Abstract
Isoprenoids are a large family of compounds with essential functions in all domains of life. Most eubacteria synthesize their isoprenoids using the methylerythritol 4-phosphate (MEP) pathway, whereas a minority uses the unrelated mevalonate pathway and only a few have both. Interestingly, Brucella abortus and some other bacteria that only use the MEP pathway lack deoxyxylulose 5-phosphate (DXP) reductoisomerase (DXR), the enzyme catalyzing the NADPH-dependent production of MEP from DXP in the first committed step of the pathway. Fosmidomycin, a specific competitive inhibitor of DXR, inhibited growth of B. abortus cells expressing the Escherichia coli GlpT transporter (required for fosmidomycin uptake), confirming that a DXR-like (DRL) activity exists in these bacteria. The B. abortus DRL protein was found to belong to a family of uncharacterized proteins similar to homoserine dehydrogenase. Subsequent experiments confirmed that DRL and DXR catalyze the same biochemical reaction. DRL homologues shown to complement a DXR-deficient E. coli strain grouped within the same phylogenetic clade. The scattered taxonomic distribution of sequences from the DRL clade and the occurrence of several paralogues in some bacterial strains might be the result of lateral gene transfer and lineage-specific gene duplications and/or losses, similar to that described for typical mevalonate and MEP pathway genes. These results reveal the existence of a novel class of oxidoreductases catalyzing the conversion of DXP into MEP in prokaryotic cells, underscoring the biochemical and genetic plasticity achieved by bacteria to synthesize essential compounds such as isoprenoids.
Keywords: Brucella, DXR, fosmidomycin, methylerythritol, phylogenetic
Isoprenoids, one of the largest groups of natural compounds, have a variety of roles in respiration, photosynthesis, membrane structure, allelochemical interactions, and growth regulation (1–3). All free-living organisms synthesize isoprenoids from the five carbon precursors isopentenyl diphosphate (IPP) and its double-bond isomer dimethylallyl diphosphate (DMAPP). For decades it was believed that IPP was exclusively synthesized from acetyl coenzyme A by the mevalonate (MVA) pathway and then converted into DMAPP by a IPP/DMAPP isomerase (IDI) enzyme (4). However, in the early nineties of the last century it was discovered that IPP and DMAPP could be formed simultaneously from pyruvate and glyceraldehyde 3-phosphate by an alternative route currently known as the methylerythritol 4-phosphate (MEP) pathway (5, 6). It is now well established that most organisms only use one of the two pathways for isoprenoid biosynthesis. Thus, archaea (archaebacteria), fungi, and animals synthesize IPP from MVA, whereas most bacteria (eubacteria) only use the MEP pathway for the production of isoprenoid precursors. Plants employ both pathways, but in different cell compartments: The MVA pathway synthesizes cytosolic isoprenoid precursors whereas the MEP pathway is located in plastids (7).
Because the MEP pathway is absent from animals (including humans) but it is essential in a large number of major bacterial pathogens, it has been proposed as a promising new target for the development of novel antiinfective agents (8, 9). However, information on possible mechanisms for resistance to a block of the MEP pathway is scarce. Antibiotic resistance can be caused by an active export or blocked import of the drug, by its inactivation inside the cell, by the genetic modification of its protein target, or by the use of an alternative pathway not affected by the inhibitor, to mention just a few possibilities. The best characterized inhibitor of the MEP pathway is fosmidomycin (FSM), first identified as a natural antibiotic effective against a wide bacterial spectrum. FSM is a specific competitive inhibitor of deoxyxylulose 5-phosphate (DXP) reductoisomerase (DXR), the enzyme catalyzing the NADPH-dependent production of MEP from DXP in the first committed step of the pathway (10). The uptake of FSM by bacterial cells is an active process involving a cAMP-dependent glycerol 3-phosphate transporter (GlpT) protein (11). A defective glpT gene in Escherichia coli mutants or the absence of a GlpT homologue in other bacteria such as Mycobacterium tuberculosis leads to FSM resistance (11, 12). Overexpression of the E. coli fsr gene, encoding a protein with similarity to bacterial drug-export proteins, also results in FSM resistance, likely because this protein facilitates the export of the inhibitor (13). Furthermore, a number of independent mutations have been shown to rescue the survival of E. coli strains defective in the first two genes of the MEP pathway, suggesting that bacteria can respond to a block of DXP synthase (DXS) or DXR activities by using other enzymes that produce DXP or MEP when mutated (14). Alternative pathways and metabolic intermediates have been proposed to be used for the biosynthesis of isoprenoid precursors in the cyanobacterium Synechocystis PCC 6803, which lacks the MVA pathway and does not use the MEP pathway under photosynthetic conditions (15, 16). These results illustrate how limited is still our knowledge of the alternative pathways that can be used by bacteria to synthesize their isoprenoids. In this context, we noticed that the completely sequenced genomes of a number of bacteria, including the pathogenic Brucella abortus 2308 (17), contain the genes of the MEP pathway with the only exception of that encoding DXR, suggesting that these bacteria may use an alternative enzyme to produce MEP. Here we report the cloning of the gene encoding such an alternative enzyme, demonstrate its biochemical activity both in vivo and in vitro, and examined its phylogenetic distribution across bacteria.
Results
B. abortus Cells Expressing GlpT Become Sensitive to FSM.
The completely sequenced genome of B. abortus lacks a DXR-encoding gene but it contains homologues of the rest of MEP pathway enzymes (Fig. 1 and Table S1). This suggests that a B. abortus protein showing no overall homology to DXR might be involved in transforming DXP into MEP. However, growth of B. abortus cells was not inhibited by concentrations of FSM up to 1 mg/mL. This result suggested that the putative DXR-like (DRL) enzyme might not be inhibited by FSM or, alternatively, that the inhibitor was degraded, expelled, or not taken by living cells. Consistent with the last possibility, the B. abortus genome contains a fsr gene (BAB1_0676) but no glpT homologue. To investigate whether a defective uptake of FSM was the cause of the resistance phenotype of this bacterium, the E. coli gene encoding the GlpT transporter was expressed in B. abortus cells. As shown in Fig. S1, transformants became FSM sensitive with a minimal inhibitory concentration of 4 μg/mL, confirming that the resistance phenotype of wild-type cells resulted solely from the absence of an appropriate uptake mechanism. These data also suggested that the activity of the putative B. abortus DRL protein was actually inhibited by FSM, consistent with the hypothesis that the biochemical mechanism used by this alternative enzyme to produce MEP from DXP might be similar to that used by DXR.
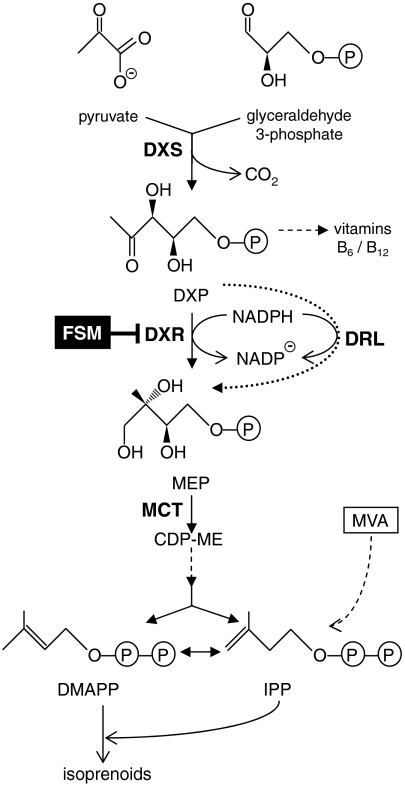
Fig. 1.
The MEP pathway in bacteria. DXP, deoxyxylulose 5-phosphate; MEP, methylerythritol 4-phosphate; CDP-ME, cytidine diphosphomethylerythritol; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate. Enzymes are indicated in bold: DXS, DXP synthase (EC 2.2.1.7); DXR, DXP reductoisomerase (EC 1.1.1.267); MCT, MEP cytidylyltransferase (EC 2.7.7.60). The step inhibited by FSM is indicated. Dashed closed arrows represent several steps. The E. coli strains defective in DXS (EcAB4-2), DXR (EcAB4-10), or MCT (EcAB4-7) activities used here were engineered to utilize exogenously supplied MVA for the biosynthesis of IPP (dashed open arrow). The dotted closed arrow marks the reaction catalyzed by the DRL enzyme identified in this work.
Complementation of E. coli Strains Defective in DXR Leads to the Identification of DRL.
To identify the gene encoding DRL in B. abortus, we constructed a genomic library of this bacterium and used it to complement a DXR-deficient E. coli mutant. The genome of E. coli strain EcAB4-10 harbours a deletion of the dxr gene and a synthetic MVA operon that allows the production of IPP and DMAPP (and therefore the survival of the cells) when MVA is supplied to the growth medium (14). Competent EcAB4-10 cells were transformed with the B. abortus genomic library and plated in the absence of MVA. Plasmids from positive transformants that grew without exogenous MVA were sequenced and shown to contain overlapping B. abortus genomic fragments containing the genes BAB2_0264 and BAB2_0265 (Fig. S2A). BAB2_0265 codes for a hydrolase whereas BAB2_0264 codes for a protein annotated as a putative oxidoreductase (a family that includes DXR) and was therefore selected for further experiments. Transformation of EcAB4-10 cells with a vector harbouring only BAB2_0264 led to full complementation of MVA auxotrophy (Fig. 2), suggesting that the encoded protein (Q2YIM3) was the predicted B. abortus DRL enzyme. The same construct was used in complementation experiments with the E. coli strains EcAB4-2 and EcAB4-7, defective in DXS and MEP cytidylyltransferase (MCT) activities respectively (14). As shown in Fig. 2, expression of BAB2_0264 in these strains did not rescue their MVA auxotrophy. These results are strong in vivo evidence that this gene encodes a DXR-like enzyme that requires a supply of DXP (or a derived metabolite synthesized by DXS) to produce MEP (or a precursor that could be transformed into MEP or used by MCT activity) for isoprenoid biosynthesis (Fig. 1).
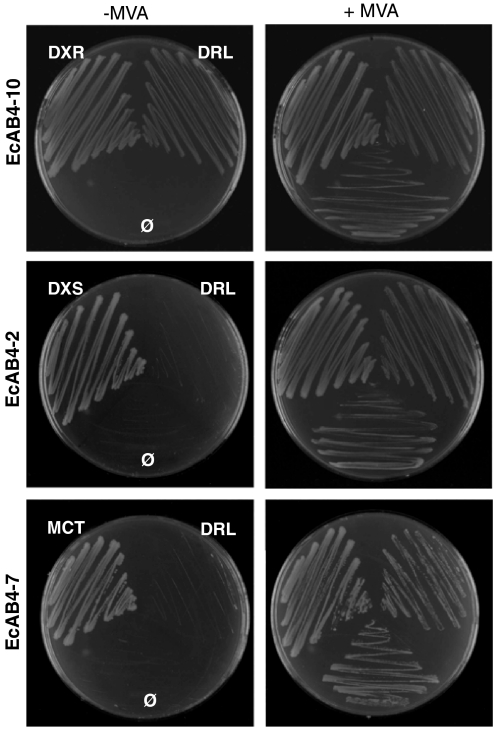
Fig. 2.
Complementation of mutant E. coli strains with the B. abortus gene encoding DRL. E. coli strains defective in DXR (EcAB4-10), DXS (EcAB4-2), or MCT (EcAB4-7) activities were transformed with a plasmid expressing the BAB2_0264 gene (DRL). Control experiments were made with plasmids for the expression of the E. coli genes encoding DXR, DXS, or MCT (33), as well as empty vectors (Ø). Ability of the cloned gene to rescue growth of the corresponding mutant strains was ascertained by streaking single colonies on plates either supplemented (+) or not (-) with 1 mM MVA as indicated.
DRL and DXR Catalyze the Same Biochemical Reaction.
The identified B. abortus DRL protein sequence showed no overall homology to DXR but to homoserine dehydrogenase (HD) enzymes (18). A search for functional domains detected the existence of an N-terminal NAD(P)-binding domain (Fig. S2B) arranged forming a modified Rossman fold similar to that found in numerous dehydrogenases. This, together with the in vivo results (Fig. 2 and Fig. S1), suggested that DRL might use NADPH to catalyze a reaction very similar (or even identical) to that catalyzed by DXR. To verify this possibility, a recombinant DRL protein fused to a polyhistidine tail was produced in E. coli and purified (Fig. S3) to be used in in vitro DXR activity assays. Control assays were performed with recombinant DXR protein from E. coli, and the reaction products were analyzed by UPLC-MS(/MS). Samples containing either DRL or DXR were found to produce MEP from DXP in a similar fashion (Fig. 3A). The identity of the reaction product in the DRL sample was confirmed by comparing its fragmentation pattern with that of a MEP standard (Fig. 3B). When reaction mixtures were prepared without enzyme or without NAPDH, only peaks for m/z 213/97 (DXP) transition were detected (Fig. 3A). Furthermore, the activity of the recombinant DRL enzyme was sensitive to inhibition with FSM (Fig. 3C). Size-exclusion chromatography (Fig. S3) showed that the molecular weight of the active enzyme is ca. 80 kDa (i.e., about twice the size deduced from the protein sequence), suggesting that DRL is a homodimer, like DXR (10, 19). Also similarly, DRL activity has a pH optimum in the range 7.5–8 and a temperature optimum of 40–45 °C (Fig. S4). Lineweaver–Burk plot of enzyme parameters (Fig. S4) showed a Vmax = 0.083 μmol NADPH min-1 mg-1, a kcat = 0.065 s-1, and a Km(DXP) = 109 μM for the recombinant DRL protein. Under the same experimental conditions the Km(DXP) for E. coli DXR was 211 μM, of the same order as the values reported in the literature for different DXR enzymes (19). By contrast, the kcat values for DXR are between two and three orders of magnitude higher than that calculated for the B. abortus recombinant DRL (19). Together, DRL defines a new class of NADPH-dependent enzymes related to HD-like oxidoreductases that catalyze the formation of MEP from DXP almost identically to DXR, even though the rate of conversion appears to be lower in the case of DRL.
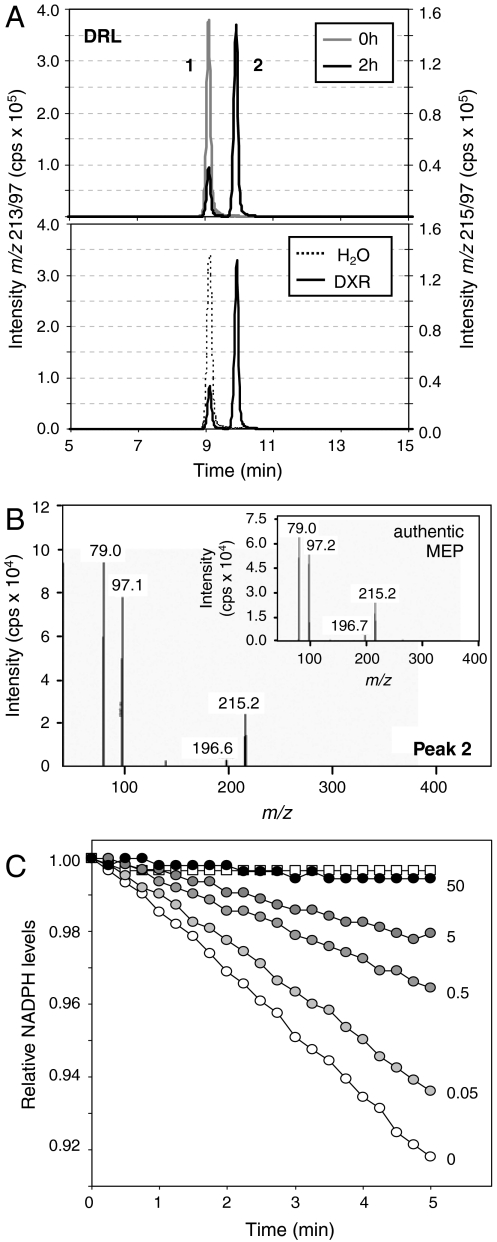
Fig. 3.
Biochemical characterization of a recombinant DRL protein. (A) MRM chromatograms of a reaction mixture containing MgCl2, DTT, NADPH, and DXP before (0 h) and 2 h after adding recombinant DRL. (Lower) Result after incubation for 2 h with either no enzyme or purified DXR instead of DRL. Peak 1 corresponds to DXP (m/z 213/97, 9.15 min). Peak 2 corresponds to MEP (m/z 215/97, 9.90 min), as demonstrated by comparing its mass spectra with that of a MEP standard (B). (C) Changes in NADPH levels monitored by absorbance at 340 nm in reaction mixtures like those described in (A) after adding recombinant DRL (circles). The DXR inhibitor FSM was incorporated in the mixtures at the indicated concentration (μM). A control without DXP (white squares) is shown.
The Distribution of DRL and DXR Sequences Is Not Mutually Exclusive in All Organisms.
BLAST searches of Universal Protein Resource (UNIPROT) databases using the B. abortus DRL sequence (Q2YIM3) as a query retrieved 185 hits (Table S2). Most bacteria only had one putative DRL homologue, but 17 strains had two and 3 strains had three (Table S1 and Table S2). When the distribution of putative DRL sequences was compared to that of MEP pathway enzymes, three classes were established. The first one (class A) was formed by bacteria with DRL instead of DXR sequences in their genomes. Most of these DRL sequences (including the B. abortus protein identified here) were from alpha-proteobacteria, but some were also found in firmicutes (Table S1). In particular, the single DRL sequences found in the genomes of the alpha-proteobacterium Bartonella henselae (Q6G2D9) or the firmicute Finegoldia magna (B0S038) were found to be active in complementation experiments with the EcAB4-10 strain (Table 1). The same approach confirmed the activity of the duplicated DRL sequences present in the genome of the alpha-proteobacteria Ochrobactrum anthropi (A6WYQ0 and A6X6G6) and Mesorhizobium loti (Q98FT2 and Q989B6). The second class (B) was formed by bacteria with sequences encoding both DRL and DXR enzymes (Table S1). From these, the DRL sequences from the alpha-proteobacteria Agrobacterium tumefaciens (A9CES2) and Candidatus Pelagibacter ubique (Q1V2P9), the beta-proteobacterium Burkholderia cepacia (B4EB12), the actinobacterium Mycobacterium smegmatis (A0QQV9), and the cyanobacteria Nostoc punctiforme (B2IVC2) and Anabaena variabilis (Q3ME48) failed to rescue the loss of DXR activity in E. coli cells (Table 1). By contrast, sequences from the alpha-proteobacterium Roseobacter litoralis (A9HDV1) and the firmicutes Listeria monocytogenes (Q723A4) and Bacillus halodurans (Q9KES5) were shown to be active in complementation assays (Table 1), suggesting that these organisms might have redundant enzymes catalyzing the production of MEP. However, only the DXR homologues from Listeria monocytogenes (Q720A5) and Bacillus halodurans (Q9KA69) were found to complement the EcAB4-10 mutant (Table 1 and Fig. S5). The DXR sequence from Roseobacter litoralis (A9GU34) was found to be inactive, likely as a consequence of the high number of amino acid changes in positions highly conserved across functional DXR enzymes (Fig. S6). These results indicate that some class B strains are functionally equivalent in terms of the MEP pathway to those in class A (i.e., have an active DRL enzyme but lack DXR activity). A third class (C) was formed by bacteria with DRL but no MEP pathway enzymes. This group included archea and bacteria that do not use the MEP pathway for isoprenoid biosynthesis (Table S1).
Table 1.
Complementation of the DXR-defective E. coli strain EcAB4-10 with the indicated sequences
| DRL |
DXR | ||||||||
| Accession | Length | Identity | Organism (Strain) | Group | Class | Clade | C-DRL | C-DXR | |
| 1 | Q2YIM3 | 438 | 100 | Brucella abortus 2308 | Alphaproteobacteria | A | + | + | np |
| 2 | A6WYQ0 | 438 | 90 | Ochrobactrum anthropi LMG3331 | Alphaproteobacteria | A | + | + | np |
| 3 | A6X6G6 | 439 | 75 | Ochrobactrum anthropi LMG3331 | Alphaproteobacteria | A | + | + | np |
| 4 | Q98FT2 | 442 | 73 | Mesorhizobium loti MAFF303099 | Alphaproteobacteria | A | + | + | np |
| 5 | Q989B6 | 433 | 67 | Mesorhizobium loti MAFF303099 | Alphaproteobacteria | A | + | + | np |
| 6 | Q6G2D9 | 445 | 65 | Bartonella henselae str Houston | Alphaproteobacteria | A | + | + | np |
| 7 | A9HDV1 | 443 | 62 | Roseobacter litoralis Och 149 | Alphaproteobacteria | B | + | + | − |
| 8 | Q9KES5 | 439 | 43 | Bacillus halodurans C-125 | Firmicutes | B | + | + | + |
| 9 | Q723A4 | 416 | 40 | Listeria monocytogenes F2365 | Firmicutes | B | + | + | + |
| 10 | B0S038 | 430 | 33 | Finegoldia magna ATCC 29328 | Firmicutes | A | + | + | np |
| 11 | B2IVC2 | 437 | 36 | Nostoc punctiforme PCC 73102 | Cyanobacteria | B | − | − | nt |
| 12 | Q3ME48 | 437 | 35 | Anabaena variabilis PCC 7937 | Cyanobacteria | B | − | − | nt |
| 13 | A9CES2 | 489 | 35 | Agrobacterium tumefaciens C58 | Alphaproteobacteria | B | − | − | nt |
| 14 | Q1V2P9 | 430 | 29 | Candidatus Pelagibacter ubique HTCC1002 | Alphaproteobacteria | B | − | − | nt |
| 15 | A0QQV9 | 454 | 32 | Mycobacterium smegmatis mc(2)155 | Actinobacteria | B | − | − | nt |
| 16 | B4EB12 | 442 | 27 | Burkholderia cepacia J2315 | Betaproteobacteria | B | − | − | nt |
Identity values (%) are relative to the B. abortus DRL protein (Q2YIM3). Class column indicates whether sequences similar to both DRL and DXR (class B) or only to DRL but not to DXR (class A) are present in the genome of the same strain. Clade column indicates the sequences belonging (+) or not (-) to the DRL clade shown in Fig. 4. C-DRL column indicates the DRL sequences complementing (+) or not (-) the DXR-defective E.coli strain. C-DXR column indicates the DXR sequences complementing (+) or not (-) the DXR-defective E.coli strain; np, DXR sequence not present in the genome; nt, DXR sequence not tested in complementation experiments.
DRL Homologues Cluster in a Monophyletic Group.
When putative DRL amino acid sequences resulting from BLAST searches were subjected to Maximum Likelihood (ML) phylogenetic analysis, all DRL sequences shown to be active in complementation experiments could be grouped in a clade (Fig. 4). Sequence identity of the proteins within the DRL clade ranged from 33–100% (Table S2). The DRL clade was supported by relatively low bootstrap values (64), but was consistently retrieved through two additional independent phylogenetic reconstruction methods [Neighbor Joining (NJ) and Maximum Parsimony (MP)]. All the organisms from class A had at least one DRL sequence from this clade (Table S1). The only exception was protein B5J045 from the strain 307 of the alpha-proteobacterium Octadecabacter antarcticus. However, a DRL sequence belonging to the clade (B5K941) was found in the genome of strain 238 (Table S1). Chloroflexus aurantiacus (chloroflexi) was the only class C organism harbouring a sequence from the DRL clade (Table S1). The rest of sequences in the DRL clade were from organisms belonging to class B (i.e., also had a DXR sequence in their genomes although not necessarily encoding an active DXR enzyme; Table 1). Most of these organisms were firmicutes but sequences from the alpha-proteobacterium Roseobacter litoralis, the beta-proteobacterium Verminephrobacter eiseniae and the marine actinobacterium PHSC20C1 were also included in the clade (Fig. 4 and Table S1). All the sequences tested from class B organisms that were not included in the DRL clade failed to complement the EcAB4-10 strain (Table 1), indicating that they were not true DRL enzymes. Taken together, these results strongly suggest that sequences showing identity to DRL but excluded from the DRL clade might not be functional DRL enzymes. Although some residues and short motifs are only found in active DRL sequences (Fig. S2B), future work will be necessary to ascertain their importance for enzymatic activity.
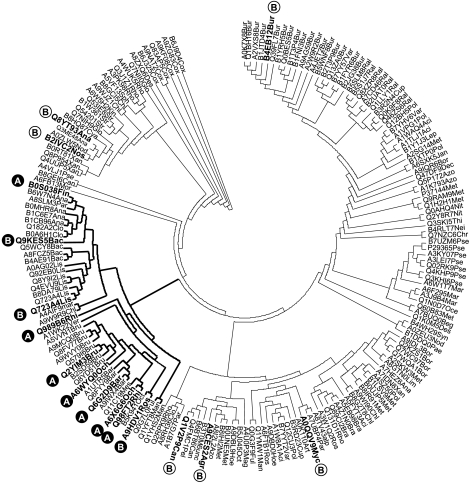
Fig. 4.
Phylogenetic analysis of putative DRL homologues. Unrooted ML phylogenetic tree depicting evolutionary relationships among DRL sequences resulting from BLAST searches with B. abortus DRL as a query (Table S2). Sequences complementing the E. coli EcAB4-10 strain are indicated with a black circle, whereas those that do not complement the mutant are indicated with a white circle. The letter inside the circles indicates whether the sequence belongs to an organism from class A (lacking a DXR sequence) or class B (harbouring a DXR sequence). The DRL clade grouping all active DRLs is highlighted.
Discussion
Phylogenetic analyses suggest that eukaryotes have inherited their pathways for IPP and DMAPP production from prokaryotes (20, 21). These studies proposed that the MVA pathway is germane to archaea (where it is required for the production of membrane ether-linked isoprenoid lipids) whereas the MEP pathway is the ancestral route in bacteria, serving for the production of precursors for quinones, carotenoids, and membrane stabilizers such as hopanoids. Although the majority of bacteria exclusively use the MEP pathway for IPP and DMAPP biosynthesis, there are some exceptions (20–22). For example, several species of parasitic Rickettsia and Mycoplasma bacteria have no genes encoding enzymes of the MVA or the MEP pathway, probably because they obtain their isoprenoids from host cells. Some bacteria have been confirmed to use the MVA pathway instead of the MEP pathway for IPP and DMAPP synthesis, whereas others possess the two full pathways (21–24). Strikingly, related species may use different pathways for isoprenoid biosynthesis as well as unrelated bacteria may use the same pathway. Here we show that some bacteria that synthesize their isoprenoids by the MEP pathway do not use DXR but DRL, either because they lack a copy of DXR in their genomes (those belonging to class A) or because their DXR sequence is not functional (such as that of Roseobacter litoralis; Table 1). Other class B strains might use both DXR and DRL (Table 1). Interestingly, some bacteria such as Listeria monocytogenes have complete and functional MVA and MEP pathways (24), including an active DXR protein, and additionally possess an active DRL enzyme (Table 1). Others such as Chloroflexus aurantiacus, which only uses the MVA pathway, might have a functional DRL (A9W9R9) as deduced for its position in the DRL clade (Fig. 4). Although functional DRL sequences have been mainly found in groups of alpha-proteobacteria and firmicutes, isolated instances of likely active DRL enzymes (as deduced by their grouping within the DRL clade) can be found in specific beta-proteobacteria, chloroflexi, and actinobacteria strains (Table S1). The scattered distribution of genes from both the MVA and the MEP pathways observed in living organisms was explained as a result of lateral gene transfer (LGT) among eukaryotes, bacteria, and archaea (20, 21). The outstanding discontinuity found in DRL distribution might be also explained by LGT occurring at different times during bacterial evolution (25). Further support for LGT as the most parsimonious explanation for the patchy distribution of DRL enzymes is provided by the discrepancy between DRL-based phylogenetic trees and organism phylogenies. Subsequent gene duplication and/or loss in specific bacterial evolutionary lineages might also have contributed to shape the puzzling phylogenetic distribution of DRL.
The use of alternative enzymes for the same biosynthetic step is not new in the isoprenoid field. For example, the interconversion of the universal IPP and DMAPP precursors is catalyzed by two types of IDI enzymes in bacteria: type I (similar to that found in animals, fungi, and plants) and type II, an unrelated enzyme (26). Although IDI activity is only essential in organisms that rely exclusively on the MVA pathway for IPP synthesis, a recent phylogenetic analysis found that many bacteria using the MEP pathway possess either a type I or a type II isomerase, whereas some bacteria have both (22). Similar to that proposed for bacteria with both type I and type II IDI enzymes, the presence of different enzymes catalyzing the conversion of DXP into MEP in the same organism (as it is the case of DXR and DRL in the firmicutes Listeria monocytogenes or Bacillus halodurans; Table 1) might confer an advantage in critical situations where isoprenoid biosynthesis is absolutely required. The existence of other class B organisms with a nonfunctional copy of any of those two enzymes would imply that such redundancy is not always favored, resulting in the mutation and inactivation of either DRL or DXR in that particular strain. This scenario provides an invaluable natural model to examine the evolutionary mechanisms responsible for the co-occurrence of homologous and nonhomologous genes encoding functionally redundant enzymes. All together, these results underscore the biochemical and genetic plasticity achieved by bacteria to synthesize essential compounds such as isoprenoids, a crucial point that should be further investigated before considering the MEP pathway as a convenient target for new antiinfective drugs.
Materials and Methods
Bacterial Strains, Media, and Reagents.
The Brucella abortus 2308 strain was grown in Brucella broth or Brucella agar plates (Pronadisa). Escherichia coli strains were grown in LB broth or plates. When indicated, the growth medium was supplemented with different concentrations of fosmidomycin (Molecular Probes) or 1 mM MVA prepared as described (27). Unless stated otherwise, chemicals and reagents were obtained from Sigma–Aldrich. Restriction enzymes and DNA modifying enzymes were purchased from Promega. Oligonucleotides were synthesized by Sigma–Aldrich, and are shown in Table S3.
Construction and Screening of a B. abortus Library.
Genomic DNA was extracted from B. abortus 2308 cells as previously described (28). Following partial digestion with Sau3A, fragments ranging from 3–6 kb were gel-purified and ligated into pUC19 previously digested with BamHI and dephosphorilated. The ligation mixture was transformed into E. coli DH5α and plated in LB medium containing 100 μg/mL ampicillin to amplify the library. Plasmid DNA was obtained from cells scrapped from the plates and pooled. For the screening, 1 μg of this DNA was electroporated into the DXR-defective EcAB4-10 strain and transformants were selected on LB plates supplemented with 20 μg/mL chloramphenicol (to select for the disruption of the dxr gene), 25 μg/mL kanamycin (to select for the presence of the MVA operon) and 100 μg/mL ampicillin (to select for the incorporation of library plasmids). Plasmids isolated from transformants were sequenced to check the identity of the inserts.
Cloning of DRL Sequences and Complementation Assays.
Genomic DNA from bacteria containing putative DRL sequences was PCR-amplified with the sets of oligonucleotides described in Table S3. DNA fragments of the expected size were purified and analyzed by restriction enzyme digestion to confirm their identity. Positive fragments were cloned into pJET1.2 (Fermentas). Plasmid DNA from two independent clones of each construct was used to transform E. coli EcAB4-10 cells. The cloned fragments were considered to encode functional DRL enzymes when they allowed growth of transformant EcAB4-10 cells in the absence of MVA. When indicated, E. coli strains EcAB4-2 and EcAB4-7 were also used for complementation assays as described (14).
Production of Recombinant B. abortus DRL Protein.
Oligonucleotides DRL_NdeI.F and DRL_XhoI.R (Table S3) were used to amplify BAB2_02264 without the stop codon from B. abortus 2308 genomic DNA. The amplified DNA fragment was cloned into the expression vector pET23b (Novagen) after digestion with NdeI and XhoI. Following transformation of E. coli BL21(DE3)pLys cells with the resulting construct (pET-DRL), the production of a chimeric DRL protein with a C-terminal tag of six histidine residues was induced by adding 0.4 mM IPTG to cultures with an OD600 = 0.5. After growth for 14 h at 28 °C, bacterial cells were recovered by centrifugation and the recombinant DRL protein was purified as described in SI Materials and Methods. Fractions containing DRL were pooled and stored at -20 °C in 50% glycerol.
Biochemical Characterization of the B. abortus DRL Enzyme.
The purified B. abortus DRL protein was assayed for potential DXR activity as follows. Reaction mixtures were prepared with 15 μg of recombinant DRL in 100 mM Tris-HCl pH 7.5, 2 mM MgCl2, 1 mM DTT, 0.78 mM DXP (Echelon), and 1 mM NADPH. Control reactions were prepared with either 15 μg of recombinant E. coli DXR protein or water. After incubation at 37 °C for 2 h, samples were diluted 2∶1 with water and analyzed by ultraperformance liquid chromatography coupled to mass spectrometry (UPLC-MS) in tandem mode as described in SI Materials and Methods. For the FSM inhibition assay and calculation of kinetic parameters, DXR activity was monitored by the change in absorbance at 340 nm as NADPH was oxidized.
Sequence and Phylogenetic Analysis.
Genes encoding isoprenoid biosynthetic enzymes were retrieved using the genomic BLAST tool at the National Center for Biotechnology Information database. BLAST searches for putative DRL sequences were performed in the UNIPROT database. Only hits returning scores corresponding to E-values < 0.001 were considered significant. Phylogenetic analyses were performed from protein sequence alignments obtained with CLUSTALW (29) by the ML, NJ, and MP methods. ML analyses were performed in PHYML v2.4.5, using the Jones, Taylor, and Thornton model of protein evolution (30, 31). NJ and MP were implemented in MEGA 4.0, using the default settings (32). To provide statistical confidence for the retrieved topology, a bootstrap analysis with 1,000 replicates was performed in each case.
Supplementary Material
Acknowledgments.
We thank L. M. Lois and J. F. Martínez-García for critical reading of the manuscript; Ana M. Cayón for technical help; and Juan Sanjuán (Estación Experimental del Zaidín CSIC, Granada, Spain), Héctor D. de Paz (University of Cantabria, Santander, Spain), Cristina Viadas and Ignacio López-Goñi (University of Navarra, Pamplona, Spain), M. Pilar Garcillán (University of Cantabria-IBBTEC, Santander, Spain), Annette Vergunst (Université de Montpellier, Nimes, France), Carlos Martín (University of Zaragoza, Zaragoza, Spain), Jose A. Vazquez-Bolland (University of Edinburgh, Edinburgh, UK), Takatsugu Goto (Gifu University, Gifu, Japan), Hideto Takami (Institute of Biogeosciences-JAMSTEC, Yokosuka, Japan), Amke den Dulk-Ras (Leiden University, Leiden, The Netherlands) Marie Johnson and Stephen Giovannoni (Oregon State University, Corvallis, OR), Yanhong Liu (US Department of Agriculture Agricultural Research Service, Pennsylvania), and Thorsten Brinkhoff (University of Oldenburg, Germany) for generously providing us with bacterial strains or DNA samples. The Spanish Ministerio de Ciencia e Innovación provided a PhD fellowship to J.P.-G., a Juan de la Cierva contract to L.C.-P. (cofinanced by the European Social Fund), research Grants BIO2007-63656 to F.J.S. and BIO2008-00432 to M.R.-C., as well as funding through a Consolider program (CSD2007-00036). This work was also supported by grants from the Fundación Marqués de Valdecilla (API 07/01) to F.J.S., and the Generalitat de Catalunya (SGR and XaRBa) to M.R.-C.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001962107/-/DCSupplemental.
References
- 1.Bouvier F, Rahier A, Camara B. Biogenesis, molecular regulation, and function of plant isoprenoids. Prog Lipid Res. 2005;44:357–429. doi: 10.1016/j.plipres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 3.Croteau R, Kutchan T, Lewis N. Natural products (secondary metabolites) In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Biologists; 2000. pp. 1250–1268. [Google Scholar]
- 4.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 5.Phillips MA, Leon P, Boronat A, Rodriguez-Concepcion M. The plastidial MEP pathway: Unified nomenclature and resources. Trends Plant Sci. 2008;13:619–623. doi: 10.1016/j.tplants.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenthaler HK, Rohmer M, Schwender J. Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol Plant. 1997;101:643–652. [Google Scholar]
- 7.Rodríguez-Concepción M, Boronat A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002;130:1079–1089. doi: 10.1104/pp.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Concepcion M. The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. Curr Pharm Des. 2004;10:2391–2400. doi: 10.2174/1381612043384006. [DOI] [PubMed] [Google Scholar]
- 9.Rohdich F, Bacher A, Eisenreich W. Isoprenoid biosynthetic pathways as anti-infective drug targets. Biochem Soc Trans. 2005;33:785–791. doi: 10.1042/BST0330785. [DOI] [PubMed] [Google Scholar]
- 10.Steinbacher S, et al. Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol 4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria drug development. J Biol Chem. 2003;278:18401–18407. doi: 10.1074/jbc.M300993200. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto Y, Furukawa S, Ogihara H, Yamasaki M. Fosmidomycin resistance in adenylate cyclase deficient (cya) mutants of Escherichia coli. Biosci Biotechnol Biochem. 2003;67:2030–2033. doi: 10.1271/bbb.67.2030. [DOI] [PubMed] [Google Scholar]
- 12.Brown AC, Parish T. Dxr is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake. BMC Microbiol. 2008;8:78. doi: 10.1186/1471-2180-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujisaki S, et al. Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene. 1996;175:83–87. doi: 10.1016/0378-1119(96)00128-x. [DOI] [PubMed] [Google Scholar]
- 14.Sauret-Güeto S, Uros EM, Ibanez E, Boronat A, Rodriguez-Concepcion M. A mutant pyruvate dehydrogenase E1 subunit allows survival of Escherichia coli strains defective in 1-deoxy-D-xylulose 5-phosphate synthase. FEBS Lett. 2006;580:736–740. doi: 10.1016/j.febslet.2005.12.092. [DOI] [PubMed] [Google Scholar]
- 15.Poliquin K, et al. Inactivation of sll1556 in Synechocystis strain PCC 6803 impairs isoprenoid biosynthesis from pentose phosphate cycle substrates in vitro. J Bacteriol. 2004;186:4685–4693. doi: 10.1128/JB.186.14.4685-4693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ershov YV, Gantt RR, Cunningham FX, Jr, Gantt E. Isoprenoid biosynthesis in Synechocystis sp. strain PCC6803 is stimulated by compounds of the pentose phosphate cycle but not by pyruvate or deoxyxylulose-5-phosphate. J Bacteriol. 2002;184:5045–5051. doi: 10.1128/JB.184.18.5045-5051.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chain PS, et al. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect Immun. 2005;73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLaBarre B, Thompson PR, Wright GD, Berghuis AM. Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases. Nat Struct Biol. 2000;7:238–244. doi: 10.1038/73359. [DOI] [PubMed] [Google Scholar]
- 19.Proteau PJ. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase: an overview. Bioorg Chem. 2004;32:483–493. doi: 10.1016/j.bioorg.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boucher Y, Doolittle WF. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol Microbiol. 2000;37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 22.Laupitz R, et al. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways. Eur J Biochem. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuzuyama T, Seto H. Diversity of the biosynthesis of the isoprene units. Nat Prod Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 24.Begley M, et al. The interplay between classical and alternative isoprenoid biosynthesis controls gammadelta T cell bioactivity of Listeria monocytogenes. FEBS Lett. 2004;561:99–104. doi: 10.1016/S0014-5793(04)00131-0. [DOI] [PubMed] [Google Scholar]
- 25.Boucher Y, et al. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 26.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc Natl Acad Sci USA. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez-Concepción M, et al. Genetic evidence of branching in the isoprenoid pathway for the production of isopentenyl diphosphate and dimethylallyl diphosphate in Escherichia coli. FEBS Lett. 2000;473:328–332. doi: 10.1016/s0014-5793(00)01552-0. [DOI] [PubMed] [Google Scholar]
- 28.Sangari FJ, Aguero J. Identification of Brucella abortus B19 vaccine strain by the detection of DNA polymorphism at the ery locus. Vaccine. 1994;12:435–438. doi: 10.1016/0264-410x(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 31.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 33.Sauret-Güeto S, Ramos-Valdivia A, Ibanez E, Boronat A, Rodriguez-Concepcion M. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem Biophys Res Commun. 2003;307:408–415. doi: 10.1016/s0006-291x(03)01211-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.