Abstract
Spider dragline silk is a remarkably strong fiber that makes it attractive for numerous applications. Much has thus been done to make similar fibers by biomimic spinning of recombinant dragline silk proteins. However, success is limited in part due to the inability to successfully express native-sized recombinant silk proteins (250–320 kDa). Here we show that a 284.9 kDa recombinant protein of the spider Nephila clavipes is produced and spun into a fiber displaying mechanical properties comparable to those of the native silk. The native-sized protein, predominantly rich in glycine (44.9%), was favorably expressed in metabolically engineered Escherichia coli within which the glycyl-tRNA pool was elevated. We also found that the recombinant proteins of lower molecular weight versions yielded inferior fiber properties. The results provide insight into evolution of silk protein size related to mechanical performance, and also clarify why spinning lower molecular weight proteins does not recapitulate the properties of native fibers. Furthermore, the silk expression, purification, and spinning platform established here should be useful for sustainable production of natural quality dragline silk, potentially enabling broader applications.
Keywords: metabolic engineering, glycyl-tRNA, silk fiber, Nephila clavipes, spinning
Spider dragline silk, used by spiders as the safety line and the web frame, is exceptionally strong and elastic; it is five times stronger by weight than steel, three times tougher than the top quality man-made fiber Kevlar (1, 2). The dragline silk is primarily composed of two proteins, the major ampullate spidroins 1 (MaSp1) and 2 (MaSp2) (3, 4). These spidroins are highly modular, each with a long repetitive sequence that is flanked on both sides by nonrepetitive amino- and carboxy-termini of approximately 100 amino acids (5). The repetitive sequence is rich in glycine and alanine, and characterized by stretches of alanine that are interrupted by glycine-rich repeats (4). The poly alanine regions form hydrophobic crystalline domains that are responsible for the high tensile strength, whereas the glycine-rich regions are hydrophilic and responsible for the links between crystalline domains as well as the elasticity of dragline fiber (6).
Due to the unique mechanical properties, spider dragline silk has received much attention as a promising material for numerous industrial applications such as parachute cords, protective clothing, and composite materials in aircrafts. Also, many biomedical applications are envisioned due to its biocompatibility and biodegradability. For example, silk-based materials have been developed as sutures for wounds, coatings for biomedical implants, drug carriers for drug delivery, and scaffolds for cell culture and organ replacement (7–9). Unfortunately, natural spider dragline silk cannot be conveniently obtained by farming spiders because they are highly territorial and aggressive. Thus, many attempts have been made to produce recombinant dragline silk proteins (10–13) followed by their spinning into artificial fibers (14–17). The fiber with best mechanical properties was derived from a 60 kDa recombinant protein of the spider Araneus diadematus (14). The fiber exhibited toughness and modulus values comparable to those of native dragline silk but with 4.2-fold lower tenacity. It might be expected to explore the relation between recombinant protein size and fiber mechanical properties and obtain superior artificial fibers. However, spinning of larger recombinant proteins was not reported although a 140 protein was also expressed in the mammalian BHK cells but at a much lower level than that of the 60 kDa protein (14).
The size of a recombinant dragline silk protein is hypothesized to be a key factor in controlling the mechanical properties of the spun fiber. First, from an evolutionary point of view, all the spiders studied have evolved natural dragline silk proteins having very high Mw in the range of 250–320 kDa (5, 18). Second, given the fact that dragline silk proteins contain iterated peptide motifs (3, 6), the larger proteins possess more repeating units than the smaller ones, allowing increased interchain and intrachain interactions. Finally, the smaller proteins result in more chain end defects in fibers, reducing mechanical properties (19).
To test the hypothesis, the relationship between recombinant protein size and the mechanical properties of the spun fibers was examined in the present study. We report production of recombinant Nephila clavipes dragline silk proteins up to 284.9 kDa, spinning into fibers, and characterization of the mechanical properties (Fig. S1). Production of these recombinant proteins, which are extremely rich in glycine (43–45%), was greatly enhanced in the metabolically engineered expression host within which the glycyl-tRNA pool was elevated. The fibers spun with the native-sized recombinant spider silk protein showed tenacity, elongation, and Young’s modulus of 508 MPa, 15%, and 21 GPa, respectively, comparable to those of native spider dragline silk. Lower Mw recombinant silk proteins did not yield similar material properties.
Results and Discussion
Expression of Silk Proteins of Different Sizes.
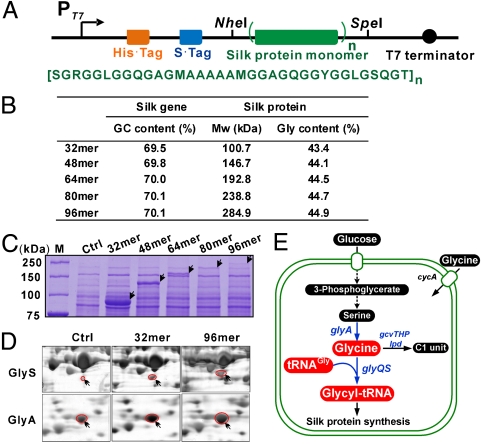
To explore the relationship between protein size and mechanical properties of the spun fibers, recombinant major ampullate spidroin I (MaSp1) of the spider Nephila clavipes was used as a model. MaSp1, whose partial cDNA sequence has been sequenced (3), is the major protein component of the spider dragline silk (18). To express recombinant silk proteins of different sizes, several gene constructs were generated containing varying numbers of the synthetic gene that encodes the iterated peptide motif of MaSp1 (Fig. 1A). Multimerization of the synthetic gene was achieved through the “head-to-tail” strategy employing two compatible yet nonregenerable restriction enzyme sites (NheI and SpeI) (Fig. S1A) (20). By doing so, gene constructs for producing recombinant silk proteins having 32–96 repeats of the monomer could be developed. These recombinant proteins have predicted Mw of 100.7 to 284.9 kDa (Fig. 1B), and the largest one has a similar size to that of natural MaSp1 found in the major ampullate silk gland of a spider (18). However, the highly repetitive nature of the gene constructs, high GC content (∼70%) of the genes, and the glycine-rich characteristics of the silk proteins, make the proteins difficult to express.
Fig. 1.
Recombinant expression of spider dragline silk proteins in Escherichia coli. (A) Recombinant spider silk protein expression constructs (Upper) (see SI Text) and amino acid sequence of the silk monomer (Lower). (B) The GC content of the silk genes and molecular weight (Mw) along with glycine content of the encoded silk proteins. (C) Coomassie-stained 10% SDS-PAGE gel analysis of lysates of E. coli BL21(DE3) cells transformed with empty vector (Ctrl) or plasmids encoding indicated silk proteins. (D) Silver-stained two-dimentional electrophoresis gel analysis of the proteomes of the cells indicated as in C. Shown are enlarged sections of β subunit (GlyS) of glycyl-tRNA synthetase and serine hydroxymethyltransferase (GlyA), the two host proteins upregulated upon silk expression. (E) The glycyl-tRNA metabolic pathways in E. coli. Glycyl-tRNA is synthesized by attaching glycine to tRNAGly by glycyl-tRNA synthetase. Intracellular glycine level is affected by the uptake of extracellular glycine, biosynthesis and degradation by the cleavage system.
Initially, the silk proteins were expressed in the host Escherichia coli BL21(DE3) by performing flask cultures in a complex medium. As the size of the recombinant proteins increased, the expression level (% total cellular proteins) sharply decreased (Fig. 1C). This might be attributed to lower gene dosage (Fig. S2), inefficient transcription or limitations of the translation machinery when expressing the larger proteins (11, 14, 21).
Metabolic Engineering of E. coli for Improved Silk Protein Production.
To produce sufficient amounts of recombinant spider silk proteins of varying sizes, metabolic engineering of the expression host was conducted. To identify relevant gene targets to be engineered, comparative proteomic analysis was carried out to understand how silk expression influences the synthesis of host cellular proteins (Fig. S3). Many stress response proteins were found to be upregulated, suggesting that silk protein expression caused stress to the host cell (Table S1). Importantly, the glycine biosynthetic enzyme, serine hydroxymethyltransferase (GlyA), and the β subunit (GlyS) of glycyl-tRNA synthetase were upregulated (Fig. 1D), suggesting an increased cellular demand for glycyl-tRNA upon expression of the glycine-rich silk proteins (SI Text).
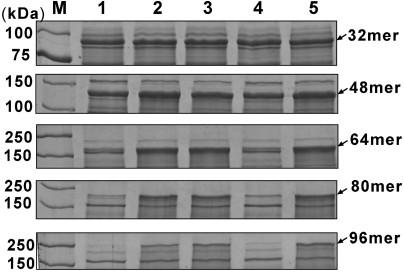
To elevate the glycyl-tRNA pool for more efficient silk protein synthesis, possible strategies include overexpression of glycyl-tRNA synthetase and elevating metabolic pools of tRNAGly and glycine (Fig. 1E). An increase in the gene dosage of glycyl-tRNA synthetase did not further enhance silk production in our preliminary study. This indicated that self-upregulation of the glycyl-tRNA synthetase upon recombinant spider silk expression, as revealed by the proteomic analysis, was sufficient. Then, we elevated the tRNAGly pool. Analysis of the silk genes revealed there are two dominant glycine codons, GGU and GGC. Therefore, the genes encoding the tRNAs that recognize GGU and GGC, E. coli glyVXY, were overexpressed in a compatible plasmid. This plasmid enabled higher expression of the four larger proteins (48- to 96-mer) except the 32-mer protein (Fig. 2, compare lanes 1 and 2). However, in all instances, elevating the tRNAGly pool enhanced cell growth by 30–50% (Fig. S4). Interestingly, the expression levels of the top two largest proteins (80- and 96-mer) were further increased by duplicating the glyVXY expression cassette (Fig. 2, compare lanes 2 and 3), suggesting that further increase of tRNAGly pool was beneficial for the production of high Mw spider silk proteins.
Fig. 2.
Expression of spider dragline silk proteins in metabolically engineered Escherichia coli. Coomassie-stained 10% SDS-PAGE gel analysis of lysates of E. coli BL21(DE3) cells cotransformed with a silk expression plasmid indicated at the right and a compatible plasmid for each lane. Lane 1, pACYC184 as control; lane 2, pTetglyVXY for overexpression of glyVXY that encode tRNAGly; lane 3, pTetgly2 containing two glyVXY expression cassettes; lane 4, p184glyAn for glyA overexpression to increase glycine pool; lane 5, either pTetgly-glyAn (for 32-, 48-, and 64-mer) or pTetgly2-glyAn (for 80- and 96-mer) that allow glyVXY and glyA overexpression simultaneously.
Having been able to enhance the silk protein production by increasing the tRNAGly pool, several strategies were designed to elevate the glycine pool. These included addition of glycine to the culture medium, inactivation of glycine cleavage system, and glyA overexpression (Fig. 1E and Fig. S5). The first two strategies were not desirable (see SI Text for further details). Thus, glyA overexpression was employed and found to enhance the expression levels of the top three largest proteins (Fig. 2, compare lanes 1 and 4). Finally, it was examined whether the beneficial effects of increasing the tRNAGly pool and glycine pool can be synergistic. Indeed, the synergistic effects were observed on the expression levels of 64-mer (Fig. 2, compare lanes 2, 4, and 5), 80- and 96-mer silk proteins (Fig. 2, compare lanes 3–5). Collectively, systematic metabolic engineering of the host strain allowed ∼10 to 35-fold higher level production of the three largest recombinant silk proteins.
Production and Purification of Different Sizes Silk Proteins.
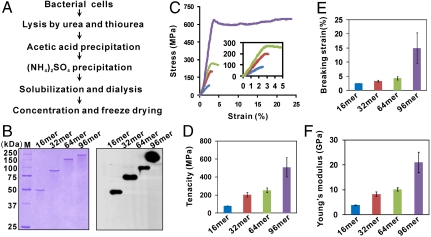
To further evaluate the applicability of the expression system, high cell density cultivation (HCDC) of the engineered strains was performed using a glucose mineral salts medium. In all instances, the strains could be grown to high cell densities (> 42 g l-1) with maximal silk protein titres estimated in the range of 0.5 to 2.7 g l-1 (minimum estimates; see SI Text and Fig. S6 for the details). Following production of the recombinant silk proteins, the next challenge is efficient purification. We focused on the purification of 96-mer silk protein because of its largest size and the best mechanical properties of the spun fiber (see results below). Cells collected from 1 L of HCDC broth were lysed and the cell lysate was acid precipitated (Fig. 3A). Most of the acid precipitates were host cell proteins (Fig. S7A, lane 2); the recombinant spider silk protein was soluble under the acidic condition because it was highly positively charged, as revealed by the protein charge analysis (Fig. S7B). Fractional ammonium sulphate precipitation further enriched and purified the target silk protein (Fig. S7A, compare lanes 3 and 5). Subsequent solubilization, dialysis and freeze drying yielded 1.2 g of the protein with a purity of ∼90% (Fig. S7A, lane 7). This simple chromatography-free method was used to purify other recombinant silk proteins with similar purities. All the purified proteins were confirmed by western blot (Fig. 3B), ruling out the possible involvement of protein truncation during silk protein production (14) or purification (18).
Fig. 3.
Purification of recombinant spider dragline silk proteins and the mechanical properties of the spun fibers. (A) Flow chart of purification. (B) The purified silk proteins [the dialysis supernatants in (A)] were separated on 10% SDS-PAGE gels and analyzed by Coomassie staining (Left) and Western blot using murine monoclonal anti-polyHistidine, Peroxidase conjugate (Right). (C)–(F) Analysis of fibers spun from 20% (w/v) recombinant silk protein solutions. (C) Typical stress-strain curves of the 16-mer (blue), 32-mer (red), 64-mer (green), and 96-mer (purple) fibers. All error bars represent s.d. (n = 10).
Relationship Between Protein Size and Fiber Mechanical Properties.
For a broader size range analysis, the 54.6 kDa 16-mer protein was also included. Because fiber properties depend partially on spinning and drawing conditions (14), the recombinant silk proteins of different sizes (54.6–284.9 kDa) were spun and drawn under the same experimental conditions. The silk proteins were dissolved in hexafluoroisopropanol (HFIP), a commonly used spinning solvent (15, 22), and spun at a spider silk protein concentration of 20% (w/v), which was the maximum operational concentration for the native-sized 96-mer protein due to the solubility and viscosity (see also SI Text and Fig. S8 for the results obtained with different concentrations of smaller proteins). Upon spinning, each of the fibers was hand-drawn to 5 times the original length. It is known that the mechanical properties of a fiber are best described by the shape of a stress-strain curve when the fiber is stretched (2). The maximum height of the stress-strain curve is called tenacity, which is a measure of the amount of stress that a fiber can take before being torn apart. The furthest horizontal extent of the stress-strain curve is breaking strain, representing the extensibility of the fiber. The ratio of stress to strain is Young's modulus, which is derived from the slope of the curve as a measure of the fiber’s stiffness. The stress-strain curves for the silk fibers are shown (Fig. 3C). The mechanical properties including breaking strain, tenacity and Young’s modulus of the fibers improved with increasing silk protein Mw (Fig. 3 D–F). The 96-mer fiber exhibited the tenacity of 508 ± 108 MPa and elongation of 15 ± 5%, which are comparable with the values reported for native N. clavipes dragline silk (740–1,200 MPa; 18–27%) (23–25). Notably, Young’s modulus of the 96-mer fiber was 21 ± 4 GPa, twice that of the native dragline silk (11–14 GPa) (24, 25). Previously, a 60 kDa recombinant dragline silk protein of the spider Araneus diadematus was spun into a fiber displaying respectable material properties (14). However, the average tenacity (ca. 260 MPa) was 4.2-fold lower than that of the native silk (1,100 MPa) (10), possibly due to the small size of the recombinant protein. In the present study, the tenacity of the 96-mer fiber (508 ± 108 MPa) is the highest ever reported for recombinant spider silk proteins.
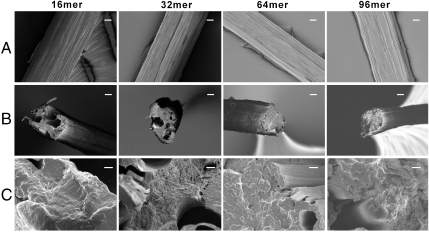
Scanning electron microscopy analysis of fiber surface revealed fibrillar structure in all the fibers (Fig. 4A). This morphology has also been observed for native spider dragline silk (26) and fibers spun from regenerated dragline silk (27). Analysis of fractured fibers showed many irregular voids in the 16- and 32-mer fibers, whereas these defects were absent in the 64- and 96-mer fibers (Fig. 4B). One possible explanation is that higher molecular weight leads to fewer chain ends and thus fewer defects in the fibers formed. Furthermore, microfilaments having a diameter of ∼0.2 μm were assembled to form hierarchical structures in the 64-mer fiber; such structure was much denser in the 96-mer fiber (Fig. 4C). Collectively, the formation of more organized structures by larger silk proteins seems to allow improved mechanical properties of the fibers. Further studies are needed to elucidate the detailed molecular level mechanisms behind this characteristic.
Fig. 4.
Field emission scanning electron micrographs of the fibers spun from 20% (w/v) protein solutions. (A) Analysis of fiber surface, illustrating fibrillar structure. (B) and (C) Analysis at break point to examine fiber interior core. Scale bars in (A) and (B), 10 μm; (C) 1 μm.
In general, the mechanical properties of a polymer rise as the molecular weight increases, but eventually level off with further increase (28). It is likely that there exists a molecular weight threshold required to yield the remarkable mechanical properties of the spider silk as well. Here, we explored the possibility of producing the native-sized spider silk protein, and found that the fiber spun from this recombinant protein gave the native silk-like mechanical properties. To explore whether higher molecular weight recombinant silk protein results in superior material properties, the same strategy was employed for the production of the 128-mer (377.0 kDa) silk protein, which is 32% longer than the native-sized 96-mer protein. It was notable that the ultra high molecular weight recombinant silk protein could be produced by E. coli (Fig. S5). Following purification, however, this 128-mer silk protein was found to be accompanied with putative truncated forms of the target protein (Fig. S7), possibly resulting from the limitation of E. coli translational machinery in expressing this ultra high molecular weight recombinant protein. The presence of mixed molecular weight proteins did not allow us to compare the fiber properties of the ultra high molecular weight protein with other proteins produced in this study.
It is unknown how the addition of the nonrepetitive N- and C-termini domains found on spider silks might alter the properties found in the present study. These domains are critical in silk processing in water (26), yet the results reported here suggest that in organic solvent this may not be the case. The roles for these domains in spider silk functions continues to be elucidated, with recent evidence in control of chain assembly driven by ionic composition, mechanical stimuli, and pH changes during transit in the glands (29–31).
Summary.
A prerequisite for the widespread application of spider silk is mass production of an artificial fiber that fully recapitulates the mechanical properties of native spider dragline silk. However, efficient production of high Mw spider silk protein has been difficult. In this paper, E. coli was metabolically engineered to overproduce recombinant spider silk proteins of up to 284.9 kDa, which is a similar size to that of natural spider silk protein, by overcoming the difficulties caused by its glycine-rich characteristics. The fibre spun with the native-sized recombinant spider silk protein showed similar mechanical properties to native spider silk, thus opening the possibility of its wide range of industrial and biomedical applications. Many other biomaterials are based on silk-like polymers such as elastin, collagen, byssus, resilin, and other repetitive proteins, which are also rich in glycine (32, 33). The general strategy of metabolic engineering reported here should be useful for their efficient biobased production and applications.
Materials and Methods
A description of plasmids construction, inactivation of glycine cleavage system, plasmid copy number analysis, two-dimensional gel electrophoresis, gel analysis, protein identification, SDS-PAGE, protein quantification, and Western blots analysis is provided in SI Text.
Silk Protein Expression in Flask Cultivation.
E. coli cells were grown at 30 °C. Except HCDC, expression experiments were conducted in a 250 mL flask containing 20 mL of LB medium in a shaking incubator at 220 rpm. Cells were induced with 1 mM IPTG (Sigma) when the OD600 reached 0.4. Samples were taken at 6 h after induction for two-dimensional gel electrophoresis, SDS-PAGE, and plasmid copy number analysis. Cells were collected and stored at -80 °C for further analysis.
Production of Silk Proteins by HCDC.
The HCDC of recombinant E. coli was performed in a 6.6 L jar fermentor (Bioflo 3000; New Brunswick Scientific Co.) containing 2 L of R/2 medium (34, 35) (pH 6.80) supplemented with 10 g l-1 of glucose. The dissolved oxygen concentration was kept at 40% of air saturation by automatically increasing the agitation speed up to 1,000 rpm and by changing the percentage of pure oxygen. A nutrient feeding solution was added by using the pH-stat feeding strategy (35). The feeding solution contained 700 g l-1 glucose and 20 g l-1 MgSO4·7H2O. The culture pH was kept at 6.80 by adding 28% (v/v) ammonia water. When the OD600 reached ∼60, cells were induced with 1 mM IPTG.
Purification of Recombinant Silk Proteins.
The 16-, 32-, 64-, and 96-mer silk proteins were purified from HCDC of E. coli BL21(DE3) cotransformed with one of the silk expression plasmids and either pTetglyVXY or pTetgly2 (96-mer). The cells induced with 1 mM IPTG for 6 h, were harvested and suspended in buffer A (1 mM Tris-HCl, pH 8.0, 20 mM NaH2PO4) supplemented with 8 M urea and 2 M thiourea. The cell suspension was agitated for 6 h, after that adjusted to pH 4.0 with glacial acetic acid, and incubated with magnetic agitation at room temperature for additional 2 h. After centrifugation, the supernatant was precipitated by adding (NH4)2SO4 to a final concentration of 1.32 M, and incubated with agitation for 6 h. The mixture was centrifuged and the resulting supernatant was further precipitated with (NH4)2SO4 to 2.80 M. The precipitate was solubilised in buffer A containing 8 M urea and dialyzed at room temperature first against buffer A containing 1.5 M urea for 12 h, then against buffer A for 12 h, and finally against water for 24 h. Following centrifugation, the supernatant was concentrated using an Amicon® Ultra-15 centrifugal filter unit and freeze dried (FD5508, Ilshin Lab).
Fiber Spinning and Mechanical Testing.
Spin dope solutions were prepared by dissolving the lyophilized silk proteins in HFIP (Sigma). The dope solutions were extruded from a 1 mL Kovax syringe through 26 G syringe needle (Korea Vaccine Co., Ltd.) into the coagulation batch containing 90% (v/v) methanol in water using a pump (KDS100; KD Scientific) at a speed of 1–2 ml h-1. After spinning, the as-spun fibers were kept in the coagulation bath for 20 min and cut into 50 mm. For hand drawing, one end of each specimen was firmly fixed and the other end stretched to 5 times the original length smoothly and continuously on a scaled plate. Fibers were then dried at room temperature under tension to prevent contraction and maintain the extended length for the measurements. Prior to the test, the specimens (n = 10) were conditioned at room temperature and a relative humidity of 50% for 24 h. The tensile test was carried out using a universal tensile tester (RB302 ML, R&B Inc.) with 100 g load cell. The gauge length was 20 mm and cross-head speed was 10 mm min-1. Mechanical properties data are shown as means ± standard deviation (n = 10). Statistical differences were determined by unpaired t-test and differences were considered statistically significant at P < 0.05.
Field Emission Scanning Electron Microscopy.
The surface and cross section of the fibers were observed by a field emission scanning electron microscope (SUPRA 55VP, Carl Zeiss) after platinum coating at an accelerating voltage of 2 kV. The fracture surfaces were observed after the tensile test for the cross section whereas the original filaments were used for the surface observation.
Supplementary Material
Acknowledgments.
We thank X. Zhang and K. Numata for preliminary tests of silk fiber spinning and material properties characterization. This work was supported by the Converging Research Center Program (2009-0082332) of the Ministry of Education, Science, and Technology (MEST) through the National Research Foundation (NRF). Further supports by the World Class University Program (R32-2008-000-10142-0) of the MEST through NRF, and the National Institutes of Health P41 Tissue Engineering Resource Center and the Air Force Office of Scientific Research (AFOSR) are appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003366107/-/DCSupplemental.
References
- 1.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: From fibroin sequence to mechanical function. J Exp Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 2.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 3.Xu M, Lewis RV. Structure of a protein superfiber: Spider dragline silk. Proc Natl Acad Sci USA. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinman MB, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. J Biol Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 5.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. Blueprint for a high-performance biomaterial: Full-length spider dragline silk genes. PLoS ONE. 2007;2:e514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 7.Lewis RV. Spider silk: Ancient ideas for new biomaterials. Chem Rev. 2006;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 8.Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Spider silks and their applications. Trends Biotechnol. 2008;26:244–251. doi: 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Hardy JG, Rőmer LM, Scheibel TR. Polymeric materials based on silk proteins. Polymer. 2008;49:4309–4327. [Google Scholar]
- 10.Heim M, Keerl D, Scheibel T. Spider silk: From soluble protein to extraordinary fiber. Angew Chem Int Ed Engl. 2009;48:3584–3596. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- 11.Fahnestock SR, Yao Z, Bedzyk LA. Microbial production of spider silk proteins. Rev Mol Biotechnol. 2000;74:105–119. doi: 10.1016/s1389-0352(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 12.Scheller J, Gührs K, Grosse F, Conrad U. Production of spider silk proteins in tobacco and potato. Nat Biotechnol. 2001;19:573–577. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- 13.Widmaier DM, et al. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol Syst Biol. 2009;5:309. doi: 10.1038/msb.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazaris A, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 15.Teulé F, et al. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat Protoc. 2009;4:341–355. doi: 10.1038/nprot.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arcidiacono S, et al. Aqueous processing and fiber spinning of recombinant spider silks. Macromolecules. 2002;35:1262–1266. [Google Scholar]
- 17.Brooks AE, et al. Properties of synthetic spider silk fibers based on Argiope aurantia MaSp2. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- 18.Sponner A, et al. Characterization of the protein components of Nephila clavipes dragline silk. Biochemistry. 2005;44:4727–4736. doi: 10.1021/bi047671k. [DOI] [PubMed] [Google Scholar]
- 19.Jin HJ, et al. In: Bionanotechnology: Proteins to Nanodevices. Renugopalakrishnan V, Lewis RV, editors. The Netherlands: Springer; 2006. pp. 189–208. [Google Scholar]
- 20.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry. 1995;34:10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 21.Fahnestock SR, Irwin SL. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl Microbiol Biotechnol. 1997;47:23–32. doi: 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- 22.Fu C, Shao Z, Fritz V. Animal silks: Their structures, properties and artificial production. Chem Commun. 2009:6515–6529. doi: 10.1039/b911049f. [DOI] [PubMed] [Google Scholar]
- 23.Elices M, et al. Mechanical behavior of silk during the evolution of orb-web spinning spiders. Biomacromolecules. 2009;10:1904–1910. doi: 10.1021/bm900312c. [DOI] [PubMed] [Google Scholar]
- 24.Swanson BO, Blackledge TA, Summers AP, Hayashi CY. Spider dragline silk: Correlated and mosaic evolution in high performance biological materials. Evolution. 2006;60:2539–2551. [PubMed] [Google Scholar]
- 25.Savage KN, Gosline JM. The effect of proline on the network structure of major ampullate silks as inferred from their mechanical and optical properties. J Exp Biol. 2008;211:1937–1947. doi: 10.1242/jeb.014217. [DOI] [PubMed] [Google Scholar]
- 26.Jin HJ, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature. 2003;424:1057–1060. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 27.Seidel A, et al. Regenerated spider silk: Processing, properties, and structure. Macromolecules. 2000;33:775–780. [Google Scholar]
- 28.Chanda M, Roy SK. In: Plastics Technology Handbook. Chanda M, Roy SK, editors. Boca Raton, FL: CRC; 2007. pp. 1–3. [Google Scholar]
- 29.Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature. 2010;465:239–242. doi: 10.1038/nature08936. [DOI] [PubMed] [Google Scholar]
- 30.Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465:236–238. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- 31.Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc Natl Acad Sci USA. 2008;105:6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Peptide-based biopolymers in biomedicine and biotechnology. Mater Sci Eng. 2008;62:125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatham AS, Shewry PR. Comparative structures and properties of elastic proteins. Philos T Roy Soc B. 2002;357:229–234. doi: 10.1098/rstb.2001.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SY. High cell density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 35.Xia XX, Han MJ, Lee SY, Yoo JS. Comparison of the extracellular proteomes of Escherichia coli B and K-12 strains during high cell density cultivation. Proteomics. 2008;8:2089–2103. doi: 10.1002/pmic.200700826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.