Abstract
Among the fundamental questions regarding cultivated plants is their geographic origin and region of domestication. The genus Cucumis, which includes cucumber (Cucumis sativus) and melon (Cucumis melo), has numerous wild African species, and it has therefore been assumed that melon originated in Africa. For cucumber, this seemed less likely because wild cucumbers exist in India and a closely related species lives in the Eastern Himalayas. Using DNA sequences from plastid and nuclear markers for some 100 Cucumis accessions from Africa, Australia, and Asia, we show here that melon and cucumber are of Asian origin and have numerous previously overlooked species-level relatives in Australia and around the Indian Ocean. The wild progenitor of C. melo occurs in India, and our data confirm that the Southeast Asian Cucumis hystrix is the closest relative of cucumber. Most surprisingly, the closest relative of melon is Cucumis picrocarpus from Australia. C. melo diverged from this Australian sister species approximately 3 Ma, and both diverged from the remaining Asian/Australian species approximately 10 Ma. The Asian/Australian Cucumis clade comprises at least 25 species, nine of them new to science, and diverged from its African relatives in the Miocene, approximately 12 Ma. Range reconstruction under maximum likelihood suggests Asia as the ancestral area for the most recent common ancestor of melon and cucumber, fitting with both having progenitor populations in the Himalayan region and high genetic diversity of C. melo landraces in India and China. Future investigations of wild species related to melon and cucumber should concentrate on Asia and Australia.
Keywords: ancestral areas, crops, economic plants, wild progenitors
Among the most fundamental and debated questions regarding the evolution of cultivated plants is their geographic origin and region of domestication (1). Recent phylogeographic and phylogenetic work on cassava, pumpkin, corn, potato, and rice (2–6) has uncovered the likely places of origin and domestication of these crops. Although many premolecular hypotheses about the domestication of particular species still require testing, it is clear that the Indo-Chinese region has produced a particularly long list of crops. These include rice (Oryza sativa), millets (Setaria spp.), beans (Vigna mungo; Vigna radiata), angled loofah (Luffa acutangula), yams (Dioscorea spp.), and taro (Colocasia esculenta) (7–9). Archaeological evidence from northern India documents these Neolithic crops from 7,000 BC onward, and by the early second millennium, there is evidence of Western crops arriving in India through trade, such as wheat, barley, lentils, grasspea, and peas (7).
One of the crops domesticated in the Indo-Gangetic plain is cucumber, Cucumis sativus. Evidence for this consists in the occurrence there of a wild progenitor, C. sativus var. hardwickii (10, 11) and in comparative linguistic evidence (7, 8). Fossil seeds of cucumber and melon cannot be reliably distinguished, and archeological reports therefore are of limited value for pinpointing areas of melon or cucumber domestication or identifying the routes by which these crops arrived in a particular region (12). Cucumber and melon today are among the 20 most important vegetable crops worldwide (13). The first complete genome of cucumber was released last year (14), and the genome of melon is being completed (15).
In contrast to cucumber, the geographic origin and region of domestication of melon (Cucumis melo) have remained unclear. Nineteenth-century taxonomists suggested that melon probably originated and was domesticated in Asia (e.g., ref. 11). This idea became discredited as workers began to study Cucumis chromosome numbers (16). C. sativus has a chromosome number of 2n = 14, whereas C. melo has a chromosome number of 2n = 24. At least 30 other species of Cucumis have had their chromosomes counted, all but one from Africa and all having 2n = 24 or multiples thereof (17). Based on the impressive species richness in Africa and the identical chromosome number of C. melo and African Cucumis, modern authors have held that C. melo is of African origin (18–22). This view has persisted even in the face of genetic data pointing to greatest genetic diversity in Indian and East Asian landraces of C. melo (23–26) and despite numerous failed attempts to produce fertile F1 offspring from crosses of C. melo and African species of Cucumis (17).
Phylogenetic studies on the genus Cucumis have been Africa-biased in terms of the included species (27–29). These studies yielded contradictory results regarding the closest relatives of C. sativus and C. melo. A 2007 study (29) inferred that South African C. sagittatus is the sister species of C. melo, although this was only observed with nuclear ribosomal internal transcribed spacer (ITS) sequences, not chloroplast sequences. Other studies that also used ITS (27, 28, 30) found C. melo isolated from the other included African species and instead sister to a small clade of five Asian and one Australian species. Although not resolving the position of C. melo, these studies clarified that the genus Cucumis in its traditional circumscription (17) was paraphyletic (28, 29, 31), with species from five other genera nested inside it. [The necessary nomenclatural changes were made previously (32, 33); here we use these up-dated species names.]
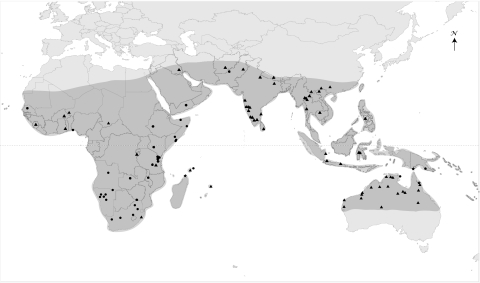
Here we investigate the evolutionary relationships and geographic origin of melon and cucumber by sampling Cucumis accessions covering the entire natural range of the genus from Africa to Southeast Asia to Australia, the Pacific and the Indian Ocean islands. The geographic origin of the plants sequenced is shown in Fig. 1. We included new collections from fieldwork in Australia and Thailand as well as old material from herbaria, including type material of long synonymized names.
Fig. 1.
The natural geographic range of Cucumis (shaded) and the geographic origin of the sequenced plant material. Triangles, Asian/Australian clade comprising cucumber and melon; circles, African grade; stars, outgroup.
Knowing the progenitors and closest relatives of melon and cucumber is important because these crops are highly susceptible to drought and pathogens, including powdery mildew and several mosaic viruses (14). Besides the interest in varieties with new taste or fruit shape, there is great interest in introducing resistance genes from wild relatives. In the case of melon, the search for these relatives has so far concentrated on Africa because of the view that C. melo is of African origin (18–22).
Results
Closest Relatives of Cucumber and Melon.
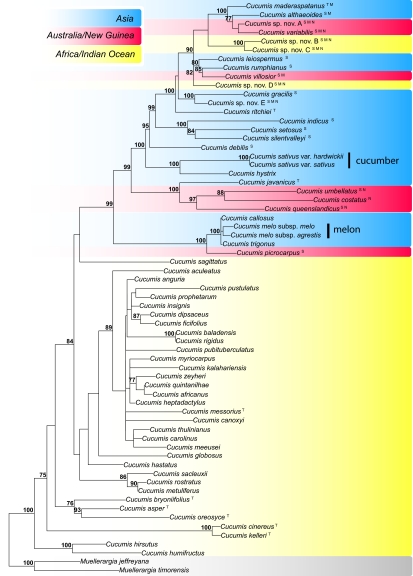
A maximum likelihood phylogeny from the combined plastid and nuclear data (Fig. 2) shows that C. sativus is sister to Cucumis hystrix, confirming previous findings (28–30). Cucumis setosus, an entity that the most recent monograph of Cucumis (17) synonymized under C. sativus, instead is closest to Cucumis silentvalleyi and Cucumis indicus from southern and southwestern India. Sequences of the Himalayan entities Cucumis trigonus and Cucumis callosus are nearly identical to those of C. melo (Fig. 2) and likely represent the wild progenitor of cultivated melon as these forms are fully crossable with C. melo (34). The sister species to C. melo populations is the Australian Cucumis picrocarpus, which had been synonymized under C. melo (17), but is genetically and morphologically highly distinct (Fig. 2; Fig. 3 shows a color photo of C. picrocarpus).
Fig. 2.
ML tree for 63 taxa of Cucumis (Fig. S3 shows a ML tree of all 113 accessions) based on combined sequences from chloroplast and nuclear data (6,202 aligned nucleotides; Table S1), analyzed under a GTR + Γ model. The tree is rooted on Muellerargia. Likelihood bootstrap values of at least 75% are given at the nodes; geographic occurrence of species is color-coded (Inset); superscript letters refer to the following: T, species transferred into Cucumis (32, 33) based on DNA sequences; S, species never before sequenced; M, specimens formerly identified as Mukia maderaspatana (Cucumis maderaspatanus); and N, species new to science. Authors of old and new names appear in Table S1.
Fig. 3.
Habits, fruits, or seeds of (A) Cucumis picrocarpus, (B) C. melo subsp. agrestis, (C) C. hystrix, (D) C. debilis, (E) C. javanicus, (F) C. costatus, (G) C. queenslandicus, (H) C. umbellatus, (I) C. ritchiei, (J) C. villosior, (K) C. sp. nov. E, and (L) Muellerargia timorensis. (Scale bar: 1 cm.) Photographs by P. Sebastian (A–C, H, J, K, L) Z. v. Herwijnen (D); B. Wannan (F and G); M. Sardesai (I); and W.J.J.O. de Wilde (reproduced from ref. 35) (E).
Cucumber and melon are part of a clade that comprises 25 poorly collected and understudied species-level entities from India, Indochina, Malesia, Australia, Africa, and Indian Ocean islands (Figs. 1 and 2). Species recently transferred to Cucumis based on morphology (32, 33) indeed belong in the genus as indicated by chloroplast and nuclear DNA sequences (Fig. 2). Our results, however, reveal another 18 close relatives (labeled in Fig. 2) of cucumber and melon that were neither accepted in the 1993 monograph of the genus by Kirkbride (17) nor included in previous phylogenetic studies (29, 30). Nine (labeled with an upper script N in Fig. 2) represent as yet undescribed species. An important factor in the unexpected Asian/Australian diversity of Cucumis is Cucumis (Mukia) “maderaspatanus.” This polymorphic taxon, revised in 2006 (35), turns out to be a highly unnatural assembly comprising at least nine species-level entities (labeled with an upper script M in Fig. 2) that are not close to each other (Fig. 2 and Table S1). Cucumis maderaspatanus was thought to range from Africa across Asia to Australia, but accessions from these three continents do not group together.
Fruit morphology, a key trait in Cucumis breeding, differs greatly among the species of the Asian/Australian clade (Fig. 3). Early-diverging species, such as the Australian sister clade of C. javanicus and several species from India and the Eastern Himalayas (C. hystrix, C. sativus var. hardwickii, Cucumis debilis, C. setosus, C. silentvalleyi, C. indicus), have more or less ellipsoid fruits that stay green or turn yellow-orange at maturity. By contrast, the Cucumis ritchiei/C. maderaspatanus clade is characterized by smooth, round fruits that turn red at maturity.
Age of Asian/Australian Cucumis Clade and Ancestral Area Reconstruction.
A relaxed molecular clock for the Asian/Australian radiation of Cucumis, calibrated with a secondary calibration from a Cucurbitaceae clock study that used three fossil and one geological calibration (36), indicates that the lineage comprising melon and cucumber split from its African ancestor 11.9 ± 2.0 Ma ago (Fig. S1). Ancestral area reconstruction under maximum likelihood yields Asia as the area of the most recent common ancestor of melon and cucumber (Fig. S2). The split between melon and its Australian sister species, C. picrocarpus, occurred approximately 2.8 ± 1.0 Ma, and that between cucumber and its sister species C. hystrix, approximately 4.6 ± 1.4 Ma.
Discussion
As per this study, Cucumis comprises some 25 Asian and Australian species (Fig. 2) in addition to its approximately 30 African species. The increase in Asian/Australian Cucumis species, compared with the 12 known in 2008 (32, 33) or the two known in 1993 (17), implies that Cucumis was much less understood than hereto thought. Few of the Asian and Australian species are in cultivation, most have never had their chromosomes counted, and little is known about their ecology and distribution. A likely reason for the lack of attention paid to Asian and Australian Cucumis is the almost dogmatic view among Cucumis specialists that the genus comprised just two Asian species-level taxa (C. hystrix and C. sativus) and that any Australian Cucumis-like plants could safely be called C. melo (17) or C. (Mukia) maderaspatanus (35).
The newly revealed Asian/Australian Cucumis radiation of 25 species completely changes the biogeography of the genus. Molecular-clock dating suggests divergence of the Asian/Australian clade from its African relatives at 11.9 ± 2.0 Ma, i.e., during a Miocene period when the African-Arabian plate joined the Asian plate, leading to a closure of the seaway that had previously separated Africa from Asia (37). The ancestor of the Asian/Australian Cucumis clade probably spread to Eurasia via this land bridge, as did numerous vertebrates (38). One species, C. prophetarum, still ranges from Africa to India and Pakistan, but based on the tree topology (Fig. 2), it dispersed to Asia independently of the ancestor of the C. sativus/C. melo clade. The seven species in Australia arose from four dispersal events into that continent, all from Southeast Asia, but at widely different times (Fig. S1).
The DNA phylogeny (Fig. 2) and ancestral area reconstruction (Fig. S2) rejects Kirkbride's (17) grouping of C. melo with the African species C. hirsutus, C. humifructus, and C. sagittatus and instead supports the view of 19th century taxonomists (11, 39) that the wild progenitor of melon would be found in India. The Himalayan entities C. callosus and C. trigonus (names often synonymized with each other and/or with C. melo) (17, 40, 41) produce fertile F1 offspring when crossed with C. melo (34) and clearly are the wild progenitor from which melon was domesticated. The melon land races occurring in South and East Asia exhibit high genetic diversity (23–26, 42) and deserve to be sampled more densely in future studies. The surprising finding that an Australian species (C. picrocarpus) is the sister of the C. melo/C. callosus/C. trigonus complex underscores how little is known of the Australian Cucurbitaceae diversity. Most native Australian cucurbits have close relatives among tropical Asian and even Eurasian lineages (e.g., Austrobryonia, Diplocyclos, Neoalsomitra, Neoachmandra, Trichosanthes) and reached Australia from the north (43). It is possible that other relatives of C. melo exist in under-collected regions between India and Australia or may be hiding among unidentified or misclassified material in herbaria.
Most of the approximately 66 species of Cucumis now known are monoecious annuals, but dioecious mating systems and a perennial habit evolved several times within the genus. We have begun bringing Asian and Australian species into cultivation to study fruit morphology and chromosomes. The evolution of smooth fruits from spiny fruits (Fig. 3), a traditional key character in Cucumis, and the mode of fruit opening are much more plastic than formerly thought (see ref. 28). Overall, the loss of spines appears correlated with a round shape and red color at maturity, probably in connection with bird dispersal. This fits with the inferred dispersal from Asia to Australia, Africa, and various Indian Ocean islands of taxa in the C. ritchiei/C. maderaspatanus clade (Fig. 2).
Analyses of the synteny between C. sativus, C. melo (melon), and Citrullus lanatus (watermelon) have revealed that five of the seven chromosomes of C. sativus arose by fusions of 10 ancestral chromosomes after the split between C. sativus and C. melo (14). To more fully understand the rearrangements, it will be useful to now study the chromosomes of other species in the newly revealed Asian/Australian cucumber/melon clade. Study of these species’ karyotypes (and other biological traits) will be key in the search for new sources of genes for melon and cucumber improvement. The data on the phylogenetic and geographic relationships of melon and cucumber provided here represent a step toward redirected breeding efforts, which should concentrate on Cucumis in Asia and Australia, instead of sub-Saharan Africa. Further population sampling of C. callosus, C. melo, and C. trigonus across Asia will be necessary to assess whether melon was domesticated multiple times.
Materials and Methods
Fig. 1 shows the locations of 97 of the 113 accessions included in this study (including outgroups). We sequenced five chloroplast markers (the trnL intron, the intergenic spacers trnL-F, rpl20-rps12, and trnS-G; and the genes rbcL and matK) and the nuclear ribosomal DNA internal transcribed spacers ITS1 and ITS2, plus the intervening 5.8 S gene using standard procedures (SI Materials and Methods). The dataset comprised 6,202 aligned positions. Sequences were edited with Sequencher (version 4.7; Gene Codes) and aligned by eye by using MacClade version 4.0.8 (44). Maximum likelihood (ML) analyses and ML bootstrap searches were performed using RAxML version 7.0.4 (45) (http://phylobench.vital-it.ch/raxml-bb/). Tree-searches relied on the GTR + Γ model, with model parameters estimated over the duration of specified runs and 100 bootstrap replicates. We carried out Bayesian time estimation with an uncorrelated-rates model, using BEAST version 1.5.3 (46), with a Yule tree prior and the GTR + Γ model with six rate categories. There are no Cucumis fossils, and we therefore used a secondary calibration from a Cucurbitaceae-wide analysis that used four calibration points (36). Mixing of the Markov chain Monte Carlo (MCMC) chain was checked using Tracer version 1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and convergence with AWTY (47). Final trees were edited in FigTree version 1.2.3 (http://tree.bio.ed.ac.uk/software/figtree/). Ancestral areas were inferred under maximum likelihood as implemented in Mesquite version 2.6 (http://mesquiteproject.org), using the Markov k-state one-parameter model, which assumes a single rate for all transitions between character states (geographic regions in this case). Further details on methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by a Waitt grant from the National Geographic Society (to S.S.R.). J. Maxwell helped with field observations in Thailand.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005338107/-/DCSupplemental.
References
- 1.Vavilov NI. Origin and Geography of Cultivated Plants. Cambridge: Cambridge Univ Press; 1935. The phyto-geographical basis for plant breeding; pp. 316–366. (English edition); trans Love D (1987) [Google Scholar]
- 2.Olsen KM, Schaal BA. Evidence on the origin of cassava: Phylogeography of Manihot esculenta. Proc Natl Acad Sci USA. 1999;96:5586–5591. doi: 10.1073/pnas.96.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanjur OI, Piperno DR, Andres CT, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: Implications for crop plant evolution and areas of origin. Proc Natl Acad Sci USA. 2002;99:10923–10928. doi: 10.1073/pnas.012577299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc Natl Acad Sci USA. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller DQ. Agricultural origins and frontiers in South Asia: A working synthesis. J World Prehist. 2006;20:1–86. [Google Scholar]
- 8.Fuller DQ. Non-human genetics, agricultural origins and historical linguistics in South Asia. In: Petraglia MD, Allchin B, editors. The Evolution and History of Human Populations in South Asia. Berlin: Springer; 2007. pp. 393–443. [Google Scholar]
- 9.Pokharia AK, Pal JN, Srivastava A. Plant macro-remains from Neolithic Jhusi in Ganga Plain: Evidence for grain-based agriculture. Curr Sci. 2009;97:546–572. [Google Scholar]
- 10.Royle JF. Illustrations of the Botany and other Branches of the Natural History of the Himalayan Mountains. London: Wm. H. Allen; 1839. [Google Scholar]
- 11.Naudin C. Essais d'une monographie des especès et des variétés du genre Cucumis. Ann Sci Nat Bot. 1859;4:5–87. [Google Scholar]
- 12.Paris HS, Janick J. What the Roman emperor Tiberius grew in his greenhouses. In: Pitrat M, editor. Cucurbitaceae 2008, Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae. Avignon, France: INRA; 2008. pp. 33–42. [Google Scholar]
- 13.FAOSTAT data Statistical database (online) of Food and Agriculture Organization of the United Nations. 2008. [Accessed July 5, 2010]. Available at http://faostat.fao.org/
- 14.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 15.Benjak A, et al. A draft sequence of the melon genome revealed by high throughput shotgun sequencing. Plant & Animal Genomes XVIII Conference. Abstracts. 2010. Available at http://www.intl-pag.org/18/abstracts/P01_PAGXVIII_053.html.
- 16.Whitaker TW. Cytological and phylogenetic studies in the Cucurbitaceae. Bot Gaz. 1933;94:780–790. [Google Scholar]
- 17.Kirkbride JHJr . Biosystematic Monograph of the Genus Cucumis (Cucurbitaceae) Boone, NC: Parkway; 1993. [Google Scholar]
- 18.Whitaker TW, Davis GN. Cucurbits: Botany, Cultivation and Utilization. New York: Interscience; 1962. [Google Scholar]
- 19.Whitaker TW, Bemis WP. Cucurbits, Cucumis, Citrullus, Cucurbita, Lagenaria (Cucurbitaceae) In: Simmonds NW, editor. Evolution of Crop Plants. London: Longman; 1976. pp. 64–69. [Google Scholar]
- 20.Kerje T, Grum M. The origin of melon, Cucumis melo: A review of the literature. Acta Hortic. 2000;510:34–37. [Google Scholar]
- 21.Périn C, et al. A reference map of Cucumis melo based on two recombinant inbred line populations. Theor Appl Genet. 2002;104:1017–1034. doi: 10.1007/s00122-002-0864-x. [DOI] [PubMed] [Google Scholar]
- 22.Luan F, Delannay I, Staub JE. Chinese melon (Cucumis melo L.) diversity analyses provide strategies for germplasm curation, genetic improvement, and evidentiary support of domestication patterns. Euphytica. 2008;164:445–461. [Google Scholar]
- 23.Akashi Y, Fukuda N, Wako T, Masuda M, Kato K. Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica. 2002;125:385–396. [Google Scholar]
- 24.Tanaka K, et al. Molecular characterization of South and East Asian melon, Cucumis melo L., and the origin of group Conomon var. makuwa and var. conomon revealed by RAPD analysis. Euphytica. 2007;153:233–247. [Google Scholar]
- 25.Dhillon NPS, et al. Diversity among landraces of Indian snapmelon (Cucumis melo var. momordica) Genet Resour Crop Evol. 2007;54:1267–1283. [Google Scholar]
- 26.Dwivedi NK, Dhariwal OP, Krishnan SG, Bhandari DC. Distribution and extent of diversity in Cucumis species in the Aravalli ranges of India. Genet Resour Crop Evol. 2010;57:443–452. [Google Scholar]
- 27.Garcia-Mas J, Monforte AJ, Arús P. Phylogenetic relationships among Cucumis species based on the ribosomal internal transcribed spacer sequence and microsatellite markers. Plant Syst Evol. 2004;248:191–203. [Google Scholar]
- 28.Renner SS, Schaefer H, Kocyan A. Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo) BMC Evol Biol. 2007;7:58. doi: 10.1186/1471-2148-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghebretinsae AG, Thulin M, Barber JC. Relationships of cucumbers and melons unraveled: Molecular phylogenetics of Cucumis and related genera (Benincaseae, Cucurbitaceae) Am J Bot. 2007;94:1256–1266. doi: 10.3732/ajb.94.7.1256. [DOI] [PubMed] [Google Scholar]
- 30.Renner SS, Schaefer H. Phylogenetics of Cucumis (Cucurbitaceae) as understood in 2008. In: Pitrat M, editor. Cucurbitaceae 2008, Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae. Avignon, France: INRA; 2008. pp. 53–58. [Google Scholar]
- 31.Kocyan A, Zhang LB, Schaefer H, Renner SS. A multi-locus chloroplast phylogeny for the Cucurbitaceae and its implications for character evolution and classification. Mol Phylogenet Evol. 2007;44:553–577. doi: 10.1016/j.ympev.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Ghebretinsae AG, Thulin M, Barber JC. Nomenclatural changes in Cucumis (Cucurbitaceae) Novon. 2007;17:176–178. [Google Scholar]
- 33.Schaefer H. Cucumis (Cucurbitaceae) must include Cucumella, Dicaelospermum, Mukia, Myrmecosicyos, and Oreosyce: A recircumscription based on nuclear and plastid DNA data. Blumea. 2007;52:165–177. [Google Scholar]
- 34.Parthasarathy VA, Sambandam CN. Taxonomy of Cucumis callosus (Rottl.) Cogn., the wild melon of India. Cucurbit Genet Coop Rpt. 1989;3:66–67. [Google Scholar]
- 35.De Wilde WJJO, Duyfjes BEE. Mukia Arn. (Cucurbitaceae) in Asia, in particular in Thailand. Thai Forest Bull (Bot.) 2006;34:38–52. [Google Scholar]
- 36.Schaefer H, Heibl C, Renner SS. Gourds afloat: A dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc Biol Sci. 2009;276:843–851. doi: 10.1098/rspb.2008.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behrensmeyer AK, Sues H-D, Potts R. Terrestrial Ecosystems Through Time: Evolutionary Paleoecology of Terrestrial Plants and Animals. Chicago: Univ of Chicago Press; 1992. [Google Scholar]
- 38.Kappelman J, et al. Oligocene mammals from Ethiopia and faunal exchange between Afro-Arabia and Eurasia. Nature. 2003;426:549–552. doi: 10.1038/nature02102. [DOI] [PubMed] [Google Scholar]
- 39.De Candolle A. Origine des Plantes Cultivées. Paris: Baillière; 1883. [Google Scholar]
- 40.Chakravarthy HL. Monograph on Indian Cucurbitaceae. Rec Bot Surv India. 1959;17:1–234. [Google Scholar]
- 41.Jeffrey C. Further notes on Cucurbitaceae V. The Cucurbitaceae of the Indian subcontinent. Kew Bull. 1980;34:789–809. [Google Scholar]
- 42.Sujatha VS, Seshadri VS, Srivastava KN, More TA. Isozyme variation in muskmelon (Cucumis melo L.) Indian J Genet Plant Breed. 1991;51:438–444. [Google Scholar]
- 43.Schaefer H, Telford IRH, Renner SS. Austrobryonia (Cucurbitaceae), a new Australian endemic genus, is the closest living relative to the Eurasian and Mediterranean Bryonia and Ecballium. Syst Bot. 2008;33:125–132. [Google Scholar]
- 44.Maddison DR, Maddison WP. MacClade 4.0.8. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- 45.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 46.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.