Abstract
In plant innate immunity, the leucine-rich repeat receptor kinase FLS2 recognizes the bacterial pathogen-associated molecular pattern (PAMP) flagellin. The molecular mechanisms underlying PAMP perception are not fully understood. Here, we reveal that the gaseous phytohormone ethylene is an integral part of PAMP-triggered immunity. Plants mutated in the key ethylene-signaling protein EIN2 are impaired in all FLS2-mediated responses, correlating with reduced FLS2 transcription and protein accumulation. The EIN3 and EIN3-like transcription factors, which depend on EIN2 activity for their accumulation, directly control FLS2 expression. Our results reveal a direct role for ethylene in regulation of an innate immune receptor.
Keywords: innate immunity, receptor kinase
The earliest events in plant immunity concern recognition of conserved elicitors called pathogen-associated molecular patterns (PAMPs) by plasma membrane receptors. The leucine-rich repeat receptor kinase (LRR-RK) FLS2 is the receptor for the bacterial PAMP flagellin or its active epitope, represented by the peptide flg22 (1, 2). Binding of flg22 to FLS2 triggers heterodimerization with and phosphorylation of the LRR-RK BAK1 (3, 4). In addition, both FLS2 and BAK1 are able to interact with and phosphorylate the cytoplasmic kinase BIK1, which seems to act as positive regulator of the FLS2 signaling pathway (5, 6). FLS2 activation leads to activation of the MAP kinase pathways MEKK-MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4, leading to transcription of defense-related genes through the WRKY transcription factors WRKY22/29 and WRKY25/33 (3). Recently, another pathway involving the calcium-dependent protein kinases (CDPK) 4/5/6/11 has been proposed to act in parallel to the MAPK pathways to control flg22-dependent gene expression (7). In addition, flg22 treatment triggers ion fluxes, RbohD-dependent oxidative burst, and callose deposition as well as ethylene biosynthesis (3). The development of Arabidopsis seedlings grown in sterile conditions is also inhibited by flg22 treatment (8). FLS2 is conserved in most plant species tested, and orthologs have been identified in Arabidopsis, Nicotiana benthamiana, tomato, and rice (1, 2). Flagellin perception is required for full immunity against bacteria, because plants deficient in FLS2 are more susceptible to adapted and nonadapted bacterial pathogens (1). Consistently, successful bacterial pathogens need to avoid or suppress PAMP-triggered immunity induced by flagellin (1). Interestingly, some virulence effectors from phytopathogenic bacteria do so by directly targeting FLS2 and/or BAK1 (9).
Many plant hormones, including auxin, salicylic acid (SA), jasmonates, and ethylene, impact immunity in complex and sometimes antagonistic relationships (10). Flagellin induces production of SA (11, 12) but also leads to repression of auxin perception (13). A classical response associated with PAMP treatment is an increase in the biosynthesis of the gaseous hormone ethylene (ET) (14–16). Here, we show a simple and direct link between the hormone ET and the earliest events in PAMP-triggered immunity (PTI) against pathogenic bacteria.
Results
To gain a better understanding of PTI signaling, we developed a forward-genetic screen to isolate Arabidopsis mutants impaired in the oxidative burst induced by flg22 from a collection of T-DNA mutagenized Arabidopsis plants (17) (Fig. S1A). Of ~35,000 Arabidopsis seedlings screened, 21 flagellin-insensitive (fin) mutants had reproducibly significant reduction in reactive oxygen species (ROS) intensity in response to flg22 (<60% compared with wild-type Col-0) (Fig. S1B). T-DNA–mediated mutagenesis often leads to missense mutations caused by failed T-DNA insertions (18). Indeed, sequencing of candidate genes revealed that fin1 carries a 1-bp deletion in FLS2 (At5g46330) itself (Fig. S1 C and D), whereas fin2 harbors a 2-bp deletion in BAK1/SERK3 (At4g33430) (Fig. S1 E and F). The identification of FLS2 and BAK1 mutants validated our genetic screen to identify PTI components.
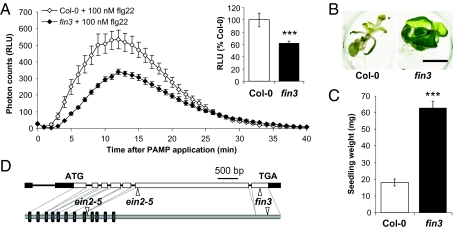
We focused on fin3, a mutant with a consistent reduction in the oxidative burst triggered by flg22 (Fig. 1A and Fig. S1B). To identify the causative mutation in fin3, we used a map-based cloning strategy based on reduced seedling-growth inhibition (SGI) in the presence of flg22, a phenotype also associated with the fin3 mutation (Fig. 1 B and C). The mutation was linked to a 38.7-kb region on chromosome V that contained 12 predicted gene loci. Sequencing of candidate genes identified a 33-bp deletion in the EIN2 (At5g03280) gene, inducing an 11-amino acid deletion in its C terminus (Fig. 1D and Fig. S1G). EIN2 encodes an integral membrane protein essential for signaling in response to the gaseous plant hormone ethylene (19). Allelism tests between fin3 and the previously characterized mutants ein2-1 and ein2-5 (19) confirmed that the three mutations are allelic (Table S1). Additionally, fin3 etiolated seedlings were as impaired as ein2-5 in the characteristic morphological triple response induced by the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) (19) (Fig. S2). Consistently, ein2-1 and ein2-5 were impaired in flg22-triggered SGI (Fig. S3). Hence, the phenotype of fin3 is caused by a mutation in EIN2.
Fig. 1.
A genetic screen for flagellin-insensitive (fin) mutants identified an ein2 allele. (A) Oxidative burst triggered by 100 nM flg22 in wild-type Col-0 and fin3 mutant plants measured in relative light units (RLU). Total ROS production (Inset) expressed as percentage of flg22-treated Col-0. Values are mean ± SE (n = 20). (B) Seedling growth inhibition triggered by flg22. Five-day-old wild-type Col-0 and fin3 seedlings were treated with 1 μM flg22 for 12 d. (Scale bar, 5 mm.) (C) Quantification of growth inhibition in wild-type Col-0 and fin3 seedlings treated with 1 μM flg22 for 12 d. Values are mean ± SE (n = 12). (D) Schematic representations of the EIN2 gene (At5g03280) and deduced protein with positions of the ein2-5 and fin3 mutations. Filled boxes and open boxes represent the 5′- and 3′-untranslated regions and exons of the gene, respectively. The ein2-5 deletion (CATGACT) is at position +1465 after start codon, and the fin3 deletion (AGACGTTGAAATGGCAATCTCTTGTAGAAAAGG) is at position +4280. The predicted transmembrane helices are shown as rectangles. Statistical significance by comparison with Col-0 was assessed using t test. ***P < 0.001. Similar results were observed in at least three independent experiments.
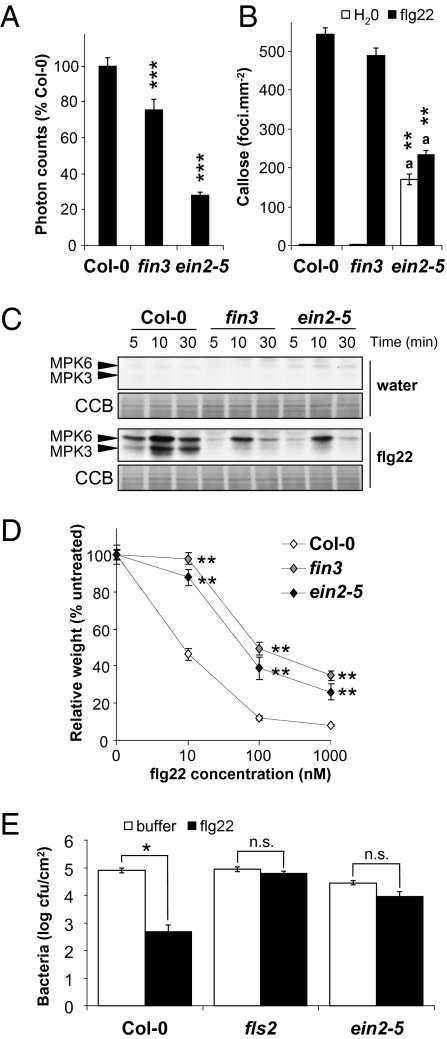
We tested if ethylene insensitivity caused by ein2 mutations affects downstream responses triggered by flg22. In addition to the oxidative burst and SGI (Figs. 1 A–C and 2 A and D and Fig. S3), fin3 and ein2-5 were also impaired in callose deposition (Fig. 2B) and MAP kinase activation induced by flg22 (Fig. 2C). Notably, the number of callose deposits in ein2-5 after water treatment was higher than in water-infiltrated wild-type leaves (Fig. 2B). Because untreated ein2-5 leaves did not exhibit callose deposition (Fig. S4), this indicates that ein2–5 is more sensitive to the wounding stress caused by water infiltration. However, treatment of ein2-5 leaves with flg22 did not further increase callose deposition to levels observed in treated wild-type leaves (Fig. 2B). Notably, the resistance to the virulent bacterium Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) triggered by flg22 treatment was also reduced in ein25 leaves compared with wild-type leaves (Fig. 2E), showing that the reduced flg22 responsiveness in this mutant impacts activation of innate immunity. In most assays, the effect of the ein2-5 mutation was stronger than fin3 (Fig. 2 A–D), indicating that fin3 is a weaker allele than the ein2-5 null mutant. The ethylene-insensitive etr1-1 mutant, which is impaired in ethylene perception (20), also showed reduced sensitivity to flg22 in SGI assays (Fig. S3). Thus, both ethylene perception and signaling seem to be important for responses triggered by flg22.
Fig. 2.
Ethylene signaling is required for flg22-induced responses. (A) Total ROS production triggered by 100 nM flg22 in Col-0, fin3, and ein2-5 mutants. Values are expressed as percentage of ROS obtained with Col-0 for 40 min and are mean ± SE (n = 20). (B) Callose deposition triggered in leaves 24 h after infiltration with water or 1 μM flg22 in Col-0, fin3, and ein2-5 plants. Results showing quantification of callose deposits (foci/mm−2) are mean ± SE (n = 4). For ein2-5, the same letter indicates statistically nonsignificant (one-way ANOVA; P > 0.05) differences between samples. (C) Activation profile of the MAP kinases MPK3 and MPK6 in response to treatment with water or 100 nM flg22 in Col-0, fin3, and ein2-5 mutants. Arrowheads indicate phosphorylated MPK6 and MPK3. Blots stained with colloidal Coomassie Blue (CCB) are presented to show equal loading. (D) Growth inhibition of Col-0, fin3, and ein2-5 seedlings treated with different concentrations of flg22 for 12 d. Values are mean ± SE (n = 12). (E) Growth of Pto DC3000 on Col-0, fls2, and ein2-5 plants pretreated with water or 1 μM flg22 for 24 h and then syringe-infiltrated with 105 cfu/mL−1 of bacteria. Bacterial growth was determined 2 d postinoculation. Values are mean ± SE (n = 8). Statistical significance by comparison with Col-0 was assessed using one-way ANOVA followed by Dunnett's test. *P < 0.05; **P < 0.01; ***P < 0.001. Similar results were observed in at least three independent experiments.
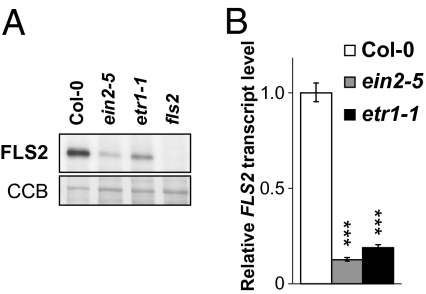
Our results showed that ethylene perception and signaling were required for all flg22-induced responses tested (Fig. 2 A–E and Fig. S3). Because some of these responses occur within a few minutes of elicitation, we hypothesized that endogenous ethylene may control FLS2 signaling at a very early step of the pathway, possibly by controlling FLS2 itself. To test this hypothesis, we measured the steady-state levels of FLS2 protein by immunoblots. Strikingly, FLS2 levels were dramatically reduced in ein2-5 and ein2-1 (Fig. 3A and Fig. S5A). The reduction of FLS2 protein levels correlated with a similar reduction of steady-state FLS2 transcript accumulation in ein2-5 and ein2-1 (Fig. 3B and Fig. S5B). A comparable reduction in FLS2 transcript and protein accumulation was also observed in etr1-1 (Fig. 3 A and B). Consistent with our previous results (Fig. 2 A–D) and the reduced FLS2 expression (Fig. 3 A and B), the transcript levels of FLS2 and SIRK/FRK1 after flg22 treatment were also reduced in ein2-5 (Fig. S6 A and B). Thus, the impaired flg22 sensitivity of ein2-5 and etr1-1 is likely caused by reduced amount of FLS2 proteins. Our results show that both ethylene perception and signaling are crucial for FLS2 expression.
Fig. 3.
Endogenous ethylene signaling regulates FLS2 expression. (A) Western blot showing accumulation of FLS2 proteins in Col-0, ein2-5, and etr1-1 13-d-old seedlings. The fls2 mutant was used as a negative control for FLS2 detection. Thirty micrograms of total protein were loaded in each lane. Blots stained with CCB are presented to show equal loading. Similar results were observed in at least three independent experiments. (B) Quantitative RT-PCR analysis of FLS2 expression in Col-0, etr1-1, and ein2-5 untreated 13-d-old seedlings. Transcript levels are normalized to the U-box housekeeping gene At5g15400 and are presented as relative to untreated Col-0. Similar results were observed in at least three independent experiments. Statistical significance by comparison with untreated Col-0 was assessed using one-way ANOVA followed by Dunnett's test. ***P < 0.001.
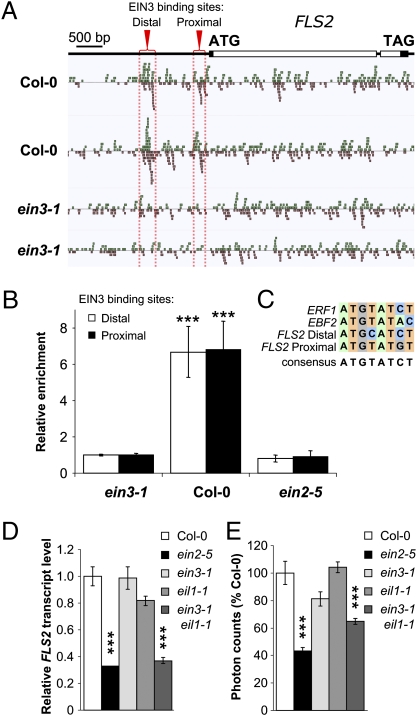
We investigated how ethylene could influence FLS2 expression. EIN2 is the only gene whose recessive loss-of-function mutation leads to complete ethylene insensitivity in the triple-response assay (19). EIN2 controls the accumulation and subsequent activity of the functionally redundant EIN3 and EIN3-like (EIL) transcription factors in response to ethylene (21, 22). We, therefore, tested if EIN3 could regulate FLS2 transcription directly. In silico motif analysis of the FLS2 promoter revealed the presence of nine potential EIN3/EILs binding sites (23, 24) (Fig. S7), suggesting that EIN3 may bind to the promoter of the FLS2 gene to influence its transcription. Regions of the Arabidopsis genome associated with EIN3 were identified by ChIP using a purified EIN3 antibody (21) and followed by Illumina sequencing (ChIP-Seq). Notably, EIN3 binds to two positions in the FLS2 promoter in wild-type etiolated seedlings treated with ethylene (Fig. 4A). EIN3 binding to these regions was absent in chromatin purified from the loss-of-function allele ein3-1, proving the specificity of the enrichment (Fig. 4A). Relative enrichments of EIN3 binding sites in the FLS2 promoter after immunoprecipitation with the EIN3 antibody were further confirmed by quantitative PCR using chromatin from light-grown seedlings (Fig. 4B). Comparison of the EIN3 binding sites experimentally determined for the ERF1 (23) and EBF2 (25) promoters reveals a close homology with the two potential sites identified in the FLS2 promoter (Fig. 4C). Whereas EIN3 is able to bind to the FLS2 promoter, FLS2 transcript levels were not significantly reduced in ein3-1 (Fig. 4D). The effect of single mutations in EIN3 and EIL transcription factors is often hindered by their homology and consequent functional redundancy (17, 26, 27). Indeed, simultaneous mutations in EIN3 and EIL1 accentuated the effect on FLS2 expression, with the double mutant ein3-1 eil1-1 accumulating as little FLS2 transcript as the ein2-5 mutant (Fig. 4D). Consequently, ein3-1 eil1-1 plants were impaired in flg22-triggered oxidative bursts to a level comparable with ein2-5 plants (Fig. 4E).
Fig. 4.
EIN3/EIL1 directly control FLS2 expression at the transcriptional level. (A) Alignment of ChIP-Seq reads across the FLS2 (At5g46330) gene and promoter region as shown with Anno-J viewer. Col-0 or ein3-1 etiolated seedlings were grown in hydrocarbon-free air for 3 d, treated with 10 ppm ethylene gas for 4 h, and then used in ChIP-Seq analysis using an anti-EIN3 polyclonal antibody. The proximal and distal regions with significant enrichment are highlighted by red brackets and dashed lines. Results from two independent experiments are presented. Reads are normalized for each sample. Gene annotation is depicted at the top; filled boxes and open boxes represent the 5′- and 3′-untranslated regions and exons of the gene, respectively. (B) Enrichment of the indicated EIN3-associated DNA fragments after ChIP-PCR. Chromatin from wild-type Col-0 and ein3-1 light-grown seedlings was immunoprecipitated with an anti-EIN3 polyclonal antibody. Enrichment of associated DNA fragments was verified by qPCR using specific primers and is presented as relative to Col-0. (C) Sequence alignment of EIN3 binding sites in ERF1, EBF2, and FLS2 promoters. EIN3 binding sites in the promoters of ERF1 (23) and EBF2 (25) are presented. Identified by ChIP-Seq, the two potential binding sites of EIN3 to the FLS2 promoter are also shown. (D) Quantitative RT-PCR analysis of FLS2 expression in Col-0, ein2-5, ein3-1, eil1-1, and ein3-1 eil1-1 seedlings. Transcript levels are normalized to the U-box housekeeping gene and are presented as relative to Col-0. Data are representative of one of two experiments. (E) Total ROS production triggered by 100 nM flg22 in Col-0, ein2-5, ein3-1, eil1-1, and ein3-1 eil1-1 mutants. Values are expressed as percentage of ROS obtained with Col-0 for 40 min and are mean ± SE (n = 20). Statistical significance by comparison with (B) ein3-1 and (C) Col-0 was assessed using one-way ANOVA followed by Dunnett's test. *P < 0.05; **P < 0.01; ***P < 0.001. Similar results were observed in at least three independent experiments.
To test if endogenous ethylene could control early responses independently of FLS2 transcription, we measured the oxidative burst induced by the general phosphatase inhibitor calyculin A that stimulates early PAMP responses downstream and independently of pattern-recognition receptors (PRRs) (4, 28). In contrast to the flg22-induced oxidative burst, the burst induced by calyculin A was not inhibited in fls2 or ein2-5 seedlings (Fig. S8). The oxidative burst induced by both elicitors was, however, completely abolished in rbohD seedlings lacking the NADPH oxidase required for PAMP-induced oxidative burst in Arabidopsis. Thus, these experiments suggest that the reduced ROS burst triggered by flg22 in ethylene perception and signaling mutants is caused by reduced FLS2 expression and is not an indirect effect on downstream responses.
Together, these results reveal that endogenous ethylene controls FLS2 expression transcriptionally through direct binding of the transcription factor EIN3 and potentially, EIL1 to the FLS2 promoter.
Discussion
An inherent difficulty in interpreting the specific contributions of ethylene to plant immunity is its role in multiple different physiological processes, often through cross-talk with other hormones. For example, ethylene controls senescence, symptom development, and abiotic resistance together with abscisic acid and gibberellic acid and resistance to insects and necrotrophic pathogens in association with jasmonic acid (29, 30). Previous reports show the role of ethylene in disease symptom development and susceptibility to bacteria conflict (29, 30). Recently, EIN3 and EIL1 were shown to repress biosynthesis of the immune hormone salicylic acid (SA) (31). Consequently, ein2-1 and ein3-1 eil1-1 plants exhibited enhanced resistance to the pathogenic bacterium P. syringae pv. tomato (Pto) DC3000, suggesting that they overaccumulate (SA) (31). However, a previous study showed that ethylene-insensitive mutant plants were more tolerant to bacteria (reduced symptoms), but no decrease in bacterial numbers could be observed (32, 33); we observed that Pto DC3000 infiltrated at low dose (105 cfu/mL−1) multiplied to lower levels in ein2-5 and ein2-1 leaves than in wild-type leaves (Fig. 2E) (34), suggesting that EIN2 may indeed play an additional role as a negative regulator of SA-mediated biosynthesis or signaling. In addition, ein3-1 eil1-1 plants exhibited deregulated callose deposition and defense gene expression in response to flg22 (31). In our conditions, we could confirm overaccumulation of SA (Fig. S9A) and increased expression of the SA-induced PR1 gene in ein3-1 eil1-1 soil-grown plants (Fig. S9B). However, we did not detect changes in SA levels in the single mutants ein3-1 or eil1-1 (Fig. S9A). Furthermore, the triple mutant ein3-1 eil1-1 sid2-2 that exhibits wild-type SA levels (31) accumulated as little FLS2 mRNA as the double mutant ein3-1 eil1-1 (Fig. S9C). Thus, the effect of the ein3-1 eil1-1 mutation on FLS2 gene is not related to alterations in SA levels. In agreement with our findings that ethylene controls FLS2 expression and thus, flg22 responsiveness, the resistance to Pto DC3000 induced by pretreatment with flg22 was reduced in ein2-5 and ein2-1 leaves (Fig. 2E) (34, 35).
Ethylene perception and signaling were recently proposed to be required for callose deposition in response to flg22 (36). However, the callose deposition observed in ein2-5 after water infiltration (Fig. 2B) excludes a requirement for EIN2 in callose biosynthesis. Furthermore, the reduced FLS2 levels in ein2-5 (Fig. 3A) explain the decreased callose deposition observed in this mutant after flg22 treatment (Fig. 2B). Our findings also explain the requirement for ethylene signaling in disruption of the association between MPK6 and the transcription factor ERF104 in response to flg22 and definitively place endogenous ethylene signaling upstream of MAP kinase activation by PAMPs (37, 38).
The data presented here reveal a surprisingly simple and direct mechanism involving ethylene that ensures optimal levels of an immune receptor. Previous reports have highlighted the complexity of ethylene signaling and its roles in development of disease symptoms, induction of antimicrobial genes as a secondary response, and cross-talk with other hormones (29). Here, we showed that mutants impaired in ethylene perception and signaling are all impaired in flg22-induced responses and that this always correlates with a corresponding reduction in FLS2 transcript levels. In addition, we revealed that the key transcription factor EIN3, which depends on ethylene perception and signaling for its accumulation and activity, binds directly to the FLS2 promoter. These results showed that endogenous ethylene controls the expression of the PRR FLS2 at the transcriptional level.
Our findings that the oxidative brust triggered by the elicitor-mimic calyculin A is not reduced in ein2-5 mutants indicate that the reduced flg22 responses in ethylene-insensitive mutants is caused by reduced FLS2 expression and not an indirect effect on downstream responses. However, future experiments, in which FLS2 gene is expressed constitutively in ethylene-insensitive mutants, are required to ascertain the causal relationship between decreased FLS2 expression and impaired FLS2-mediated responses.
Flg22 induces the MAP kinases MPK3 and MPK6 (39, 40) as well as ethylene production (16). These same MAP kinases phosphorylate the ethylene biosynthetic enzymes ACC synthases (ACS) 2 and 6 as well as EIN3, leading to its stabilization (41, 42). We, therefore, hypothesize a simple feedback model in which ethylene produced in response to flg22 treatment could maintain FLS2 levels in a positive feedback loop. This mechanism would be especially effective to maintain the plasma membrane FLS2 pool after internalization after flagellin binding and receptor activation (43). In the absence of flagellin, endogenous ethylene ensures a constitutive level of FLS2 expression. On flg22 binding, FLS2 activates MPK6 that, in turn, phosphorylates ACS2/6 and further leads to EIN3 stabilization, resulting in increased ethylene production and signaling. Congruently, flg22 perception leads to EIN3 accumulation (31). This model may explain the long-established observation that PAMP perception triggers increased ethylene biosynthesis (14–16). Our results show an unexpected layer of regulation by ethylene to maintain optimal levels of innate immune receptors.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type control. Arabidopsis plants used in this study were grown as one plant per pot at 20–21 °C with a 10-h photoperiod in environmentally controlled chambers or on plates containing Murashige and Skoog (MS) medium (including vitamins) (Duchefa) and 1% sucrose supplemented with 8% agar for the first 5 d at 22 °C and with a 16-h photoperiod. The first set of 10,000 T-DNA SALK lines (17) was obtained from the Nottingham Arabidopsis Stock Center. The mutant lines used in this study are: fls2 (SALK_093905), cerk1-2 (GK-096F09), rbohD, etr1-1, ein2-1 ein2-5, ein3-1, eil1-1, sid2-2, ein3-1 eil1-1, and ein3-1 eil1-1 sid2-2.
Elicitor.
Flg22 peptide was purchased from Peptron and solubilized in sterile water.
Identification of fin Mutants.
To identify flg22 insensitive (fin) mutants, we screened ~35,000 Arabidopsis seedlings from the first set of 10,000 T-DNA SALK lines produced in the Col-0 background (17). Surface-sterilized seeds were individually dispensed in 96-well plates containing 1× Gamborg B5 medium (including vitamins) (Duchefa) supplemented with 1% sucrose, pH 5.7. After 2 d of stratification at 4 °C, plates were transferred to light for 9 d. Liquid media were then removed and replaced by assay solution containing 10 μg/mL peroxidase from horseradish Type VI-A (Sigma), 100 μM luminol (Sigma), and 100 nM flg22. ROS production was measured immediately and during the 15-min period using a Varioskan microplate luminometer (Thermo Scientific). Seedlings with a ROS production comparable with flg22-insensitive fls2 mutant were rescued by rinsing in water and transferred to the greenhouse. Three and four weeks later, production of oxidative burst was tested again with two leaf discs and 100 nM flg22 as described in ref. 44. SALK lines were considered as fin candidates when they produce repeatedly, in the leaf disk assay, less than 60% of the ROS production obtained with wild-type Col-0.
Map-Based Cloning.
The fin3 mutation (in Col-0 background) was identified by map-based cloning as described in SI Materials and Methods.
Measurement of Reactive Oxygen-Species Generation.
Oxidative-burst measurement was performed as previously described (44). ROS was elicited with 100 nM flg22, and elicitation in the absence of any PAMP (water treatment) was included in all experiments as negative control. Twenty leaf discs from 10 5-wk-old plants were used for each condition. ROS was also triggered by 500 nM calyculin A (NEB) using whole 7-d-old sterile seedlings grown in 1× Gamborg B5 liquid medium (including vitamins) supplemented with 1% sucrose (pH 5.7). Luminescence was measured over time using an ICCD photon-counting camera (Photek).
Seedling-Growth Inhibition and Callose Assays.
Quantitative seedling-growth inhibition was assessed using 12 seedlings per condition as described in ref. 45. Callose deposition analysis was performed as described in ref. 46.
MAP Kinase Activation.
MAPK assays were performed on six 2-wk-old seedlings grown in liquid medium. Seedlings were then elicited with 100 nM flg22 or water for 5, 10, or 30 min and frozen in liquid nitrogen. MAPK activation was monitored by Western blot with antibodies that recognize the dual phosphorylation of the activation loop of MAPK (pTEpY). Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit monoclonal antibodies from Cell Signaling were used according to the manufacturer's protocol. Blots were stained with colloidal Coomassie blue (CCB) to verify equal loading.
Induced Resistance to Bacteria.
Induced resistance assays were realized as described in ref. 34. Briefly, water or 1 μM flg22 was infiltrated with a needleless syringe into leaves of 5-wk-old Arabidopsis plants. After 24 h, the same leaves were syringe-infiltrated with 105 cfu/mL of P. syringae pv. tomato DC3000. Bacterial growth was determined 2 d postinoculation.
RNA Isolation and Quantitative RT-PCR.
Total RNA was extracted from six 2-wk-old seedlings grown in liquid medium using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were treated with Turbo DNA-free DNase (Ambion) and quantified with a Nanodrop spectrophotometer (Thermo Scientific). cDNA synthesis and qRT-PCR were realized as described in SI Materials and Methods.
Protein Extraction and Immunoblot Assays.
To prepare samples for examining the accumulation of PRR proteins, five leaf discs (0.38 cm2 each) from soil-grown plants or six 13-d-old seedlings grown in sterile conditions were harvested and ground in liquid nitrogen. Total protein crude extracts were prepared with 100 μL of extraction buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 0.5 mM PMSF, 10% glycerol, 1% vol/vol protease inhibitor mixture for plant cell (Sigma)]. The lysates were cleared by centrifugation at 16,000 × g for 5 min. The supernatants were collected, and protein concentrations were determined by Bradford quantification (BioRad) according to the manufacturer's indications; 30 μg of total proteins were separated on SDS/PAGE gels. FLS2 was detected with 1:1,000 rabbit anti-FLS2 polyclonal antibodies (44). A peroxidase-conjugated goat anti-rabbit IgG (Sigma) was used as secondary antibody.
ChIP-Seq and ChIP-PCR.
ChIP was performed as described in ref. 47 with modifications using Col-0 or ein3-1 etiolated seedlings grown for 3 d in hydrocarbon-free air treated for 4 h with 10 ppm ethylene gas followed by sequencing as per Illumina specifications. Sequencing data were visualized using the Anno-J viewer (http://www.annoj.org). Enrichment values have been determined using PeakSEq (48). ChIP was performed again using 13-d-old light-grown Col-0, ein2-5, and ein3-1 seedlings followed by quantitative PCR. Actin was used as internal control for normalization. Primers used for quantitative PCR are as follows: FLS2 (At5g46330)-1251 binding site, forward 5′-CAAGTCTTCAAGTAAACATGATATGG-3′ and reverse 5′-GATTTGGACAACCTCATCTTGACCC-3′; FLS2 (At5g46330)-267 binding site, forward 5′-CAATCGCTCAAAACTAAATCGG-3′ and reverse 5′-CGGATGTGAAAAGGCCAAGACGC-3′; ACTIN (At5g09810) forward 5′-CGTTTCGCTTTCCTTAGTGTTAGCT-3′ and reverse 5′-AGCGAACGGATCTAGAGACTCACCTTG-3′ (49). In silico motif analysis of the FLS2 promoter for the presence of primary ethylene response elements (AYGWAYCT) was realized using the fuzznuc software (http://embossgui.sourceforge.net/demo/fuzznuc.html).
SA Measurement.
SA extraction followed by liquid chromatography-MS quantification was realized using 5-wk-old plants as described in ref. 50.
Statistical Analysis.
Statistical significances based on t test and one-way ANOVA analyses were performed with Prism 5.01 software (GraphPad Software).
Supplementary Material
Acknowledgments
The technical assistance of Selena Gimenez-Ibanez, Carol Moreau, Helen Chapman, and Maryam Arasteh is much appreciated. We thank Lionel Hill for performing the SA measurements. We thank Jonathan Jones (The Sainsbury Laboratory, Norwich, UK) for rbohD, ein2-1, ein2-5, and ein3-1 seeds, Silke Robatzek (The Sainsbury Laboratory, Norwich, UK) for etr1-1 seeds, and Jian-Min Zhou (National Institute of Biological Sciences, Beijing, China) for ein3-1, eil1-1, sid2-2, ein3-1 eil1-1, and ein3-1 eil1-1 sid2-2 seeds. J.P.R. is an Australian Research Council Future Fellow (FT0992129). This research was funded by National Science Foundation Grant MCB-0924871 (to J.R.E.), Department of Energy Grant DE-FG02-04ER15517 (to J.R.E.), Biotechnology and Biological Sciences Research Council Grants BB/G024936/1 “ERA-PG PRR-CROP” (to C.Z.) and BB/E017134/1 (to J.P.R.), and the Gatsby Charitable Foundation (to C.Z. and J.P.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003347107/-/DCSupplemental.
References
- 1.Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 3.Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze B, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Boudsocq M, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 9.Göhre V, Robatzek S. Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- 10.Bari R, Jones JD. Role of plant hormones in plant defense responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 11.Mishina TE, Zeier J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007;50:500–513. doi: 10.1111/j.1365-313X.2007.03067.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- 13.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 14.Paradies I, Konze JR, Elstner EF, Paxton J. Ethylene: Indicator but not inducer of phytoalexin synthesis in soybean. Plant Physiol. 1980;66:1106–1109. doi: 10.1104/pp.66.6.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauch F, Hadwiger LA, Boller T. Ethylene: Symptom, not signal for the induction of chitinase and β-1,3-glucanase in pea pods by pathogens and elicitors. Plant Physiol. 1984;76:607–611. doi: 10.1104/pp.76.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 17.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 18.Tinland B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1996;1:178–184. [Google Scholar]
- 19.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 20.Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- 21.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 22.Potuschak T, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 23.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosugi S, Ohashi Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28:960–967. doi: 10.1093/nar/28.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- 26.Binder BM, et al. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell. 2007;19:509–523. doi: 10.1105/tpc.106.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao Q, et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 28.Felix G, Regenass M, Spanu P, Boller T. The protein phosphatase inhibitor calyculin A mimics elicitor action in plant cells and induces rapid hyperphosphorylation of specific proteins as revealed by pulse labeling with [33P]phosphate. Proc Natl Acad Sci USA. 1994;91:952–956. doi: 10.1073/pnas.91.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broekaert WF, Delauré SL, De Bolle MFC, Cammue BPA. The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol. 2006;44:393–416. doi: 10.1146/annurev.phyto.44.070505.143440. [DOI] [PubMed] [Google Scholar]
- 30.van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21:2527–2540. doi: 10.1105/tpc.108.065193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bent AF, Innes RW, Ecker JR, Staskawicz BJ. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact. 1992;5:372–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- 33.Lund ST, Stall RE, Klee HJ. Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell. 1998;10:371–382. doi: 10.1105/tpc.10.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 35.Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bethke G, et al. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA. 2009;106:8067–8072. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bethke G, Scheel D, Lee J. Sometimes new results raise new questions: The question marks between mitogen-activated protein kinase and ethylene signaling. Plant Signal Behav. 2009;4:672–674. doi: 10.4161/psb.4.7.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 40.Nühse TS, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo S-D, Cho Y-H, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 45.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 46.Hann DR, Rathjen JP. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 2007;49:607–618. doi: 10.1111/j.1365-313X.2006.02981.x. [DOI] [PubMed] [Google Scholar]
- 47.Gendrel A-V, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 48.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathieu O, et al. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell. 2003;15:2929–2939. doi: 10.1105/tpc.017467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.