Abstract
We describe an innovative selection approach to generate self-reporting aptamers (SRAs) capable of converting target-binding events into fluorescence readout without requiring additional modification, optimization, or the use of DNA helper strands. These aptamers contain a DNAzyme moiety that is initially maintained in an inactive conformation. Upon binding to their target, the aptamers undergo a structural switch that activates the DNAzyme, such that the binding event can be reported through significantly enhanced fluorescence produced by a specific stacking interaction between the active-conformation DNAzyme and a small molecule dye, N-methylmesoporphyrin IX. We demonstrate a purely in vitro selection-based approach for obtaining SRAs that function in both buffer and complex mixtures such as blood serum; after 15 rounds of selection with a structured DNA library, we were able to isolate SRAs that possess low nanomolar affinity and strong specificity for thrombin. Given ongoing progress in the engineering and characterization of functional DNA/RNA molecules, strategies such as ours have the potential to enable rapid, efficient, and economical isolation of nucleic acid molecules with diverse functionalities.
Keywords: G-quadruplex, systematic evolution of ligands by exponential enrichment, sensor, binding induced folding, molecular recognition
Nucleic acid-based aptamers (1, 2) can be selected in vitro against a wide range of targets (3–8), chemically modified and synthetically produced, thereby making them a promising class of molecules for many applications including diagnostics (9, 10), in vivo imaging (11, 12), and targeted therapeutics (13, 14). Unlike most conventional affinity reagents (e.g., antibodies), aptamers can also be engineered to perform complex molecular functions beyond binding. For example, our group has recently demonstrated the use of an aptamer that undergoes target-binding-induced conformational change for continuous, real-time detection of a small molecule in undiluted blood serum (15). However, the selection process required to directly isolate aptamers with the desired function poses a significant technical challenge. One common approach entails selection for an aptamer on the basis of target binding, followed by postmodification and optimization through rational design in order to link the molecular binding capability to the desired functions (16, 17). Unfortunately, such approaches rely upon prior knowledge of the three-dimensional aptamer structure and require lengthy optimization steps that often compromise aptamers’ affinity and specificity for their targets (18–20).
Alternately, one can directly isolate aptamers with the desired function by combining a specifically designed nucleic acid library and a suitable selection process. For example, pioneering work by Nutiu and Li (21) described the isolation of structure-switching aptamers using a DNA library that was designed to incorporate two short randomized stretches sandwiching a central fixed-sequence motif, with binding sites for two PCR primers at the ends. The randomized segments participated in target recognition, whereas the central fixed-sequence motif was designed to hybridize to a complementary, 5′-biotinylated DNA (BDNA) strand, enabling immobilization of the DNA library onto streptavidin-coated beads. In this arrangement, aptamers capable of target-binding-induced folding are released from the beads into the solution due to disruption of the BDNA duplex. Although the aptamers obtained by this approach have an encoded capability for structure switching upon target binding, fluorescent signaling of this binding event requires the use of additional, modified DNA helper strands.
In this work, we describe an improved in vitro selection strategy for the efficient isolation of self-reporting aptamers (SRAs), which are capable of performing a biosensor function directly following isolation without the need for additional modification, optimization, or the use of DNA helper strands. More specifically, SRAs are selected to bind to a specific protein target, triggering folding and activation of a DNAzyme moiety that is initially inactive within the SRA structure. The DNAzyme sequence we used is a guanine-rich sequence that Sen and coworkers selected for binding to the small molecule dye N-methylmesoporphyrin IX (NMM) (22), and which was later found to catalyze porphyrin metalation (23). Accordingly, subsequent addition of NMM enables the simple detection of binding events based on significant enhancement of fluorescence. As a model, we demonstrate the isolation of SRAs that specifically recognize human α-thrombin with low nanomolar affinities, and which produce a strong fluorescent readout exclusively in response to specific target-binding events in either buffer or serum.
Results
Structure and Function of SRA Molecules.
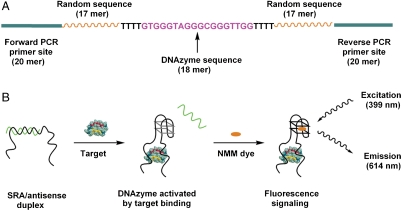
In order to directly select for structure-switching aptamers that self-report a specific target-binding event, we designed a structured DNA library in which each molecule includes three functional domains (Fig. 1A). The central, 18-base guanine-rich sequence is the DNAzyme domain (Fig. 1A, purple), which is able to form a supramolecular G-quadruplex structure with NMM, yielding significantly enhanced fluorescence at 614 nm (22, 24). The DNAzyme domain is flanked by two 17-base stretches of randomized sequence (Fig. 1A, orange), which participate in target binding during in vitro selection. The DNAzyme and randomized domains are separated by flexible linkers consisting of four thymine residues. Finally, two 20-base PCR primer-binding sites are located at the 5′ and 3′ ends (Fig. 1A, green).
Fig. 1.
The structure and function of SRAs. (A) The DNA library design features a central DNAzyme segment between a pair of 17-base random sequences, flanked by two 20-base PCR primer sites. (B) Specific binding between the SRA probe and its target protein induces the release of the antisense strand that maintains the DNAzyme moiety in an inactive state. Once unmasked, the DNAzyme moiety is able to form an active G-quadruplex structure and, upon binding to NMM, emits an enhanced fluorescence signal to report the specific binding event.
The central DNAzyme domain is able to adopt two distinct conformations: a low-fluorescence DNA duplex and a high-fluorescence aptamer-target complex. In the absence of target, the DNAzyme domain forms a stable duplex with an antisense strand (Fig. 1B, Left), prohibiting formation of the G-quadruplex structure; as a result, interaction with NMM is inhibited, yielding minimal background fluorescence. We found that a 12-base antisense strand is sufficient to effectively inhibit G-quadruplex formation. Upon target binding, however, the DNA duplex undergoes a large conformational change, dissociating from the antisense strand and forming the G-quadruplex structure (Fig. 1B, Center). This state enables the specific stacking interaction between NMM and the G-quadruplex structure (24, 25) to take place, yielding significantly enhanced fluorescence and thereby providing an effective readout for target-binding events (Fig. 1B, Right).
In Vitro Selection of SRAs.
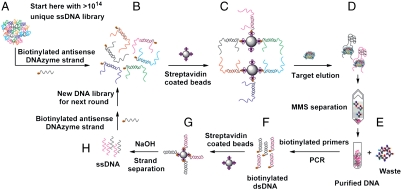
The SRA selection process (Fig. 2) begins with the preparation of the DNA library (∼1 nmol total, approximately 1014 random molecules) for the selection process. First, we inactivated the central DNAzyme domain via hybridization with the 12-base biotinylated antisense DNAzyme strand (Fig. 2 A and B). During this step, we also added two short DNA strands to block the PCR primer sites and limit their involvement in the folding of eventual SRAs. We then conjugated the resulting biotinylated DNA duplexes to streptavidin-coated magnetic beads (Fig. 2C). We employed the bead-conjugated library to isolate SRA molecules, which undergo target-binding-induced folding and are subsequently eluted from those molecules that remain bound to the bead surface (Fig. 2D). We performed this separation in two steps: first in a microcentrifuge tube with a conventional magnetic particle concentrator (MPC), after which the collected supernatant was further purified in the MicroMagnetic Separation (MMS) chip (Fig. 2E). The large magnetic field gradients within the MMS chip enabled high recovery (∼99.5%) of the captured beads and significantly reduced the presence of bead-associated DNA background in the supernatant (26, 27), yielding SRAs with exceptional purity in an efficient and rapid manner. After MMS separation, we PCR amplified the selected aptamers with biotinylated reverse primers (Fig. 2F) using an optimized cycle number to eliminate undesired PCR by-products (28). We purified the PCR products with streptavidin-coated magnetic beads and subsequently prepared single-stranded SRAs (Fig. 2G), which were in turn hybridized to the antisense DNAzyme- and primer site-blocking oligonucleotides in preparation for the next round of selection (Fig. 2H).
Fig. 2.
Overview of the SRA selection process. (A) Preparation of the DNA library. (B) Incubation of the DNA library with a 12-base biotinylated antisense DNAzyme strand and PCR primer site-blocking strands. (C) Immobilization of the duplex library on streptavidin-coated magnetic beads. (D) Library-bead assemblies are challenged with human α-thrombin target, resulting in elution of SRAs from beads. (E) Magnetic separation of eluted SRAs from bead-bound DNA in the MMS chip. (F) PCR amplification of the eluted SRAs with biotinylated reverse primers, followed by purification on streptavidin-coated magnetic beads. (G) Single-stranded DNA is generated from double-stranded PCR amplicons. (H) Incubation of enriched SRAs with biotinylated antisense DNAzyme strand and PCR primer site-blocking strands in preparation for the next round of selection.
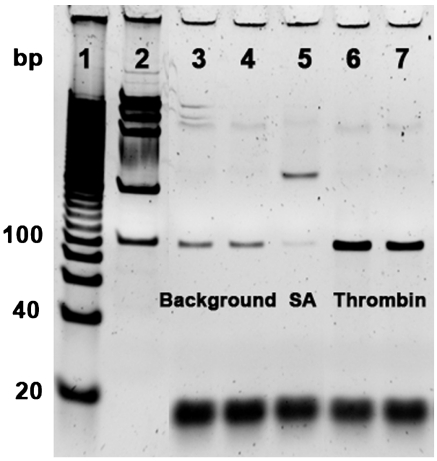
Initially, we performed eight rounds of positive selection to obtain SRAs with affinity for human α-thrombin. In order to further increase the specificity of the SRAs, we performed negative selection against streptavidin in rounds four through eight before positive selection. We chose streptavidin as the target for negative selection because it was used to immobilize the library DNA onto the beads and to generate single-stranded products after PCR. To monitor the convergence of the selection process, we used real-time PCR (RT-PCR) to measure the enrichment ratio of DNA molecules eluted by thrombin over those eluted by the selection buffer after each round. We amplified fractions of these DNA samples through an optimized PCR cycle, visualized them by gel electrophoresis, and calculated the enrichment ratio from the intensity of bands. After eight rounds of selection, the obtained aptamer pool clearly exhibited specific binding to thrombin. For example, aptamers eluted with 500 nM or 1 μM thrombin (Fig. 3, lanes 6 and 7) produced bands of ∼360% higher intensity than those eluted with target-free selection buffer (Fig. 3, lanes 3 and 4), confirming that selection was based on the induced conformational change of aptamers upon target binding. In contrast, we observed that a significantly smaller amount of aptamers were eluted with 1 μM streptavidin (Fig. 3, lane 5).
Fig. 3.
Gel electrophoresis image showing the affinity and specificity of the selected SRAs (100 bp product) for thrombin after eight rounds of selection. Lanes: 1, 100 bp ladder; 2, DNA mass standard (intensity of the 100 bp band is equivalent to 10 ng DNA); 3, DNA sample eluted with the selection buffer overnight; 4, DNA sample eluted with the selection buffer for 2 h; 5, DNA sample eluted with 1 μM streptavidin; 6, DNA sample eluted with 500 nM thrombin; 7, DNA sample eluted with 1 μM thrombin. Thrombin and streptavidin incubations were each performed for 2 h.
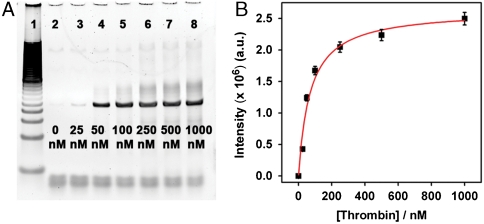
In order to further increase the affinity and specificity of the SRAs, we performed seven additional rounds of selection where each round involved a negative selection against streptavidin followed by a positive selection against thrombin. We also applied higher selection stringency by systematically decreasing thrombin concentrations from 1 μM to 25 nM over the course of these additional rounds of selection. After a total of 15 rounds of selection, we measured the equilibrium dissociate constant (Kd) of the selected SRA pool by immobilizing the SRAs on streptavidin-coated beads and challenging them with varying concentrations of thrombin. The eluted SRAs were PCR amplified and characterized by gel electrophoresis (Fig. 4A). The resulting gel image demonstrated that the amount of released aptamers increased proportionally with thrombin concentration, with a 2,400% enrichment of DNA aptamers eluted by 1 μM thrombin relative to selection buffer alone, implying that the structure switching between the DNA duplex structure and the aptamer-target complex is also dependent on target concentration (Fig. 4A). Assuming ideal PCR conditions, the Kd value of the resulting SRA pool is estimated to be 70 ± 14 nM from the intensities of the sample bands (Fig. 4B).
Fig. 4.
The dependence of SRA elution on thrombin concentration after 15 rounds of selection. (A) Gel image of the PCR-amplified DNA samples eluted by different concentrations of thrombin ranging from 25 nM to 1 μM (lanes 3–8). Lane 1 is 100 bp ladder, and lane 2 is selection buffer only; (B) The dissociation constant (Kd) of the selected SRA pool, calculated from the intensity of the sample bands, is 70 ± 14 nM.
Characterization of Individual SRA Sequences.
We cloned individual aptamer sequences from the pool of SRAs via insertion into the pCR4-TOPO vector and transformation into competent bacterial cells (29). Sequencing of 75 randomly picked colonies revealed three dominant consensus sequences, which constituted 95% of the population (Table 1). We synthesized these sequences and measured their dissociation constants to thrombin via NMM fluorescence as well as RT-PCR.
Table 1.
Sequences of the three dominant SRAs
| Aptamer ID | Selected sequence 5′ end | DNAzyme sequence | Selected sequence 3′ end |
| SRA_1 | ACTCGGGGCGTCAAAAAT4 | GTGGGTAGGGTGGGTTGG | T4AGCACAGAGTTGTGGAT |
| SRA_2 | GAACGGGGCAAGCAAAAT4 | GTGGGTAGGGTGGGTTGG | T4GCGGAATACCGTGAGAT |
| SRA_3 | GAGAAGGCGCAGAAAATT4 | GTGGGTAGGGTGGGTTGG | T4ATGCGCGATATGGTGAA |
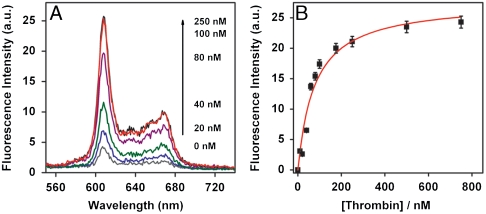
The three SRA sequences each exhibited effective biosensor function; when thrombin and NMM were added, all three antisense-hybridized SRA molecules emitted strong fluorescence that increased monotonically with increasing thrombin concentration (Fig. 5). For example, with SRA_1, fluorescence increased up to ∼700% when challenged with 100 nM thrombin (Fig. 5A). Assuming a 1∶1 Langmuir binding model, we calculated low nanomolar Kd values for all three selected SRAs based on the fluorescence data: 77 ± 14 nM, 83 ± 17 nM, and 76 ± 22 nM for SRA_1, SRA_2, and SRA_3, respectively (Fig. 5B and Figs. S1 and S2).
Fig. 5.
SRA_1 functions as an effective fluorescent reporter of target-binding events. (A) Fluorescence measurements of antisense-hybridized SRA_1 upon the addition of thrombin at varying concentrations. The intensity of fluorescence increased monotonically with increasing thrombin concentration. A 700% increase in fluorescence was observed with 100 nM thrombin compared to the sample without target. (B) Calibration curve of SRA_1 molecule derived from the fluorescence signal shows a Kd of 77 ± 14 nM.
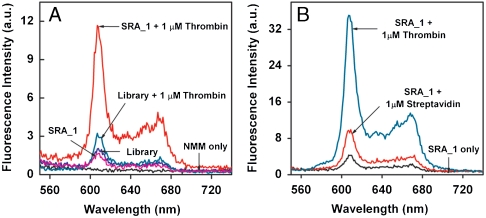
In order to verify that the enhanced fluorescence signals were indeed the result of target-binding-induced conformational change of the SRA molecules, we performed a number of control experiments. As expected, we observed that NMM alone produced negligible fluorescence background (Fig. 6A). Both the unselected library and SRA_1 emitted limited fluorescence when hybridized to the antisense DNAzyme strand in the absence of thrombin (Fig. 6A). The addition of thrombin to the antisense-hybridized unselected library yielded a slight increase in fluorescence (Fig. 6A), presumably due to low levels of nonspecific DNA-target binding. In contrast, SRA_1 showed a significant increase in fluorescence upon addition of thrombin due to specific target-induced conformational change (Fig. 6A). These results also indicate that NMM association with the SRA-target complex is essential in producing the enhanced fluorescence signal. Furthermore, the three SRA sequences demonstrated specific recognition of thrombin target; when challenged with 1 μM streptavidin, all three exhibited significantly lower fluorescence: 27%, 19%, or 35% of the fluorescence signal obtained from 1 μM thrombin-challenged SRA_1, SRA_2, or SRA_3, respectively (Fig. 6B and Fig. S3).
Fig. 6.
SRA_1 shows high specificity toward thrombin target. (A) Fluorescence measurements of target specificity of SRA_1. In comparison to the strong fluorescent signal from antisense-hybridized SRA_1 challenged with thrombin (red), we observed only limited signal in negative control experiments with NMM only (black), antisense-hybridized SRA_1 only (purple), or antisense-hybridized unselected library molecules either alone (blue) or challenged with thrombin (cyan). (B) SRA_1 duplexes challenged with 1 μM streptavidin exhibited 27% of the fluorescence signal obtained with 1 μM thrombin.
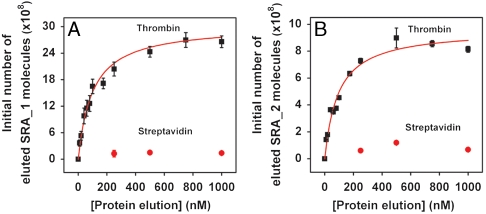
We further verified the Kd values of the three SRA sequences with RT-PCR. We challenged bead-conjugated SRA duplexes with varying concentrations of thrombin, and then determined the quantities of aptamers released as a result of target binding by measuring the threshold cycle (CT) values. The CT is calculated by determining the intersection of an amplification plot and a fixed fluorescence threshold value (30), and we note that this value is linearly related to the initial number of SRA molecules in the sample. Based on standard curves generated over four orders of magnitude, we determined that RT-PCR efficiencies were almost identical either in the presence or absence of thrombin (86% and 87%, 82% and 83%, and 83% and 84% for sequence SRA_1, SRA_2, and SRA_3, respectively) (see Fig. S4). We then used these standard curves and the obtained CT values to calculate the initial SRA quantity in each eluted sample. Finally, we subtracted the background, which was determined by the quantity of binders eluted in target-free selection buffer, and used the resulting numbers to determine the Kd of each SRA molecule. The Kd values obtained with RT-PCR correlated well with those measured by NMM fluorescence assay, yielding dissociation constants of 99.3 ± 12.9 nM and Kd = 93.6 ± 14.2 nM for SRA_1 and SRA_2, respectively (Fig. 7). Once again, both SRAs showed strong specificity for thrombin, and the respective signal magnitudes observed from SRA_1 and SRA_2 challenged with 1 μM streptavidin were only 5% and 8% of those produced by 1 μM thrombin (Fig. 7). Interestingly, SRA_3 showed slightly lower affinity (Kd = 156 ± 19.8 nM) and specificity compared to the other SRA molecules (see Fig. S5). Such variations have been previously reported in other examples of aptamer selection (31, 32).
Fig. 7.
SRA_1 and SRA_2 show specific affinity toward thrombin with nM dissociation constants and negligible binding to streptavidin. RT-PCR determination of dissociation constants for SRA_1 (A) and SRA_2 (B) yields Kd values of 99.3 ± 12.9 nM and Kd = 93.6 ± 14.2 nM, respectively.
Because the SRA signaling mechanism is based on a specific, target-induced conformation change, we hypothesized that SRA-based sensors may be relatively insensitive to the presence of contaminants. To test this, we measured the performance of antisense-hybridized SRA_1 molecules after a 10-min incubation in 50% FBS in the presence or absence of thrombin (Fig. 8). This relatively short incubation period was chosen because, unlike the signal in buffer, which increases with the increasing incubation time, the response in serum decreases after 10 min of incubation (see Fig S6). We attribute this phenomenon to DNA digestion by serum nucleases. Despite the short incubation time, the sensor yielded a 220% fluorescence increase in serum compared with the 270% fluorescence gain obtained in buffer at 750 nM thrombin concentration. We found that the background signals were noticeably larger in undoped serum compared to buffer; given that the FBS used in the experiment was harvested from calves in utero, we suspect that some activation of the blood-clotting cascade may have occurred, thereby producing detectable levels of thrombin in the serum (33). Similar background thrombin signal in serum has also been observed in related work (34).
Fig. 8.
Fluorescence measurements of antisense-hybridized SRA_1 upon addition of varying concentrations of thrombin in (A) Hepes buffer or (B) 50% FBS. The relatively larger background in serum caused by the presence of thrombin in the undoped serum. The background fluorescence of the undoped serum is subtracted from the three serum samples in B.
Discussion
We have demonstrated here an innovative in vitro selection method for the isolation of aptamer molecules capable of performing complex functions. These SRA molecules convert molecular recognition events into fluorescence readout via target-binding-induced folding of a known catalytic DNAzyme, and are thus capable of acting as ligand-specific signal reporters both in buffer and in the complex environment of serum. Unlike previous work, our combination of rational library design (i.e., incorporation of the DNAzyme domain into the library sequence) with efficient in vitro selection allows for the isolation of aptamers with desired functionality which can be directly used as biosensors without the need for postoptimization or modification.
Using this strategy, we obtained a pool of SRAs exhibiting low nanomolar affinities to thrombin with an average Kd of ∼70 nM after 15 rounds of selection. Upon sequencing 75 clones, we identified three consensus sequences that were most prevalent in the pool, two of which (SRA_1, SRA_2) showed specific affinity toward thrombin with Kd values (Kd = 77 and 83 nM, respectively) similar to the selected SRA pool. Similar Kd values were obtained from NMM fluorescence and RT-PCR assays, with minor differences that may be attributed to slight variations in buffer conditions (35). Interestingly, upon completion of the selection, we observed a single point mutation in the central DNAzyme domain (a C-to-T substitution at position 11 from the 5′ end), which produced relatively high background fluorescence. It may be possible to eliminate such phenomena via increased selection stringency (e.g., higher temperature, higher salt concentration) or through the use of modified nucleotides such as peptide nucleic acids or locked nucleic acids to create a stable, perfectly matched antisense structure for blocking the DNAzyme domain.
A number of functional DNA/RNA molecules such as cleavage DNAzymes (36), RNA riboswitches (37), and RNA ligases (38) have been recently discovered, and given the availability of efficient selection methods such as capillary electrophoresis (39–41) and microfluidic techniques (26, 27), we extrapolate that it may soon become possible to rapidly isolate nucleic acid molecules that incorporate multiple, diverse functionalities.
Materials and Methods
Reagents.
The DNA library, antisense DNAzyme strand, and primer site-blocking strands were synthesized and purified by Integrated DNA Technologies. Each library molecule contains a central 18-base DNAzyme sequence flanked by two 17-base randomized segments, with 20-base PCR primer-binding sites at each terminus. HotStarTaq Master Mix and RNase-free water were purchased from Qiagen, Inc. MyOne Streptavidin C1 Dynabeads were purchased from Invitrogen Dynal AS. Streptavidin (SA) and FBS were purchased from Sigma-Aldrich, Inc. and used without further purification. Human α-thrombin was purchased from Haematologic Technologies, Inc. (specific activity, 3,428 U mg-1). The iQ SYBR Green Supermix was purchased from Bio-Rad Laboratories for RT-PCR experiments.
Library Preparation and Antisense-Library Duplex Immobilization on Magnetic Beads.
One nanomole of ssDNA library (∼1014 unique molecules) was mixed with 1.5 nmol of each primer site-blocking strand, and 1.50 nmol of the biotinylated, antisense DNAzyme strand, and hybridized in 20 μL 1× Perfect Match buffer (PM; 1 mM phosphate buffer, 1 mM NaCl and 30 mM MgCl2, pH 6.8) by heating at 95 °C for 10 min and allowing to cool down to room temperature over 3 h. The library mixture was then incubated with 300 μL of SA-coated magnetic beads in 150 mM phosphate buffer with 1 M NaCl, 5 mM Mg2+, and 0.01% Tween-20, pH 7.5. Incubation was performed at RT overnight for optimal binding between SA and biotin. Nonspecifically bound DNA molecules were removed by washing the library-bound magnetic beads with various buffers in which NaCl concentration was gradually decreased from 1 M to 1 mM. Library-bead assemblies were resuspended in 300 μL of 1× thrombin binding buffer (TBB; 0.1 M Tris, 140 mM NaCl, 20 mM MgCl, and 20 mM KCl, pH 7.4,) and incubated overnight at room temperature.
Selection of SRAs.
The library-bead assemblies were challenged with 1 μM thrombin in 300 μL of 1× TBB for 2 h at RT. Aptamer molecules that underwent target-binding-induced conformation change were separated from bead-bound DNA molecules in two steps. The first was performed in a conventional tube with an MPC separator; the collected supernatant was then further purified in an MMS chip to achieve exceptional purity of the desired aptamers, as previously described (27). We performed negative selection against streptavidin in rounds four through eight before positive selection: the SRA-bead assemblies were incubated with 1 μM streptavidin in 1× TBB for 2 h at RT before challenged with thrombin target.
PCR Amplification.
A PCR mixture containing 50 μL HotStarTaq Master Mix, 0.25 μL of 0.1 mM forward primer, and 0.25 μL of 0.1 mM biotinylated reverse primer was prepared. This PCR mixture was then combined with 10 μL of DNA sample collected from the MMS chip and nuclease-free water to bring the total volume to 100 μL. The HotStarTaq polymerase was activated prior to PCR by heating reactions to 95 °C for 15 min, followed by 25 cycles of a rapid three-step PCR (30-s denaturation at 95 °C, 30-s annealing at 56 °C, 30-s extension at 72 °C). During the extension step of each cycle, 8 μL of PCR mixture were collected and resolved on a 10% PAGE-TBE (1× TBE: 89 mM Tris borate, 2 mM Na2EDTA, pH 8.3) gel to determine the optimal PCR amplification cycle number. Finally, the collected DNA sample was PCR amplified at the optimized cycle number.
ssDNA Generation.
The biotinylated, double-stranded PCR product was purified using the MiniElute PCR Purification Kit (Qiagen) with the manufacturer’s protocol, and the purified dsDNAs were then incubated with 150 μL of Dynabeads MyOne Streptavidin C1 for 2 h at RT. ssDNAs were generated by adding 100 mM NaOH and incubating for 4 min at RT, after which the supernatant was collected and neutralized with 1 N HCl, followed by a desalting step for the collected ssDNAs. ssDNA was quantified via UV-visible measurement at 260 nm.
Cloning and Sequencing of Selected Aptamers.
After 15 rounds of selection, the selected SRA pool was PCR amplified with unlabeled forward and reverse primers at the optimized PCR cycle number determined by the pilot PCR. The generated PCR products were purified by the MiniElute PCR Purification Kit (Qiagen) and cloned into Escherichia coli using the TOPO TA cloning kit (Invitrogen). Seventy-five colonies were randomly picked and sequenced at the GENEWIZ San Diego Laboratory.
RT-PCR Affinity Measurement.
We obtained real-time PCR standard curves to show that the threshold cycle (CT) values were linearly related to the initial number of SRA molecules over four orders of magnitude for SRA_1, SRA_2, and SRA_3, with and without thrombin. All samples were tested in triplicate, and PCR efficiencies were calculated from the slope of linear regressions through each set of data, using the equation E = (10-1/m - 1), where m is the slope of the linear regression, and E is PCR efficiency. Ten picomoles of the selected SRA pool or individual SRA sequences were immobilized on ∼2 × 108 magnetic beads by the same method used for the preparation of the ssDNA library. The DNA duplex-conjugated beads were challenged with thrombin target at varying concentrations from 10 to 1,000 nM in 100 μL of 1× TBB for 2 h at RT. Aptamers released as a result of target-binding-induced conformational change were subsequently quantified by RT-PCR. Each reaction contained 10 μL iQ SYBR Green Supermix, 8.8 μL PCR water, 0.1 μL of 0.1 mM forward primer, 0.1 μL of 0.1 mM reverse primer, and 1 μL DNA template, with fluorescence signal monitored using the iQ™ 5 multicolor RT-PCR Detection System (Bio-Rad Laboratories). CT values were subsequently determined for each target concentration. We then used the standard curves and the obtained CT values of the eluted SRA molecules to calculate the initial number of molecules in each sample. After subtracting background measurements obtained from samples eluted with target-free selection buffer, we used the initial numbers calculated for the eluted thrombin-challenged samples to determine the Kd of the SRA molecules.
Fluorescence Affinity Measurement in Buffer.
The affinities of the three dominant SRA sequences for thrombin were determined by fluorescence measurement using a Cary Eclipse spectrophotometer (Varian, Inc.) with quartz fluorescence cuvettes (4 × 10 mm; Submicro, 50 μL) with the following settings: excitation wavelength = 399 nm, excitation slit = 10 nm, emission slit = 5 nm, and photomultiplier tube voltage = 800 V. We hybridized 1 μM synthesized SRA or library DNA with primer site-blocking strands (1.5 μM) and antisense DNAzyme strand (1.5 μM) as described above in 60 μL of 25 mM Hepes buffer containing 20 mM MgCl2, 140 mM NaCl, and 100 mM KCl, pH 8.5. This mixture was challenged by thrombin target at varying concentrations ranging from 5 to 1,000 nM for 2 h at RT, after which 1 μL of 6 μM NMM dye was added, and incubated overnight. Fluorescent emission profiles were monitored in wavelength range from 450 to 760 nm, and the largest emission peak for each target concentration was determined.
Fluorescence Affinity Measurement in Serum.
Fifty percent FBS was prepared by diluting FBS with 50 mM Hepes buffer containing 40 mM MgCl2, 280 mM NaCl, and 200 mM KCl (pH 8.5) (total volume of 60 μL), and thrombin was added into the 50% FBS solution at RT to a final concentration of 0, 80, or 750 nM. We subsequently added 6 μL of 10 μM antisense-hybridized SRA_1 (in 1× PM buffer), followed immediately by 0.3 μL of 20 μM NMM (in DMSO). After a 10-min incubation, fluorescent emission profiles were monitored in the wavelength range from 450 to 760 nm, and the largest emission peak for each target concentration was determined. The measurement settings were the same as described above.
Supplementary Material
Acknowledgments.
We are grateful for the financial support of Office of Naval Research, National Institutes of Health, and the Institute for Collaborative Biotechnologies through the US Army Research Office.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009172107/-/DCSupplemental.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 4.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee J-H, et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Natl Acad Sci USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami J, Imanaka H, Yokota Y, Sugimoto N. In vitro selection of aptamers that act with Zn2+ J Inorg Biochem. 2000;82:197–206. doi: 10.1016/s0162-0134(00)00158-6. [DOI] [PubMed] [Google Scholar]
- 7.Gopinath SCB, et al. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J Gen Virol. 2006;87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 8.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JW, Cao ZH, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 11.Chen XL, et al. Using aptamer-conjugated fluorescence resonance energy transfer nanoparticles for multiplexed cancer cell monitoring. Anal Chem. 2009;81:7009–7014. doi: 10.1021/ac9011073. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 14.Farokhzad OC, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swensen JS, et al. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nutiu R, Li YF. Structure-switching signaling aptamers: Transducing molecular recognition into fluorescence signaling. Chem Eur J. 2004;10:1868–1876. doi: 10.1002/chem.200305470. [DOI] [PubMed] [Google Scholar]
- 17.Wang KM, et al. Molecular engineering of DNA: Molecular beacons. Angew Chem Int Edit. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 19.Jhaveri SD, et al. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J Am Chem Soc. 2000;122:2469–2473. [Google Scholar]
- 20.Jhaveri S, Rajendran M, Ellington AD. In vitro selection of signaling aptamers. Nat Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 21.Nutiu R, Li YF. In vitro selection of structure-switching signaling aptamers. Angew Chem Int Edit. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 22.Li YF, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 23.Li YF, Sen D. A catalytic DNA for porphyrin metallation. Nat Struct Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 24.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 1998;26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paramasivan S, Bolton PH. Mix and measure fluorescence screening for selective quadruplex binders. Nucleic Acids Res. 2008;36:e106. doi: 10.1093/nar/gkn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou XH, et al. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci USA. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian JR, Lou XH, Zhang YT, Xiao Y, Soh HT. Generation of highly specific aptamers via micromagnetic selection. Anal Chem. 2009;81:5490–5495. doi: 10.1021/ac900759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tok JBH, Fischer NO. Single microbead SELEX for efficient ssDNA aptamer generation against botulinum neurotoxin. Chem Commun. 2008:1883–1885. doi: 10.1039/b717936g. [DOI] [PubMed] [Google Scholar]
- 29.Shuman S. Novel-approach to molecular-cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J Biol Chem. 1994;269:32678–32684. [PubMed] [Google Scholar]
- 30.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 31.Baldrich E, Restrepo A, O’Sullivan CK. Aptasensor development: Elucidation of critical parameters for optimal aptamer performance. Anal Chem. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- 32.Cho EJ, Collett JR, Szafranska AE, Ellington AD. Optimization of aptamer microarray technology for multiple protein targets. Anal Chim Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 33.Aronson DL, Stevan L, Ball AP, Franza BR, Finlayson JS., Jr Generation of the combined prothrombin activation peptide (F1-2) during the clotting of blood and plasma. J Clin Invest. 1977;60:1410–1418. doi: 10.1172/JCI108902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Edit. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 35.Haynes SR, editor. RNA-Protein Interaction Protocols. Totowa, NJ: Humana Press; 1999. pp. 105–114. [Google Scholar]
- 36.Willner I, Shlyahovsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem Soc Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 37.Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 38.Robertson MP, Ellington AD. Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res. 2000;28:1751–1759. doi: 10.1093/nar/28.8.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendonsa SD, Bowser MT. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 40.Berezovski M, et al. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J Am Chem Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 41.Drabovich A, Berezovski M, Krylov SN. Selection of smart aptamers by equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM) J Am Chem Soc. 2005;127:11224–11225. doi: 10.1021/ja0530016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.