Abstract
Pax transcription factors are involved in a variety of developmental processes in bilaterians, including eye development, a role typically assigned to Pax-6. Although no true Pax-6 gene has been found in nonbilateral animals, some jellyfish have eyes with complex structures. In the cubozoan jellyfish Tripedalia, Pax-B, an ortholog of vertebrate Pax-2/5/8, had been proposed as a regulator of eye development. Here we have isolated three Pax genes (Pax-A, Pax-B, and Pax-E) from Cladonema radiatum, a hydrozoan jellyfish with elaborate eyes. Cladonema Pax-A is strongly expressed in the retina, whereas Pax-B and Pax-E are highly expressed in the manubrium, the feeding and reproductive organ. Misexpression of Cladonema Pax-A induces ectopic eyes in Drosophila imaginal discs, whereas Pax-B and Pax-E do not. Furthermore, Cladonema Pax-A paired domain protein directly binds to the 5′ upstream region of eye-specific Cladonema opsin genes, whereas Pax-B does not. Our data suggest that Pax-A, but not Pax-B or Pax-E, is involved in eye development and/or maintenance in Cladonema. Phylogenetic analysis indicates that Pax-6, Pax-B, and Pax-A belong to different Pax subfamilies, which diverged at the latest before the Cnidaria–Bilateria separation. We argue that our data, showing the involvement of Pax genes in hydrozoan eye development as in bilaterians, supports the monophyletic evolutionary origin of all animal eyes. We then propose that during the early evolution of animals, distinct classes of Pax genes, which may have played redundant roles at that time, were flexibly deployed for eye development in different animal lineages.
Keywords: biodiversity, Cladonema radiatum, Cnidaria, evo-devo, gene duplication
The Pax gene family encodes transcription factors involved in a variety of developmental processes in metazoans (1–3). It can be subdivided into five subfamilies—Pax-4/6, Pax-2/5/8, Pax-1/9, Pax-3/7, and pox neuro (poxn)—on the basis of structural comparison and phylogenetic relationship (4). These subfamilies diverged from each other at the latest before the separation of cnidarians and bilaterians (4, 5).
It is widely accepted that Pax-6, a member of Pax-4/6 subfamily, is one of the most significant components of the gene network that controls eye development in many bilaterians (e.g., refs. 6, 7); mutations in Pax-6 genes cause severe eye defects both in mammals and in Drosophila (8, 9), and Pax-6 genes cloned from diverse bilaterians can ectopically initiate eye development both in Drosophila and in Xenopus when they are misexpressed (6, 10, 11).
Cnidaria are the earliest branching animal phylum containing species with multicellular eyes, which sometimes show complex structures such as lens, iris, pigmented layer, and photosensitive layer (12). Among the five classes of the phylum Cnidaria (Anthozoa, Hydrozoa, Cubozoa, Scyphozoa, and recently recognized Staurozoa), four of them (Hydrozoa, Cubozoa, Scyphozoa, and Staurozoa) include eye-bearing species (12). However, no bona fide Pax-6 has been identified in cnidarians to date.
Kozmik et al. (13) have proposed that Pax-B, a member of the Pax-2/5/8 subfamily, is responsible for eye development in Tripedalia cystophora, a cubozoan jellyfish. Tripedalia Pax-B is expressed in the rhopalia, which are batteries of sensory organs including eyes. This gene is also able to transactivate the promoters of a Tripedalia crystallin gene and a Drosophila rhodopsin gene in cell culture, and it induces ectopic eyes in Drosophila when misexpressed in imaginal discs. These data suggest that Pax-B is responsible for eye development in Tripedalia. The authors have further proposed that Pax-6 diverged from Pax-B (Pax-2/5/8) by gene duplication in the bilaterian lineage after the separation from cnidarians and was independently recruited for controlling eye development in the bilaterian lineage (13, 14). Accordingly, they have hypothesized that cnidarian and bilaterian eyes arose independently.
Besides the class Cubozoa, the class Hydrozoa includes several species with complex eyes (12). Sun et al. (15) have investigated Cladonema californicum, a hydrozoan jellyfish bearing eyes. However, they have found only a Pax-B gene, whose function has not been well studied in Cladonema.
In this study, we have isolated and characterized three Pax genes from Cladonema radiatum, which possesses eyes with elaborate structures including lens, pigmented cell layer, and photosensitive cell layer (16, 17) (Fig. 1A). Our data suggest that Pax-A, a member of the poxn subfamily, rather than Pax-B, plays a major role in the Cladonema eye. We argue that our results support a hypothesis that all of the animal eyes have a single evolutionary origin (6). We propose that, for development and/or maintenance of eyes, distinct lineages of animals flexibly selected different classes (corresponding to subfamilies) of Pax genes, the ancestors of which may have had redundant roles in the common ancestor of cnidarians and bilaterians.
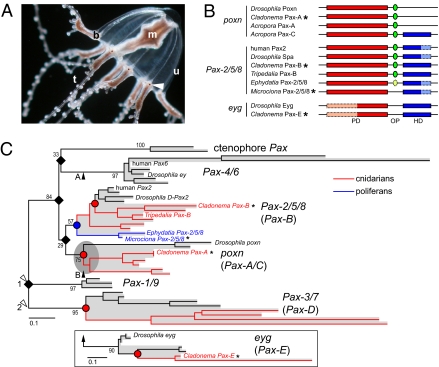
Fig. 1.
Pax genes cloned from Cladonema and Microciona, and molecular phylogenetic tree of the Pax family. (A) Medusa of C. radiatum. Arrowhead indicates an eye. b, tentacle bulb; m, manubrium; t, tentacle; u, umbrella. Photo courtesy of Claudia List. (B) Structures of Pax proteins. Those cloned in this study are marked by asterisks. Red box, green ellipse, and blue box represent the PD, octapeptide (OP), and HD, respectively. Octapeptide-like motifs of the poxn subfamily are found at amino acid positions 360–367 for Drosophila Poxn, 435–442 for Cladonema Pax-A, 324–331 for Acropora Pax-A, and 153–160 for Acropora Pax-C. Symbols with dashed line and pale color indicate highly divergent sequences. (C) The ML tree inferred from a comparison of the whole PD amino acid sequences. See Fig. S1 for details. Two possible root positions are indicated according to previous publications (4, 5, 22) (white arrowheads). The eyg subtree derived from an additional ML analysis (Fig. S2B), which is performed on the basis of comparison of the latter halves (RED subdomains) of the PD sequences, is shown in a box. Black arrowheads indicate two possible ML positions of the eyg subtree suggested by further ML analyses based either on a comparison of the RED subdomain sequences (arrowhead A; Fig. S3A) or on that of the RED subdomain plus the whole HD sequences (arrowhead B; Fig. S3B). Because complete HDs are present only in Pax-C among the members of the poxn subfamily, Pax-A and poxn were excluded from the latter analysis. The position of the eyg subtree is therefore ambiguously shown at the root of poxn subtree (gray ellipse). Subtrees corresponding to distinct subfamilies are shaded gray and the subfamily names are shown on the right, with the names of cnidarian members in parentheses. Cnidarian sequences and poriferan sequences are shown in red and blue, respectively. Red and blue circles indicate the cnidarians–bilaterians and the poriferans–eumetazoans splits, respectively. Filled rhombi indicate the gene duplications that gave rise to the subfamilies. The extended local bootstrap probability is shown at each branch that separates subfamilies.
Results
Identification of Pax Genes from Hydrozoan Jellyfish and Marine Sponge.
By performing degenerate PCR with multiple primer combinations, we obtained three cDNAs encoding Pax proteins from C. radiatum. Two of the cloned Pax genes show high sequence similarities and identical domain structures (Fig. 1B) to previously identified cnidarian Pax genes, Pax-A and Pax-B; C. radiatum Pax-A and Pax-B (designated CrPax-A and CrPax-B) show 100% and 83% amino acid identity in the paired domain (PD) to Hydra magnipapillata Pax-A and Pax-B, respectively. The third gene (CrPax-E) does not show a particularly close relationship to any of the cnidarian Pax genes identified thus far. Interestingly, an identical domain structure is shared by CrPax-E and Drosophila Eyegone (Eyg), both of them having a highly divergent N-terminal subdomain (called PAI) of PD, and a complete homeodomain (HD), but lacking an octapeptide sequence (Fig. 1B).
To gain further insights into the diversity of Pax genes in basal metazoans, we investigated several additional organisms. Among metazoans, poriferans (sponges) show the simplest body plans, lacking nervous system and true organs. Although some sponge larvae show photosensitive responses (18, 19), they seem to use different cell- and molecular-level mechanisms of photoreception from those of eumetazoans (18–21). All of the Pax genes identified so far from various species of demosponges are of the Pax-2/5/8 type. Only one Pax cDNA that we could obtain from the marine sponge Microciona prolifera by degenerate PCR is closely related to the Pax-2/5/8 of the freshwater sponge Ephydatia fluviatilis (22) (92% amino acid identity in the PD). The only Pax gene found in the complete genome sequence of the marine sponge Amphimedon queenslandica (named AmqPaxB) (23), and another Pax gene recently found in another marine sponge, Chalinula loosanoffi (24), also belong to the Pax-2/5/8 subfamily. From the genome sequence of the choanoflagellate Monosiga brevicollis, a protist thought to be the closest relative to metazoans (25), no Pax gene was found.
Phylogenetic Tree Analyses of the Pax Gene Family.
We inferred a phylogenetic tree based on a comparison of the whole PD sequences by the maximum likelihood (ML) method (Fig. 1C). The phylogenetic tree provides supports to the previous classification of the Pax genes into five subfamilies: Pax-4/6, Pax-2/5/8, Pax-1/9, Pax-3/7, and pox neuro (poxn) (4, 5). The ctenophore Pax genes form an independent cluster, being away from the other metazoan Pax subfamilies. It remains unclear whether they represent a novel subfamily or it is simply an artifact caused by the high evolutionary rate (4).
In Fig. 1C, CrPax-A and CrPax-B are classified into the poxn and Pax-2/5/8 subfamilies, respectively. The classification of CrPax-E was carried out separately because its PD is incomplete. The identical domain structure shared by CrPax-E and Drosophila Eyg suggests their close evolutionary relationship. Searches of the genome sequences of Anopheles, two species of nematodes, and Hydra revealed in each organism the presence of a Pax gene that lacks a PAI subdomain or has a highly divergent one, as happens in CrPax-E and Drosophila eyg. Phylogenetic analyses using the sequences of the second half of PD (RED subdomain) provides a strong support to the independent clustering of all those Pax genes that have the truncated PDs (Fig. S2; Bayesian posterior probability of 1.0 and extended local bootstrap probability of 90%). We therefore propose that CrPax-E and eyg compose a novel subfamily (eyg subfamily) together with the other Pax genes bearing the degenerated PDs. To determine the branching position of the eyg subfamily, we exhaustively evaluated all of the possible branching positions of the eyg subtree on the ML tree in Fig. 1C by log-likelihood values. These analyses using (i) only the RED subdomains and (ii) the RED subdomains plus the whole HDs indicated two different branching positions (Fig. 1C; arrowheads A and B, respectively) as most likely, although none of them excluded the other possibilities statistically significantly (Fig. S3).
Consistent with previous reports (4, 26), our phylogenetic tree analysis strongly suggests that the majority of gene duplications (subfamily-generating duplications; indicated by black rhombi in Fig. 1C) that gave rise to different Pax subfamilies occurred before the separation of cnidarians and bilaterians (Fig. 1C, red circles), no matter where the tree root is situated (white arrowheads) (4, 5, 22). The affiliation of the sponge Pax genes to the Pax-2/5/8 subfamily (4, 22) suggests an even earlier occurrence (before the separation of poriferans and eumetazoans) of these duplications (4, 22). It is thus likely that at the latest the common ancestor of cnidarians and bilaterians once possessed a gene belonging to the Pax-4/6 subfamily, which was subsequently lost or has not yet been found in the cnidarian lineage.
Cladonema Pax-A Is Expressed in the Eye, Whereas Pax-B Is Expressed in the Oocytes.
In the cubozoan jellyfish T. cystophora, Pax-B is highly expressed in the eyes (13). This supports the hypothesis that Pax-B is involved in eye development in this species. To identify a Pax gene that is potentially involved in eye development in the hydrozoan jellyfish C. radiatum, we performed expression analyses of the Pax genes by real-time quantitative RT-PCR (Fig. 2 A–C). The data unexpectedly showed that CrPax-B is mainly expressed in the manubrium, the feeding and reproductive organ, but not at the bases of the tentacles (tentacle bulbs) where the eyes develop. CrPax-E shows a similar expression profile to CrPax-B. Instead, CrPax-A turned out to be highly expressed in the tentacle bulbs.
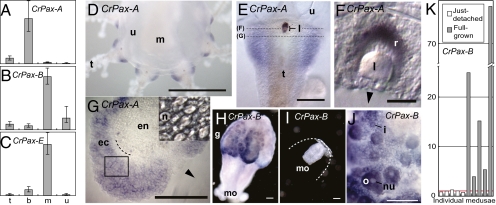
Fig. 2.
Expression of Cladonema Pax genes. Cladonema medusae were dissected into four body parts, tentacles (t), tentacle bulbs with eyes (b), manubrium (m), and umbrella (u) (Fig. 1A) and expression levels of CrPax-A (A), CrPax-B (B), and CrPax-E (C) were quantified by real-time RT-PCR analysis. The expression level in each body part relative to the whole body (1.0) was calculated and subsequently normalized to the expression level of elongation factor 1α (EF1α). The quantifications were performed three times on different cDNAs generated independently, and geometric means were calculated. The y axes are arbitrary. Error bars represent SDs. (D–J) Whole-mount in situ hybridization analyses with anti-sense RNA probes for CrPax-A (D–G) and CrPax-B (H–J). Cross-sections of the eye (F) and the whole tentacle (G) of the CrPaxA probe-hybridized jellyfish are shown. The cutting planes are indicated by the dashed lines in E. Note that the natural eye pigment disappears during the hybridization procedure. (G) A phase contrast microscopic picture of the ectoderm tissue, which corresponds to the box, is shown in the Inset. Dashed line indicates the border between endoderm and ectoderm. Arrowheads indicate the frontal side of a tentacle. (H and I) Excised manubriums. The original position of the umbrella is indicated by a dashed line in I. ec, ectoderm; en, endoderm; g, gonads; i, immature oocyte; l, lens; m, manubrium; mo, mouth; n, nematoblast; nu, nucleus; o, mature oocyte; r, retina; t, tentacle; u, umbrella. (Scale bars, 500 μm in D, 100 μm in E and G–I, 10 μm in F.) (K) CrPax-B expression levels quantified independently for five just-detached and five full-grown medusae by real-time RT-PCR. Their expression levels relative to the average (red line) of the just-detached medusae are presented. Data normalized to EF1α.
The expression pattern of CrPax-A (Fig. 2 D–G), analyzed by whole-mount in situ hybridization, is consistent with the result of real-time RT-PCR analysis. CrPax-A is expressed both in the ocellus and in the peripheral tissue of the tentacle bulb (Fig. 2 D and E). Cryosections clearly show that the staining is localized in the retina (Fig. 2F) and in the ectodermal tissue of the tentacle bulb (tentacle bulb ectoderm; TBE) (Fig. 2G), where numerous differentiating nematoblasts (developing stinging cells) (27) are observed (Fig. 2G, Inset).
CrPax-B transcripts were detected by in situ hybridization in the aboral region of the manubrium, where gonads develop (Fig. 2H). In a young medusa with no apparent gonads, the expression was undetectable (Fig. 2I). The CrPax-B expression was observed in the cytoplasm of oocytes at various stages of development (Fig. 2J). We further quantified the CrPax-B expression levels in 10 medusae at two different growth stages: just-detached and full-grown (see Materials and Methods for definitions). The expression of CrPax-B in the full-grown medusae with gonads was 3.9–77 times higher than that in the just-detached medusae that lack gonads (Fig. 2K). These data indicate that CrPax-B is active in the maturing oocytes.
Cladonema Pax-A Induces Ectopic Eyes in Drosophila, Whereas Pax-B and Pax-E Do Not.
Tripedalia Pax-B can ectopically initiate eye development in Drosophila when it is misexpressed in imaginal discs by activating the intrinsic gene network that directs compound eye development (13). We tested the ability of Cladonema Pax genes to induce ectopic eyes in Drosophila by using the yeast Gal4/UAS system (28). The targeted expression of CrPax-A in imaginal discs using the dppblink-Gal4 driver induced ectopic eyes, but only in a limited portion of the screened flies. However, the additional use of UAS-Gal4, which increases Gal4 expression, resulted in ectopic eye inductions on more than 50% of the screened flies (Fig. 3A). Most of the induced eyes were observed in the notum, which develops from the wing imaginal disk. The induced eyes clearly displayed ommatidia when analyzed by SEM (Fig. 3B). In contrast to CrPax-A, CrPax-B and CrPax-E did not induce ectopic eyes either with the dppblink-Gal4 driver alone or with the combination of that driver and the UAS-Gal4 transgene.
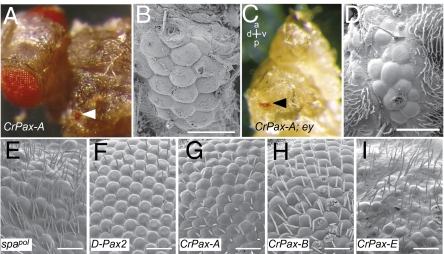
Fig. 3.
Ectopic eye induction in Drosophila and rescue of the spapol mutant phenotype by Cladonema Pax genes. (A) CrPax-A was expressed under the control of dppblink-Gal4 driver with UAS-Gal4. Arrowhead indicates the induced eye. (B) SEM picture of the induced eye. (C) Misexpression of CrPax-A under the control of dppblink-Gal4 driver in a homozygous ey null mutant (eyJ5.71) background induced ectopic eyes (arrowhead). The anteroposterior (a-p) and dorsoventral (d-v) axes are shown. Note that the natural compound eye is absent. The genotype of ey mutant was confirmed by the absence of the second exon of ey (30) by PCR. (D) SEM picture of the induced eye. (E) Eye phenotype of the spapol homozygous fly. (F–I) Rescue experiments of the spapol mutant phenotype. UAS-D-Pax2 (F; positive control), UAS-CrPax-A (G), UAS-CrPax-B (H), and UAS-CrPax-E (I) transgenic lines were crossed with the spa-Gal4 driver line in a spapol homozygous background. (Scale bars, 30 μm.)
Cladonema Pax-A Can Activate the Drosophila Eye Determination Gene Network in the Absence of ey.
Initiation of eye development in Drosophila requires expression of a set of genes that compose a transcriptional regulatory network, sometimes referred to as the retinal determination gene network (RDGN) (7). This network contains a positive feedback transcriptional loop comprising eyeless (ey; Drosophila Pax-6), sine oculis, eyes absent, and dachshund (7, 29). Initially, this transcriptional loop was thought to be ignited only by the activation of ey by twin of eyeless (toy; another Pax-6 gene) (29). However, it has been shown that toy alone is able to initiate the transcriptional loop in the absence of ey by directly activating another loop member, sine oculis (30). To examine how the jellyfish Pax gene activates the Drosophila RDGN, we tested whether CrPax-A can still induce ectopic eyes in the absence of ey. Indeed, the expression of CrPax-A, driven by the dppblink enhancer, induced ectopic eyes in an ey null mutant (eyJ5.71) background (Fig. 3 C and D). Our data indicate that the CrPax-A initiates the Drosophila RDGN by activating the components of the transcriptional loop, even in the absence of ey, as toy does.
Both Cladonema Pax-A and Pax-B Can Substitute for Pax-2 in the Drosophila Eye.
In the Drosophila spapol mutant (Fig. 3E), the expression of D-Pax2, a member of the Pax-2/5/8 subfamily, in cone and primary pigment cells is abolished, resulting in a severely disturbed development of ommatidial cells (31). We used this mutant to test whether the Cladonema Pax genes can substitute for the D-Pax2 functions in ommatidial cells, by performing rescue experiments. Interestingly, not only CrPax-B, an ortholog of D-Pax2, but also CrPax-A significantly rescued the spapol eye phenotype when they were expressed under the control of the cone- and pigment cell–specific enhancer (spa enhancer) (31, 32) of D-Pax2. CrPax-E, however, did not rescue the phenotype. SEM pictures clearly show that the hexagonal shape of each ommatidium and the regular arrangement of interommatidial bristles were largely recovered both by the expression of CrPax-A and by that of CrPax-B in the developing eye (Fig. 3 F–I).
CrPax-A PD Directly Binds to the Upstream Regions of Eye-Specific Opsin Genes.
Previous publications had proposed that Drosophila Ey directly regulates the expression of rhodopsin genes through its HD (33, 34). Likewise, Tripedalia Pax-B can transactivate a lacZ reporter gene under the control of a Drosophila rhodopsin promoter, presumably through its HD (13). Cladonema Pax-A, however, lacks an HD. We therefore tested the possibility that CrPax-A can still directly bind the promoter regions of Cladonema eye-specific opsin genes, which had been characterized in our previous study (17), in the absence of an HD. We screened ≈1 kb of 5′ promoter fragments by performing EMSA with PD proteins using sets of probes that cover the fragments. In two examined opsin genes (CropG1 and CropN1), one and two CrPax-A–specific PD binding sites (red vertical lines in Fig. 4A) were identified, respectively (Fig. 4B). No or very faint binding was detected when the putative binding sites were mutated (Fig. 4B). The sequences of the identified CrPax-A PD binding sites moderately match the chordate Pax-6 PD consensus binding sequence (35) (Fig. S4). In contrast, CrPax-B PD showed little, if any, binding to any of the tested probes (Fig. 4B; only the three probes that showed the CrPax-A binding are shown). These results are consistent with the notion that Pax-A, but not Pax-B, is involved in eye development and/or maintenance in Cladonema.
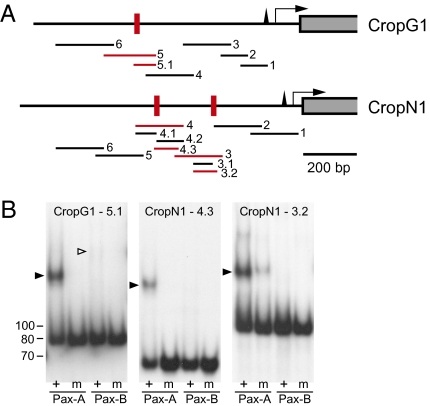
Fig. 4.
CrPax-A PD binding sites in the upstream region of eye-specific Cladonema opsins. (A) Schematic drawing showing the positions of the probes generated for EMSA. Probes to which CrPax-A PD bound are shown in red. Red vertical bars indicate the positions of the CrPax-A binding sites, which were precisely identified by the use of mutated probes. Black triangles, arrows, and gray boxes represent TATA boxes, transcription starting sites, and protein coding regions, respectively. (B) EMSA for the three positive probes: probe 5.1 of CropG1, and probe 4.3 and 3.2 of CropN1. +, wild-type probes; m, mutated probe. The size (bp) of the probe is shown at left. Arrowheads indicate the band shifts caused by the binding of proteins. Note that the bindings of CrPax-B PD are very faint or undetectable (white arrowhead), even though the same amount of the proteins as CrPax-A PD were used. CrPax-B PD was separately proven to be active by the use of a control probe carrying D-Pax2 binding sites (Materials and Methods).
Discussion
Functional Diversity of Cladonema Pax Genes.
In this study, we have characterized three Pax genes (CrPax-A, CrPax-B, and CrPax-E) from C. radiatum, a hydrozoan jellyfish. From the expression analyses, we assume that CrPax-A is involved in eye development and/or maintenance, whereas CrPax-B is required for the oocyte maturation process. Although CrPax-E is predominantly expressed in the manubrium, attempts to detect its transcripts by in situ hybridization were not successful, probably owing to its low expression level. Interestingly, the dramatic gene up-regulation observed for CrPax-B upon gonad formation (Fig. 2K) was not observed for CrPax-E, suggesting that they exert different functions in the manubrium.
The expression of CrPax-A in the TBE, in addition to the retina, suggests its involvement in nematogenesis because the TBE is the specialized site for the tentacular nematocyte differentiation in hydrozoan jellyfish (27, 36, 37). Like bilaterian Pax genes (2, 3, 38), CrPax-A seems to be involved in multiple aspects of developmental processes.
Involvement of Pax-A in Cladonema Eye Development and/or Maintenance.
By targeted expression experiments, we have shown that CrPax-A is able to ectopically initiate eye development in Drosophila, whereas CrPax-B and CrPax-E are not. This agrees with the results of our expression analyses, which show that only CrPax-A is highly expressed in the eye (Fig. 2).
The Drosophila ey gene includes an eye-specific enhancer that contains Pax-6 protein binding sites (29). It is possible that an introduced Pax gene simply augments the endogenous Ey protein level, which then induces the ectopic eyes. By showing the ability of CrPax-A to induce ectopic eyes in Drosophila in an ey null mutant, however, we have demonstrated that the induction of ectopic eyes by CrPax-A is not solely the effect of this jellyfish Pax gene-mediated induction of the Drosophila endogenous ey. It seems that, even in the absence of Drosophila Ey protein, the RDGN components expression is directly or indirectly induced by the CrPax-A protein.
The ability of the Cladonema Pax-A PD to bind to the 5′ upstream regions of two eye-specific opsin genes raises the possibility that CrPax-A directly regulates the expression of opsin genes during eye development and/or maintenance. In contrast, CrPax-B PD is incapable of binding to the same sequences. These data are consistent with the expression analyses and the ectopic eye induction experiments. The direct interaction of Pax proteins with opsin promoters might have contributed to the evolution of ancestral animal eyes (13). Intercalation of other transcription factors into this simple regulatory cascade may explain how the complex gene network regulating eye development evolved (39). It should be noted, however, that DNA–protein interactions demonstrated by EMSA do not always reflect the same interactions in vivo. Studies on other species of jellyfish bearing eyes should allow testing of our hypothesis.
Flexible Choice of Distinct Pax Genes for Eye Development in Different Animal Lineages.
Pax-6 has been characterized as one of the central components of the gene network that controls eye development in most of the bilaterians studied to date (6). In Tripedalia, a cubozoan jellyfish that seems to lack a bona fide Pax-6, Pax-B has been implicated in eye development (13). We have now shown that Pax-A, rather than Pax-B, seems to be involved in eye development and/or maintenance in Cladonema, a hydrozoan jellyfish. Pax-6, Pax-B, and Pax-A belong to different Pax subfamilies, which diverged from each other by gene duplication at the early stage of animal evolution, at the latest before the separation of cnidarians and bilaterians. It is thus very likely that for eye development and/or maintenance, three distinct animal lineages use three distantly related Pax genes, which were generated by gene duplication before the three animal lineages separated from each other.
It can be argued that these data provide evidence in favor of three independent origins of animal eyes during evolution (13). If it is the case, however, completely different sets of transcription factors could have been recruited for eye development in different animal lineages. The observation that the three animal lineages use genes that belong to the same gene family (i.e., Pax family) in their eyes rather supports the hypothesis of the monophyletic origin of all animal eyes (6). This hypothesis is further supported by the functional conservation of the Six family genes that are involved in eye development and/or regeneration both in Cladonema and in several bilaterians studied to date (40–44). In addition, we recently cloned the Cladonema homolog of eyes absent, one of the main components of Drosophila RDGN (45), and detected its expression in the eye. Recent publications revealed that not only the transcription factor network controlling eye development but also its potential downstream targets that indeed constitute the eye, such as genes involved in photoreception, phototransduction, and pigmentation, are also well conserved between cnidarians and bilaterians (17, 46). The common ancestry of cnidarian eyes and bilaterian ones seems to be the most reasonable interpretation of these data.
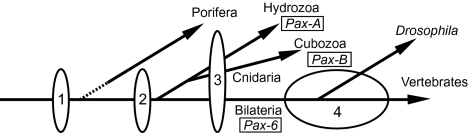
In our present model, gene duplications that gave rise to distinct subfamilies occurred most likely before the separation of poriferans and eumetazoans (1 in Fig. 5), as suggested by the statistical tests in refs. 4 and 22. We cannot, however, completely eliminate the possibility that some (or all) of these duplications postdate the poriferans–eumetazoans split (dashed line in Fig. 5) (4) until more sponge Pax genes that do not belong to the Pax-2/5/8 subfamily are found. When the ancestral animal eye evolved in the common ancestor of cnidarians and bilaterians, Pax genes may have been recruited as components of the gene network responsible for eye development (2 in Fig. 5). We assume that, at this stage, several classes (corresponding to subfamilies) of Pax genes were redundantly involved in this network. After the divergence of bilaterians and cnidarians on one hand, and hydrozoans and cubozoans on the other hand, the three distinct animal lineages selected different classes of Pax genes for the roles in eye development and/or maintenance (3 in Fig. 5). Such molecular-level opportunism is often observed in evolution (e.g., lens crystallins) (47). Interestingly, formation of some bilaterian eyes seems to be Pax-6 independent (ref. 14 for review). This suggests that the gene network directing eye development can be anomalously modified, making Pax-6 dispensable for eye development in some bilaterian lineages (4 in Fig. 5).
Fig. 5.
Evolution of Pax genes deployed for animal eye development. 1: Gene duplications that gave rise to distinct Pax classes (corresponding to subfamilies) occurred. 2: The ancestral animal eye evolved and different classes of Pax genes were redundantly recruited for eye development. 3: In each of three different animal lineages, a specific Pax gene was selected for the eye development. 4: Pax genes responsible for eye development were altered in some bilaterians. See text for detailed description.
Our model predicts that genes from different Pax subfamilies may still retain to some extent the ability to perform each other's function redundantly. The ability of Cladonema Pax-A (poxn subfamily), Pax-B (Pax-2/5/8 subfamily), and Drosophila ey and toy (Pax-4/6 subfamily) to rescue the D-Pax2 (Pax-2/5/8 subfamily) mutant spapol in the Drosophila eye is in agreement with this prediction (Fig. 3 G and H and ref. 13). Similarly, misexpressed D-Pax2 induces eyes in the imaginal discs, as ey and toy do (13). Also at the molecular level, chordate Pax-6 and Pax-2 PDs recognize almost identical sequences, even though they show clear differences in their affinities to certain DNA sequences (35).
In summary, our study uncovers the diversity of the Pax genes used for development and/or maintenance of animal eyes. We propose that in the ancestral animal eye, which is likely to have evolved in the common ancestor of cnidarians and bilaterians, different classes (corresponding to the present subfamilies) of Pax genes were redundantly recruited and may then have been flexibly selected in distinct animal lineages for their roles in eye development.
Materials and Methods
Detailed descriptions of animal culture, fly strains, and all of the technical information regarding the gene cloning and sequencing, real-time PCR, molecular phylogenetic tree analysis, in silico search for Pax genes, in situ hybridization, protein expression, and EMSA are found in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Noll (University of Zurich, Zurich) for fly strains; P. Callaerts (Katholieke Universiteit Leuven, Louvain, Belgium) and J. Blanco (Insitute of Medical Biology, Singapore, Singapore) for the PD control probe; and M. M. Burger (Friedrich Miescher Institute, Basel, Switzerland) for Microciona cDNA; J. Blanco and D. Papadopoulos for critically reading the manuscript; and D. Duboule for allowing P.T. to continue the project. H.S. was supported by Yamada Science Foundation for a long-term visit. This work was supported by the Swiss National Foundation and the Kantons of Basel-Stadt and Basel-Landschaft.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequence data from this report have been deposited in the GenBank/European Molecular Biology Laboratory/DNA Data Base in Japan database under accession nos. AB379656–AB379659, AB332437, and AB439133.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008389107/-/DCSupplemental.
References
- 1.Mansouri A, Goudreau G, Gruss P. Pax genes and their role in organogenesis. Cancer Res. 1999;59(7, Suppl):1707s–1709s. discussion 1709s–1710s. [PubMed] [Google Scholar]
- 2.Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 3.Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: Roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Hoshiyama D, Iwabe N, Miyata T. Evolution of the gene families forming the Pax/Six regulatory network: Isolation of genes from primitive animals and molecular phylogenetic analyses. FEBS Lett. 2007;581:1639–1643. doi: 10.1016/j.febslet.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol Dev. 2007;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 6.Gehring WJ. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol. 2004;48:707–717. doi: 10.1387/ijdb.041900wg. [DOI] [PubMed] [Google Scholar]
- 7.Silver SJ, Rebay I. Signaling circuitries in development: Insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 8.Hill RE, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 9.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 10.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 11.Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci USA. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin VJ. Photoreceptors of cnidarians. Can J Zool. 2002;90:1703–1722. [Google Scholar]
- 13.Kozmik Z, et al. Role of Pax genes in eye evolution: A cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- 14.Kozmik Z. The role of Pax genes in eye evolution. Brain Res Bull. 2008;75:335–339. doi: 10.1016/j.brainresbull.2007.10.046. [DOI] [PubMed] [Google Scholar]
- 15.Sun H, Dickinson DP, Costello J, Li WH. Isolation of Cladonema Pax-B genes and studies of the DNA-binding properties of cnidarian Pax paired domains. Mol Biol Evol. 2001;18:1905–1918. doi: 10.1093/oxfordjournals.molbev.a003731. [DOI] [PubMed] [Google Scholar]
- 16.Weber C. Structure, histochemistry, ontogenetic development, and regeneration of the ocellus of Cladonema radiatum Dujardin (Cnidaria, Hydrozoa, Anthomedusae) J Morphol. 1981;167:313–331. doi: 10.1002/jmor.1051670306. [DOI] [PubMed] [Google Scholar]
- 17.Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. Curr Biol. 2008;18:51–55. doi: 10.1016/j.cub.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 18.Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biol Bull. 2001;201:323–338. doi: 10.2307/1543611. [DOI] [PubMed] [Google Scholar]
- 19.Maldonado M, Durfort M, McCarthy DA, Young CM. The cellular basis of photobehavior in the tufted parenchymella larva of demosponges. Mar Biol. 2003;143:427–441. [Google Scholar]
- 20.Björn LO, Rasmusson AG. Photosensitivity in sponge due to cytochrome c oxidase? Photochem Photobiol Sci. 2009;8:755–757. doi: 10.1039/b904988f. [DOI] [PubMed] [Google Scholar]
- 21.Leys SP, Cronin TW, Degnan BM, Marshall JN. Spectral sensitivity in a sponge larva. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002;188:199–202. doi: 10.1007/s00359-002-0293-y. [DOI] [PubMed] [Google Scholar]
- 22.Hoshiyama D, et al. Sponge Pax cDNA related to Pax-2/5/8 and ancient gene duplications in the Pax family. J Mol Evol. 1998;47:640–648. doi: 10.1007/pl00006421. [DOI] [PubMed] [Google Scholar]
- 23.Larroux C, et al. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- 24.Hill A, et al. Origin of Pax and Six gene families in sponges: Single PaxB and Six1/2 orthologs in Chalinula loosanoffi. Dev Biol. 2010;343:106–123. doi: 10.1016/j.ydbio.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 25.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller DJ, et al. Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): Implications for the evolution of the Pax gene family. Proc Natl Acad Sci USA. 2000;97:4475–4480. doi: 10.1073/pnas.97.9.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denker E, Manuël M, Leclère L, Le Guyader H, Rabet N. Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria) Dev Biol. 2008;315:99–113. doi: 10.1016/j.ydbio.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Czerny T, et al. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- 30.Punzo C, et al. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004;131:3943–3953. doi: 10.1242/dev.01278. [DOI] [PubMed] [Google Scholar]
- 31.Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao R, et al. Headless flies generated by developmental pathway interference. Development. 2001;128:3307–3319. doi: 10.1242/dev.128.17.3307. [DOI] [PubMed] [Google Scholar]
- 33.Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- 34.Papatsenko D, Nazina A, Desplan C. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech Dev. 2001;101:143–153. doi: 10.1016/s0925-4773(00)00581-5. [DOI] [PubMed] [Google Scholar]
- 35.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouillon J. In: Compendium of Zoology, Cnidaria, Ctenophora, Part 2. Grassé PP, editor. III. Paris: Masson; 1994. pp. 165–209. (in French) [Google Scholar]
- 37.Campbell RD. In: The Biology of Nematocysts. Hessinger DA, Lenhoff HM, editors. San Diego: Academic Press; 1988. pp. 123–142. [Google Scholar]
- 38.Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 39.Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 40.Cheyette BN, et al. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 41.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes—structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Pineda D, et al. Searching for the prototypic eye genetic network: Sine oculis is essential for eye regeneration in planarians. Proc Natl Acad Sci USA. 2000;97:4525–4529. doi: 10.1073/pnas.97.9.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo HC, Drivenes Ø, Ellingsen S, Fjose A. Expression of two zebrafish homologues of the murine Six3 gene demarcates the initial eye primordia. Mech Dev. 1998;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- 44.Stierwald M, Yanze N, Bamert RP, Kammermeier L, Schmid V. The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: Development and eye regeneration. Dev Biol. 2004;274:70–81. doi: 10.1016/j.ydbio.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 45.Pignoni F, et al. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 46.Kozmik Z, et al. Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc Natl Acad Sci USA. 2008;105:8989–8993. doi: 10.1073/pnas.0800388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wistow G. Lens crystallins: Gene recruitment and evolutionary dynamism. Trends Biochem Sci. 1993;18:301–306. doi: 10.1016/0968-0004(93)90041-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.