. One reporter construct (promoter-luciferase) and one effector construct (35S:transcription factor) were cotransfected into protoplasts as described (34). A reference construct containing the Renilla luciferase gene driven by the 35S promoter was also cotransfected to determine the transfection efficiency. Luciferase activities were determined using the dual-luciferase reporter assay system (Promega). The firefly luciferase activity was calculated by normalizing against the Renilla luciferase activity in each transfection event. Data are presented as averages ± SD of three biological replicates.
Dex-Inducible System to Show Direct Activation of F5H by SND1.
SND1 minus the stop codon was cloned into p2GW7, and the GR domain (17) cloned at the C terminus of SND1. Protoplasts transfected with this construct were incubated with 10 μM Dex (Sigma-Aldrich) for 6–8 h to allow SND1 to transfer from cytoplasm to nucleus. The same amount of ethanol used to dissolve Dex was applied to the control protoplasts. A total of 2 μM cycloheximide was applied 30 min before addition of Dex to inhibit new protein synthesis. The treated protoplasts were then used for dual-luciferase reporter activity assay or real-time quantitative PCR analysis of luciferase transcripts. The expression of F5H-luciferase in the control protoplasts was set to 1.0 and the data are presented as averages ± SD of three biological replicates.
Real-Time PCR.
cDNA samples were used for quantitative real-time PCR (qRT-PCR) with technical duplicates. The 10-μL reaction included 2 μL primers (0.5 μM of each primer), 5 μL Power Sybr (Applied Biosystems), 2 μL 1:20 diluted cDNA from the reverse transcription step, and 1 μL water. qRT-PCR data were analyzed using SDS 2.2.1 software (Applied Biosystems). PCR efficiency was estimated using the LinRegPCR software (36) and transcript levels determined by relative quantification (37) using the M. truncatula actin gene as a reference.
EMSA.
To express the recombinant NAC domain of the SND1 protein, a 191-amino-acid peptide was fused in frame with maltose binding protein (MBP) and purified using amylose resin. The resulting protein was used for EMSA with the F5H promoter fragments. The promoter fragments were PCR amplified with one 5′ biotin-labeled primer and purified using a PCR purification kit (Invitrogen). The biotin-labeled DNA fragments were incubated for 20 min with 100 ng of SND1-MBP in EMSA binding buffer (Pierce). Reaction mixtures were resolved by polyacrylamide gel electrophoresis, and DNA was electroblotted onto nitrocellulose membrane and detected by chemiluminescence. Generation of nonlabeled promoter fragments for competition assays is described in SI Materials and Methods.
Homology Searching.
The following hidden Markov models (HMMs) were used to search against the translated peptides of the 21 species in Table S1 using the hmmsearch command of the HMMER package (38). For MYB proteins, the Pfam Myb_DNA-binding domain HMM model (PF00249) was used; for NAC proteins, the A to E subdomain HMM model was built as described previously (33); and for F5H proteins, a HMM model was generated using the hmmbuild and hmmcalibrate commands of the HMMER package based on multiple sequence alignment of known F5H protein sequences. If the plant genome was sequenced, the automated predicted peptides were used; otherwise the translated peptides in six frames of the assembled EST UniGenes by plantGDB were used (39).
Phylogenetic Analyses.
The homologs found in the three searches under the E-value cutoff <0.01 were collected, respectively, and subjected to multiple sequence alignments using MAFFT v6.717 program (40) with the L-INS-I method. The phylogenies were further reconstructed using the FastTree v2.1.1 program (41). The local support values beside the nodes were computed by resampling the site likelihoods 1,000 times and performing the Shimodaira Hasegawa test to show the confidence levels with regard to the clustering of relevant proteins into one group.
The Arabidopsis MYB genes reported previously (20), the two Arabidopsis F5H genes listed at http://cellwall.genomics.purdue.edu/families/1-3.html, and the previosuly reported Arabidopsis NAC genes (12, 19, 20) were identified in the phylogenies, and the subclades containing these marker genes were taken out and displayed using the Interactive Tree of Life (iTOL) Web server (42).
Note Added in Proof.
A recent review (43) observed that S lignin is present in a few gymosperm species but that its evolutionary/biochemical origin is not yet clear. It will be interesting to determine whether it arose from a NAC-controlled classical F5H system that was then lost in later gymnosperm lineages or whether it has arisen independently as in Selaginella.
Supplementary Material
Acknowledgments
We thank Drs. Rui Zhou and Zeng-Yu Wang for critical reading of the manuscript. This work was supported by grants to R.A.D. from the state of Oklahoma (through the Oklahoma Bioenergy Center) and the Department of Energy Bioenergy Feedstock Genomics Program (Award DE-FG02-06ER64303) and to R.A.D. and Y.X. from the Department of Energy Bioenergy Research Centers, through the Office of Biological and Environmental Research in the Department of Energy Office of Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009170107/-/DCSupplemental.
References
- 1.Ralph J, et al. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev. 2004;3:29–60. [Google Scholar]
- 2.Weng JK, et al. Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell. 2010;22:1033–1045. doi: 10.1105/tpc.109.073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA. 1999;96:10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Popko JL, Umezawa T, Chiang VL. 5-hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem. 2000;275:6537–6545. doi: 10.1074/jbc.275.9.6537. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goicoechea M, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 7.Patzlaff A, et al. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 8.Patzlaff A, et al. Characterisation of Pt MYB1, an R2R3-MYB from pine xylem. Plant Mol Biol. 2003;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- 9.Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuda N, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Q, et al. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 2010;3:100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
- 15.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 16.Kurzer MS, Xu X. Dietary phytoestrogens. Annu Rev Nutr. 1997;17:353–381. doi: 10.1146/annurev.nutr.17.1.353. [DOI] [PubMed] [Google Scholar]
- 17.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 18.Weng JK, Li X, Stout J, Chapple C. Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci USA. 2008;105:7887–7892. doi: 10.1073/pnas.0801696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong R, Ye ZH. Regulation of cell wall biosynthesis. Curr Opin Plant Biol. 2007;10:564–572. doi: 10.1016/j.pbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhong R, Ye ZH. Transcriptional regulation of lignin biosynthesis. Plant Signal Behav. 2009;4:1028–1034. doi: 10.4161/psb.4.11.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 22.Martone PT, et al. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol. 2009;19:169–175. doi: 10.1016/j.cub.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 23.Li L, et al. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell. 2001;13:1567–1586. doi: 10.1105/TPC.010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke R, et al. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000;22:223–234. doi: 10.1046/j.1365-313x.2000.00727.x. [DOI] [PubMed] [Google Scholar]
- 26.Ruegger M, Meyer K, Cusumano JC, Chapple C. Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 1999;119:101–110. doi: 10.1104/pp.119.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): Effects on lignin composition in specific cell types. New Phytol. 2008;179:738–750. doi: 10.1111/j.1469-8137.2008.02502.x. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- 29.Wuyts N, Lognay G, Swennen R, De Waele D. Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J Exp Bot. 2006;57:2825–2835. doi: 10.1093/jxb/erl044. [DOI] [PubMed] [Google Scholar]
- 30.Lange BM, Lapierre C, Sandermann H., Jr. Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins) Plant Physiol. 1995;108:1277–1287. doi: 10.1104/pp.108.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menden B, Kohlhoff M, Moerschbacher BM. Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry. 2007;68:513–520. doi: 10.1016/j.phytochem.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima K, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 33.Shen H, Yin Y, Chen F, Xu Y, Dixon RA. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. BioEnergy Res. 2009;2:217–232. [Google Scholar]
- 34.Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- 35.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 36.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 39.Dong QF, et al. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 43.Weng J-K, Chapple C. The origin and evolution of lignin biosynthesis. New Phytol. 2010;187:283–285. doi: 10.1111/j.1469-8137.2010.03327.x. 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.