Abstract
Jawless vertebrates such as lamprey and hagfish lack T-cell and B-cell receptors; instead, they have unique antigen receptors known as variable lymphocyte receptors (VLRs). VLRs generate diversity by recombining highly diverse leucine-rich repeat modules and are expressed clonally on lymphocyte-like cells (LLCs). Thus far, two types of receptors, VLRA and VLRB, have been identified in lampreys and hagfish. Recent evidence indicates that VLRA and VLRB are expressed on distinct populations of LLCs that resemble T cells and B cells of jawed vertebrates, respectively. Here we identified a third VLR, designated VLRC, in the lamprey. None of the ≈100 VLRC cDNA clones subjected to sequencing had an identical sequence, indicating that VLRC can generate sufficient diversity to function as antigen receptors. Notably, the C-terminal cap of VLRC exhibits only limited diversity and has important structural differences relative to VLRA and VLRB. Single-cell PCR analysis identified LLCs that rearranged VLRC but not VLRA or VLRB, suggesting the presence of a unique population of LLCs that express only VLRC.
Keywords: antigen receptors, immune system evolution, jawless vertebrates, leucine-rich repeats, somatic rearrangement
Jawless vertebrates such as lamprey and hagfish produce specific agglutinins when immunized with particulate antigens (1–7). Hence, they were long thought to have immunoglobulins. Accumulated evidence indicates, however, that jawless vertebrates have neither immunoglobulins nor T cell receptors but instead have unique antigen receptors called variable lymphocyte receptors (VLRs) (8–15). Whereas immunoglobulins and T cell receptors generate repertoire diversity by recombining variable (V), diversity (D), and joining (J) gene segments, VLRs achieve comparable diversity through the rearrangement of highly diverse leucine-rich repeat (LRR) modules (16, 17). The germline VLR gene has an incomplete structure incapable of encoding functional proteins; it is, however, flanked by a large number of LRR-encoding modules exhibiting remarkable sequence diversity. During the development of lymphocyte-like cells (LLCs), these modules are incorporated into the VLR gene by a gene conversion-like process presumably mediated by cytidine deaminases of the AID-APOBEC family (18–20). Crystallographic analysis of VLR monomers showed that they adopt a horseshoe-shaped structure and suggested that they most likely bind antigens through the hypervariable concave surface composed of β-strands (21). This suggestion was confirmed recently by in vitro mutagenesis experiments (22) and the crystal structure analysis of VLR monomer–antigen complexes (23, 24).
Two VLR genes, designated VLRA and VLRB, have been identified in hagfish and lampreys (19, 25, 26). In hagfish, VLRA and VLRB are encoded by two separate loci (27), suggesting that the two VLR genes function as independent recombination units; the same seems to be the case with the lamprey genes (19). Surprisingly, recent evidence indicates that lamprey VLRA and VLRB are expressed on distinct populations of LLCs that resemble T cells and B cells of jawed vertebrates, respectively (28). When challenged with antigen, LLCs expressing specific VLRB molecules undergo blast transformation, expand clonally, and begin to secrete VLRB molecules in a manner analogous to the secretion of immunoglobulins by B cells (29). Secreted VLRB molecules, which form pentamers or tetramers of dimers (22), function as strong agglutinins, thus accounting for the earlier observations suggesting the existence of antibodies in jawless vertebrates (1–7). By contrast, VLRA molecules are expressed on a population of VLRB− LLCs and apparently occur only in a membrane-bound form; this population of LLCs responds to a T cell mitogen and up-regulates the expression of interleukin-17 (28), indicating that lampreys have humoral and cellular arms of adaptive immunity analogous to those of jawed vertebrates.
Here we analyzed publicly available sea lamprey (Petromyzon marinus) EST sequences and identified a previously undescribed VLR gene, which we propose to call VLRC.
Results
Identification of the VLRC Gene.
We collected sea lamprey EST sequences from the National Center for Biotechnology Information (NCBI) trace archive and constructed an EST database. TBLASTN searches of this database using hagfish VLRA and VLRB (accession nos. ABB59067 and ABB59026, respectively) as query proteins identified an EST clone that predicted a VLR-like protein with a 3′ terminus distinct from those of known VLRs.
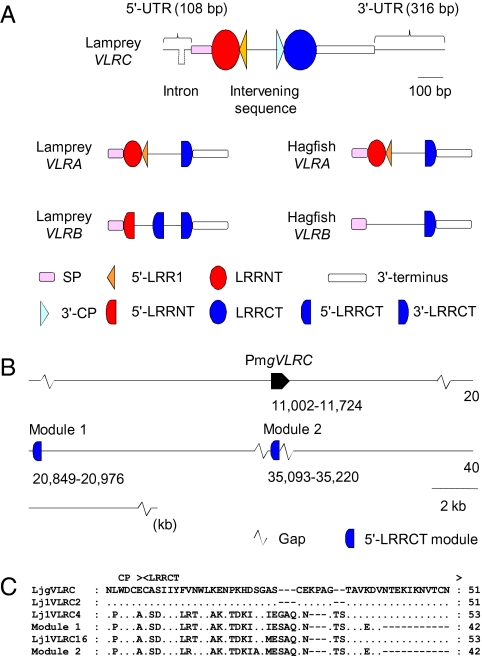
To establish the identity of this clone (accession no. EC382912.1), we isolated the corresponding cDNA fragment from the Japanese lamprey (Lethenteron japonicum) by 3′-RACE. The Japanese lamprey leukocyte cDNA library was then screened using this fragment as a probe. We chose a cDNA clone that seemed to contain a full-length insert and determined its complete sequence (accession no. AB507271). This clone encoded a protein consisting of a 24-residue signal peptide (SP), 36-residue N-terminal cap (LRRNT), 25-residue LRR1, 24-residue LRR, 24-residue LRRVe, 16-residue connecting peptide (CP), 49-residue C-terminal cap (LRRCT), and 74-residue 3′ terminus (Fig. 1). The top 50 BLAST hits to this protein were all agnathan VLRs, with inshore hagfish (Eptatretus burgeri) VLRA encoded by clone Es6VLRA.H5 showing the greatest similarity (49% amino acid identity). The SP and 3′ terminus of this protein were, however, only weakly similar to those of known sea lamprey VLRs, suggesting that it is a unique member of the VLR family. Isolation of Japanese lamprey VLRA and VLRB cDNAs confirmed the existence of three types of VLRs in a single lamprey species (Fig. 1). Therefore, the VLR identified in this study was named VLRC. In all known VLRs, LRRNT and LRRCT contain a characteristic four-cysteine motif CXnCXCXnC (where X stands for any amino acid other than cysteine). These motifs, known to be involved in the formation of two sets of disulfide bridges, are also present in VLRC.
Fig. 1.
VLRC is a member of the VLR family. Deduced amino acid sequences of sea lamprey (Pm, P. marinus), Japanese lamprey (Lj, L. japonicum), inshore hagfish (Eb, E. burgeri), and Pacific hagfish (Es, Eptatretus stoutii) VLRs. C, conserved cysteine residues in LRRNT and LRRCT; #, highly variable inserts (23). Conserved residues are shaded: light-blue, ≥90% identity; green, 70–89% identity; and yellow, 69–50% identity. “-” indicates a gap. Accession nos. are as follows: LjVLRA, AB507269; PmVLRA, EF094821; EbVLRA, AY964726; EsVLRA, AY964825; LjVLRB, AB507270; PmVLRB, AY577957; EbVLRB, AY965530; EsVLRB, AY965561; and LjVLRC, AB507271.
We constructed 3D models of VLRC and VLRA using the crystal structure of lamprey VLRB specific for the H-trisaccharide of human blood group O antigen (23) as a template (Fig. S1). VLRC was predicted to have a structure similar to that of lamprey VLRB but to lack protrusions located in the LRRCT of VLRB. By contrast, the LRRCT of some lamprey VLRA molecules was predicted to form a protrusion.
Germline Structure of the VLRC Locus.
To determine the germline structure of the VLRC gene, Japanese lamprey genomic DNA was subjected to PCR using a set of primers designed to amplify the region spanning from 5′-UTR to 3′-UTR. Only a single DNA fragment of 1,154 bp was obtained, indicating that VLRC is a single-copy gene. This fragment was sequenced completely (accession no. AB507272) and its sequence compared with the full-length VLRC cDNA sequence. Like other VLR genes, the germline VLRC gene lacks sequences coding for LRR modules and contains only the sequences coding for 5′-UTR, SP, LRRNT, 5′-part of LRR1, 3′-part of CP, LRRCT, 3′ terminus, and 3′-UTR (Fig. 2A); 5′-part of LRR1 and 3′-part of CP are separated by a sequence of 161 bp with no apparent similarity to any known sequence. Similar to other VLR genes, VLRC contained an intron (35 bp) in its 5′-UTR.
Fig. 2.
Organization of the germline VLRC locus. (A) Structure of the germline VLRC gene of the Japanese lamprey (drawn to scale) and the schematic germline structures of other known VLR genes (not drawn to scale). (B) Organization of the sea lamprey VLRC locus. Two 5′-LRRCT-encoding modules, designated modules 1 and 2, are located downstream of the germline VLRC gene. This information is based on the analysis of contig 377 in the Pre Ensembl genome database. (C) The 5′-LRRCT of VLRC shows only limited diversity; basically only two major types of sequences were observed. One type represented by Lj1VLRC2 has a sequence identical or nearly identical to the 5′-LRRCT sequence located in the germline VLRC gene (LjgVLRC). The other type falls into two closely related subtypes represented by Lj1VLRC4 and Lj1VLRC16; the sequences of these clones are very similar to those of modules 1 and 2, respectively. “.” and “-” indicate identity with the top sequence and absence of residues, respectively. Clones are identified by the species name, followed by the animal number, gene name, and clone number. Thus, Lj1VLRC2 indicates a VLRC clone 2 isolated from Japanese lamprey individual 1.
Analysis of the sea lamprey genome assembly revealed that this lamprey species also has a single copy of VLRC and that the organization of the germline VLRC gene is conserved between the two lamprey species. Two 5′-LRRCT-encoding modules, designated modules 1 and 2, respectively, were identified downstream of the VLRC gene (Fig. 2B). BLAST searches of the sea lamprey genome identified no other 5′-LRRCT-encoding modules with significant similarity to those observed in Japanese lamprey VLRC transcripts. Among known VLR genes, the structure of germline VLRC is unique in that it contains LRRNT- and LRRCT-modules in their entirety (Fig. 2A).
VLRC Generates Diversity Comparable to That of VLRA and VLRB, but Its LRRCT Shows Only Limited Diversity.
To assess the extent of diversity generated by the VLRC gene, we cloned the region spanning from LRRNT to LRRCT by RT-PCR from the leukocytes of five Japanese lampreys and sequenced a total of 101 clones (Fig. S2). None of the clones had an identical nucleotide sequence, and the number of 24-residue LRR modules varied from zero to four. Of the 302 LRR modules encoded by these clones, 70% had unique sequences. Similarly, 66% of LRR1, 53% of CP, and 75% of LRRVe had unique sequences. These figures are comparable to those previously reported for hagfish VLRA (25), hagfish VLRB (25), lamprey VLRB (17), and lamprey VLRA molecules (19). Thus, VLRC seems to be capable of generating diversity comparable to that of VLRA and VLRB.
Consistent with the observation that LRRCT is involved in antigen recognition (23, 24), the 5′-LRRCT of lamprey VLRA and VLRB exhibits high levels of sequence diversity. By contrast, only two major types of 5′-LRRCT sequences were detected in Japanese lamprey VLRC (Fig. 2C and Fig. S2). One type completely or closely matched the 5′-LRRCT sequence encoded by the germline VLRC gene; another type fell into two closely related subtypes, with sequences almost identical to those of modules 1 and 2 located downstream of the sea lamprey VLRC gene, respectively (Fig. 2C). These observations suggest that, similar to sea lampreys, Japanese lampreys have 5′-LRRCT-encoding modules corresponding to modules 1 and 2 and that they use such modules for VLRC assembly, along with the 5′-LRRCT module encoded by the germline gene. The 5′-LRRCT sequence encoded by the germline gene was so divergent from that encoded by module 1 or 2 that VLRC clones formed distinct clusters (Fig. S3) depending on whether their 5′-LRRCTs were derived from the germline gene (marked with filled circles) or from module 1 or 2 (marked with open circles).
Phylogenetic Analysis Indicates That VLRC Is More Closely Related to VLRA Than to VLRB.
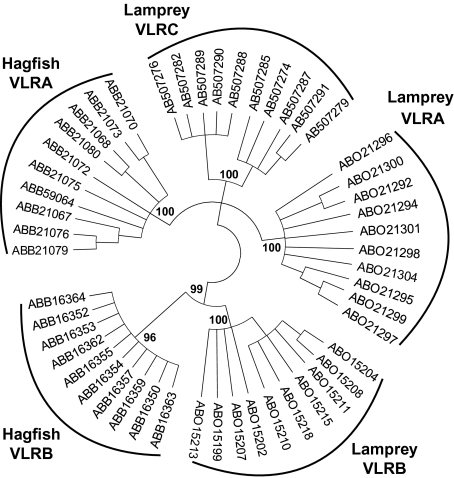
To examine the relationship of VLRC to known VLRs, we constructed a neighbor-joining tree (Fig. 3). The tree supported the orthologous relationship of hagfish and lamprey VLRB and showed that VLRC is more closely related to hagfish and lamprey VLRA than to hagfish and lamprey VLRB. Lamprey VLRA and VLRC were almost equidistant from hagfish VLRA, and their relationship could not be resolved with high bootstrap support, thus precluding us to determine which lamprey gene is orthologous to hagfish VLRA according to sequence comparison alone.
Fig. 3.
Phylogenetic relationship of VLR genes. A neighbor-joining tree was constructed using the amino acid sequences of the diversity region that could be reliably aligned (LRRNT, LRR1, LRRVe, and LRRCT). Nodes that received bootstrap support of <70% were collapsed. Bootstrap values >90% are shown. For abbreviations of species’ names, see the legend to Fig. 1.
Identification of a Unique Population of LLCs That Rearrange Only the VLRC Gene.
We compared by RT-PCR the expression profiles of VLRA, VLRB, and VLRC in representative tissues of adult lampreys using elongation factor 1α (EF1α) as a positive control (Fig. 4A). The three VLR genes showed similar expression patterns, with transcripts detected most abundantly in peripheral blood leukocytes, followed by gills, intestine, and kidney. Real-time RT-PCR analysis showed that VLRC transcripts were ≈60–100 times less abundant than VLRB transcripts in all of the tissues examined; on the other hand, the expression levels of VLRC and VLRA were nearly the same (Fig. 4B).
Fig. 4.
Tissue distribution of VLRC transcripts in Japanese lampreys. (A) Tissue distribution of VLRA, VLRB, and VLRC transcripts. PBL, peripheral blood leukocytes. (B) Real-time RT-PCR analysis. Two animals were subjected to experiments.
To examine whether VLRC+ cells are distinct from those expressing known VLRs, we generated mAbs specific for VLRA and VLRB, respectively. The specificities of these mAbs were verified by testing against a panel of transfected mammalian cells expressing VLRA, VLRB, or VLRC (Fig. S4). Purified LLCs (≈92% LLCs as judged by forward and side scatter characteristics and Giemsa staining) were subjected to magnetic cell sorting with VLRB-specific mAb, and then the flow-through was flow-sorted into VLRA+ and VLRA−/VLRB− fractions. The latter fraction was assumed to contain VLRC+ cells. Each of the three fractions, VLRA+, VLRB+, and VLRA−/VLRB−, was subjected to single-cell sorting, and rearrangements of VLR genes were assessed by genomic PCR. Representative electrophoretic profiles of single-cell PCR products are shown for VLRA+, VLRB+, and VLRA−/VLRB− cells (Fig. 5). We typically observed a germline product along with a rearranged product. Of 24 VLRA−/VLRB− cells in which VLRC assembly was detected, 23 cells rearranged the VLRC gene and maintained the VLRA and VLRB genes in germline configuration, indicating the existence of a unique population of LLCs that exclusively express VLRC. In one cell, we detected assembly of both VLRA and VLRC genes; however, consistent with the VLRA− phenotype, the rearranged VLRA gene contained a 2-bp insertion in its diversity region, resulting in a frameshift mutation. Thus, only the rearranged VLRC gene was apparently functional in this cell.
Fig. 5.
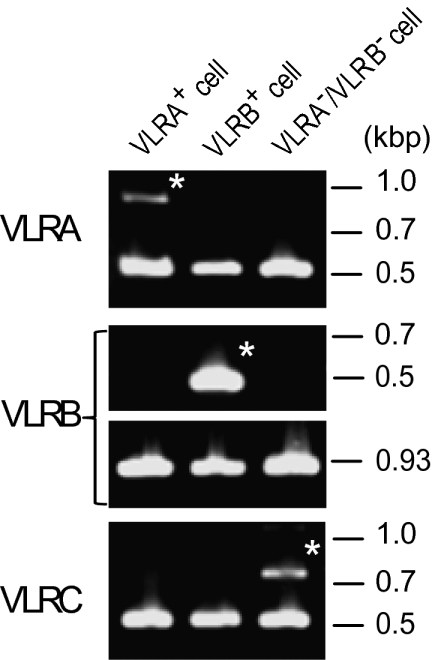
Identification of LLCs that rearrange only the VLRC gene. Single-cell genomic PCR identified LLCs that rearranged only the VLRA (Left), VLRB (Center), or VLRC (Right) gene. Bands marked with asterisks represent rearranged products, whereas unmarked bands represent germline products.
Discussion
The VLRC gene described here generates diversity comparable to that of VLRA and VLRB by rearranging highly diverse LRR modules (Figs. S2 and S3); the rearranged VLRC gene is transcribed predominantly in peripheral blood leukocytes and tissues thought to be involved in hematopoiesis (kidney and intestine) or defenses at the body surface (gill and intestine) (Fig. 4). Furthermore, single-cell PCR experiments provided convincing evidence for the presence of LLCs in which only the VLRC gene is rearranged (Fig. 5). Collectively, these observations indicate the existence of a unique population of LLCs that solely express VLRC as antigen receptors. Thus, lampreys have at least three populations of LLCs distinguished by differential expression of the three VLR loci.
The 5′-LRRCT region exhibits high levels of sequence diversity in VLRA and VLRB molecules; consistent with this, the sea lamprey genome contains 54 and 58 5′-LRRCT-encoding modules that can potentially be used for VLRA and VLRB assembly, respectively (16, 19). By contrast, besides the module encoded by the germline gene, the lamprey genome seems to contain only two 5′-LRRCT–encoding modules used for VLRC assembly (Fig. 2A). Consequently, the 5′-LRRCT region of VLRC shows only limited diversity (Fig. 2C and Fig. S2). This observation may have functional significance because, in VLRB molecules, 5′-LRRCT is known to be involved in antigen recognition (23, 24). If the same is the case with VLRC, VLRC might recognize antigens bearing conserved epitopes or antigens in association with invariant molecules, with its LRRCT and LRR interacting with conserved and variable epitopes, respectively.
Another notable feature of VLRC is that, unlike lamprey VLRA or VLRB, its 5′-LRRCT is predicted to lack the capacity to form protrusions (Fig. S1). In lamprey VLRB molecules, these protrusions are formed by a stretch of amino acid residues, known as a highly variable insert, that displays marked variations in length, amino acid composition, and secondary structure (23). In a VLRB molecule, RBC36, a 10-residue insert forms a β-hairpin protrusion that interacts with H-antigen trisaccharide (23). In another VLRB molecule, VLRB.2D, a six-residue insert forms a protrusion that extends to the catalytic cleft of hen egg lysozyme (24). Consistent with the observation that lamprey VLRA molecules have a variable insert with the average length of 12 residues (24), 3D modeling predicted protrusions in some VLRA molecules (Fig. S1). By contrast, the region of VLRC corresponding to the variable insert contains only a few residues (Fig. 1 and Fig. S2) and hence is too short to form any protrusions. This structural feature might impose additional restrictions on the nature of antigen recognized by VLRC.
Phylogenetically, VLRB is present in both hagfish and lampreys, and VLRC is more closely related to VLRA than to VLRB (Fig. 3). This observation indicates that, in a common ancestor of hagfish and lampreys, a primordial VLR gene duplicated to give rise to VLRB and a common ancestor of VLRA and VLRC, and that this was followed by a second duplication event that separated VLRA from VLRC. The observation that lamprey VLRA and VLRC are almost equally related to hagfish VLRA (Fig. 3) raises three major possibilities concerning their relationship. First, lamprey VLRA is orthologous to hagfish VLRA, and hagfish VLRC remains to be identified. Second, lamprey VLRC is orthologous to hagfish VLRA, and the lamprey counterpart of hagfish VLRA remains to be identified. Third, lamprey VLRA and VLRC diverged in a lamprey lineage soon after its separation from a hagfish lineage, and thus, neither lamprey VLRA nor VLRC is orthologous to hagfish VLRA. The germline VLRA genes of hagfish and lamprey have an identical structure, whereas the germline structure of lamprey VLRC is unique (Fig. 2A). This observation argues against the second possibility because gene structure is generally more conserved than protein sequences during evolution. Thus, the first and third possibilities remain open at the moment.
Despite the fact that VLRs are structurally unrelated to T/B cell receptors, both receptors rely on combinatorial diversity to generate a vast repertoire of binding specificities, highlighting similarities in immune defense strategies adopted by all vertebrates. Most remarkable in this regard is the recent demonstration that cells expressing VLRA and VLRB resemble T cells and B cells of jawed vertebrates, respectively (28). This naturally raises the question, “Do VLRC+ cells resemble T cells or B cells?” Previous work has shown that lamprey agglutinins generated against human erythrocytes are exclusively derived from VLRB+ cells (29). Together with our observation that VLRC is phylogenetically more closely related to VLRA (Fig. 3), this finding suggests that VLRC+ cells probably resemble T cells rather than B cells. If VLRC+ cells are indeed T cell-like, another interesting question arises: “Are VLRA+ and VLRC+ cells functionally specialized in a manner analogous to αβ and γδ T cells of jawed vertebrates?” The discovery of VLRC provides an opportunity to further examine the origin of lymphocytes and the similarities and dissimilarities of host defense strategies used by jawed and jawless vertebrates.
Materials and Methods
Animals.
Adult Japanese lampreys were purchased from local dealers and maintained in temperature-controlled tanks. All procedures of animal experimentation were approved by the Institutional Review Board of Hokkaido University.
Computational Analysis.
Sea lamprey EST sequences were retrieved from the NCBI trace archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi), and a private database was constructed with the GENETYX-PDB program package (version 5; GENETYX). This database was searched with the TBLASTN program implemented in the package. The genomic organization of the sea lamprey VLRC gene was determined using the Pre Ensembl genome browser (http://pre.ensembl.org/Petromyzon_marinus/Info/Index). 3D models of lamprey VLRA and VLRC were generated using the SWISS-MODEL server (30) and edited by PyMol (http://pymol.sourceforge.net).
Isolation of Japanese Lamprey VLR cDNA and Genomic Clones.
Partial cDNA fragments coding for Japanese lamprey VLRA, VLRB, and VLRC were obtained by RACE using the primers designed in the regions predicted to be conserved between sea lampreys and Japanese lampreys. The cDNA fragments thus obtained were used as probes to screen a λZAPII cDNA library constructed from Japanese lamprey leukocytes, which resulted in the isolation of full-length VLRB and VLRC cDNA clones. Full-length VLRA cDNA clones were isolated by 5′- and 3′-RACE. The genomic clone containing germline VLRC was isolated by PCR using Japanese lamprey genomic DNA as a template (primer sequences: 5′-TGGTCCCGTGCGAGCCGAGCC-3′ and 5′-CGCTATGCAACGGGGATGTCTAC-3′).
Phylogenetic Analysis.
Amino acid sequences were aligned with the Clustal X version 2 program (31). The distance matrix was obtained by calculating Poisson-correction distances for all pairs of sequences. Sites containing gaps were excluded from the analysis using the pairwise deletion option. Neighbor-joining trees were constructed using the MEGA version 4 program (32). The reliability of branching patterns was assessed by bootstrap analysis (500 replications).
Expression Analysis.
Tissue expression profiles of VLR genes were analyzed by conventional and real-time RT-PCR using the same primer sets. The primer sequences for VLRA, VLRB, VLRC, and a reference housekeeping gene EF1A were 5′-ACCGTGAAGAGAGAGGATTGT-3′ and 5′-CTTTTCACACGTTTTCCACGA-3′, 5′-CAGCTACAGCCACCACTGTCT-3′ and 5′-GGGACATGCTACTGCACTTT-3′, 5′-TTCATCATTCTGAAGATTATCGCTG-3′ and 5′-TTGCCGACCGCAAGGCAAGCT-3′, and 5′-GTCTACAAAATTGGCGGTATT-3′ and 5′-ACATCCTTGACAGACACGTT-3′, respectively. Cycling conditions were 1 cycle at 95 °C for 10 min, followed by 30 or 35 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min (30 cycles for VLRB and EF1A, 35 cycles for VLRA and VLRC). Real-time PCR was conducted as previously described (33) using an ABI PRISM 7000 sequence detection system (Applied Biosystems).
Magnetic Cell Sorting.
Whole blood isolated from lampreys was suspended in 60% PBS containing 100 mM EDTA and centrifuged for 5 min at 500 × g. The pellet was resuspended in the same solution and centrifuged for 5 min at 220 × g. Cells in the supernatant were collected and further purified with Histopaque-1077 (Sigma-Aldrich) to obtain LLCs. Magnetic cell sorting was performed using MACS anti-PE MicroBeads according to the instructions of the manufacturer (Miltenyi Biotec). Briefly, 1 × 107 LLCs were reacted with mAb specific for VLRB at 4 °C for 15 min. After washing, they were reacted with 1:10-diluted PE-conjugated anti-mouse IgG F(ab´)2 fragment (eBioscience) at 4 °C for 15 min. They were then washed with buffer, mixed with anti-PE MicroBeads, and applied to the MACS MS columns.
Single-Cell Genomic PCR.
LLCs were sorted and seeded at one cell per well into 96-well plates using BD FACSAria II. Each well was preloaded with 10 μL of LA-Taq PCR buffer (TaKaRa). Single-cell genomic PCR was performed as described previously (18, 20). The primer sets for VLRA were 5′-ATTGTGCATCCACAGCCACT-3′ and 5′-ATCTTGACGAGGCTGGAGAT-3′ for multiplex PCR and 5′-CCTCATCGGTGCAGATCAC-3′ and 5′-CTCACTGCCGCAGGATAAAT-3′ for nested PCR. Those for VLRC were 5′-TGGTCCCGTGCGAGCCGAGCC-3′ and 5′-CGTTCTGTGCTCATGGATTG-3′ for multiplex PCR and 5′-ATGGGGTTTGTCGTGGCGCT-3′ and 5′-TTGCATTGAGATTGGTGGTC-3′ for nested PCR. The primer sets for VLRB were described previously (18, 20).
Supplementary Material
Acknowledgments
We thank Dr. Satoshi Kusuda (Hokkaido Fish Hatchery) for his help in procuring lampreys and Dr. Fumikiyo Nagawa (University of Tokyo) for his advice on single-cell PCR. This work was supported by Grants-in-Aids from The Ministry of Education, Culture, Sports, Science and Technology of Japan and by grants from the Japan Science and Technology Agency and the NOASTEC Foundation. J.K. and Y.S. were supported by the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB507269–AB507373).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001910107/-/DCSupplemental.
References
- 1.Finstad J, Good RA. The evolution of the immune response. III. Immunologic responses in the lamprey. J Exp Med. 1964;120:1151–1168. doi: 10.1084/jem.120.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boffa GA, Fine JM, Drilhon A, Amouch P. Immunoglobulins and transferrin in marine lamprey sera. Nature. 1967;214:700–702. doi: 10.1038/214700b0. [DOI] [PubMed] [Google Scholar]
- 3.Marchalonis JJ, Edelman GM. Phylogenetic origins of antibody structure. 3. Antibodies in the primary immune response of the sea lamprey, Petromyzon marinus. J Exp Med. 1968;127:891–914. doi: 10.1084/jem.127.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linthicum DS, Hildemann WH. Immunologic responses of Pacific hagfish. 3. Serum antibodies to cellular antigens. J Immunol. 1970;105:912–918. [PubMed] [Google Scholar]
- 5.Pollara B, Litman GW, Finstad J, Howell J, Good RA. The evolution of the immune response. VII. Antibody to human “O” cells and properties of the immunoglobulin in lamprey. J Immunol. 1970;105:738–745. [PubMed] [Google Scholar]
- 6.Litman GW, Finstad FJ, Howell J, Pollara BW, God RA. The evolution of the immune response. 3. Structural studies of the lamprey immuoglobulin. J Immunol. 1970;105:1278–1285. [PubMed] [Google Scholar]
- 7.Fujii T, Nakagawa H, Murakawa S. Immunity in lamprey. II. Antigen-binding responses to sheep erythrocytes and hapten in the ammocoete. Dev Comp Immunol. 1979;3:609–620. doi: 10.1016/s0145-305x(79)80056-7. [DOI] [PubMed] [Google Scholar]
- 8.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: New perspectives. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 10.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Kasahara M, Kasamatsu J, Sutoh Y. Two types of antigen receptor systems in vertebrates. Zoolog Sci. 2008;25:969–975. doi: 10.2108/zsj.25.969. [DOI] [PubMed] [Google Scholar]
- 12.Flajnik MF, Du Pasquier L. In: Fundamental Immunology. Paul WE, editor. Philadelphia: Lippincott Williams & Wilkins; 2008. pp. 56–124. [Google Scholar]
- 13.Cooper MD, Herrin BR. How did our complex immune system evolve? Nat Rev Immunol. 2010;10:2–3. doi: 10.1038/nri2686. [DOI] [PubMed] [Google Scholar]
- 14.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm T. One problem, two solutions. Nat Immunol. 2009;10:811–813. doi: 10.1038/ni0809-811. [DOI] [PubMed] [Google Scholar]
- 16.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 17.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 18.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8:206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 19.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 20.Kishishita N, et al. Regulation of antigen-receptor gene assembly in hagfish. EMBO Rep. 2010;11:126–132. doi: 10.1038/embor.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HM, et al. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282:6726–6732. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 22.Herrin BR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105:2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009;16:725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasumi S, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106:12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasamatsu J, Suzuki T, Ishijima J, Matsuda Y, Kasahara M. Two variable lymphocyte receptor genes of the inshore hagfish are located far apart on the same chromosome. Immunogenetics. 2007;59:329–331. doi: 10.1007/s00251-007-0200-3. [DOI] [PubMed] [Google Scholar]
- 28.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008;9:319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 30.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 31.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki T, Shin-I T, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005;174:2885–2891. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.