Abstract
TAL1 plays pivotal roles in vascular and hematopoietic developments through the complex with LMO2 and GATA1. Hemangioblasts, which have a differentiation potential for both endothelial and hematopoietic lineages, arise in the primitive streak and migrate into the yolk sac to form blood islands, where primitive hematopoiesis occurs. ZFAT (a zinc-finger gene in autoimmune thyroid disease susceptibility region / an immune-related transcriptional regulator containing 18 C2H2-type zinc-finger domains and one AT-hook) was originally identified as an immune-related transcriptional regulator containing 18 C2H2-type zinc-finger domains and one AT-hook, and is highly conserved among species. ZFAT is thought to be a critical transcription factor involved in immune-regulation and apoptosis; however, developmental roles for ZFAT remain unknown. Here we show that Zfat-deficient (Zfat−/−) mice are embryonic-lethal, with impaired differentiation of hematopoietic progenitor cells in blood islands, where ZFAT is exactly expressed. Expression levels of Tal1, Lmo2, and Gata1 in Zfat−/− yolk sacs are much reduced compared with those of wild-type mice, and ChIP-PCR analysis revealed that ZFAT binds promoter regions for these genes in vivo. Furthermore, profound reduction in TAL1, LMO2, and GATA1 protein expressions are observed in Zfat−/− blood islands. Taken together, these results suggest that ZFAT is indispensable for mouse embryonic development and functions as a critical transcription factor for primitive hematopoiesis through direct-regulation of Tal1, Lmo2, and Gata1. Elucidation of ZFAT functions in hematopoiesis might lead to a better understanding of transcriptional networks in differentiation and cellular programs of hematopoietic lineage and provide useful information for applied medicine in stem cell therapy.
During embryonic development, mesodermal progenitors give rise to hemangioblasts, which have a differentiation-potential for both endothelial and hematopoietic lineages (1–3). Hemangioblasts arise in the primitive streak and then migrate into the extraembryonic yolk sac to form blood islands (4, 5). Blood islands are foci of hemangioblasts, which form a luminal layer of endothelial cells with a property of producing hematopoietic progenitor cells, and are eventually assembled into a functional vascular network to transfer nutrients from the yolk sac to the embryo proper (6, 7).
Recent studies have revealed that TAL1, a basic helix-loop-helix transcription factor, is an essential transcription factor for differentiation of hemangioblasts into hemogenic endothelium (1, 8–12). TAL1 also plays pivotal roles in vascular and hematopoietic developments through the complex with LMO2 and GATA1 (9, 13–17). LMO2 functions as a bridging molecule between TAL1 and GATA1 in the DNA-binding complex (14). GATA1 also functions as a key molecule in the differentiation process of the erythroid lineage (18, 19). However, the transcriptional regulations upstream of these genes remain elusive.
The human ZFAT gene was originally identified as a susceptibility gene for autoimmune thyroid diseases (20). The mouse Zfat gene encodes an immune-related transcriptional regulator containing 18 C2H2-type zinc-finger domains and one AT-hook and is highly conserved from fish to human (21). ZFAT is predominantly expressed in placenta, thymus, spleen, and lymph nodes (20, 21). ZFAT was a critical transcriptional regulator in immune-regulation (21) and an antiapoptotic molecule in lymphoblastic leukemia cell line (22). Recently, ZFAT was reported to be associated with IFN-β responsiveness in multiple sclerosis (23). However, developmental roles for ZFAT remain unknown.
In this study, we generated Zfat-deficient (Zfat−/−) mice and found that Zfat-deficiency results in early embryonic lethality, with the reduction in the number of blood islands and impaired differentiation of hematopoietic progenitor cells in blood islands. Furthermore, in vitro and in vivo molecular analyses revealed that ZFAT directly regulates the transcription factors including Tal1, Lmo2, and Gata1 in blood islands. Taken together, these results suggested that ZFAT plays critical roles in the development of hematopoietic system in blood islands.
Results
Zfat-Deficient Mice with Early Embryonic Lethality.
To examine developmental roles for ZFAT, we targeted the Zfat allele for disruption by homologous recombination (Fig. 1A). In construction of the targeting vector, a 1.4-kb fragment of Zfat genomic DNA including exon 8 was replaced with neomycin resistance (neo) gene (Fig. 1A). Targeted ES cell clones with homologous recombination and heterozygous (Zfat+/−) mice were confirmed by Southern blot analysis (Fig. 1B, Left) and by PCR (Fig. 1B, Right). Then, Zfat+/− mice with the genetic background of C57BL/6 were established and analyzed in this study. Zfat+/− mice were viable, fertile, and phenotypically indistinguishable from wild-type (Zfat+/+) littermates. Obvious developmental abnormalities in T or B cells from Zfat+/− mice were not observed in the thymus or spleen, where ZFAT is abundantly expressed (20, 21) (Fig. S1); however, the possibilities of altered immune-responses in peripheral T and B cells of Zfat+/− mice are not excluded and a full understanding of the ZFAT function in the immune system awaits future studies.
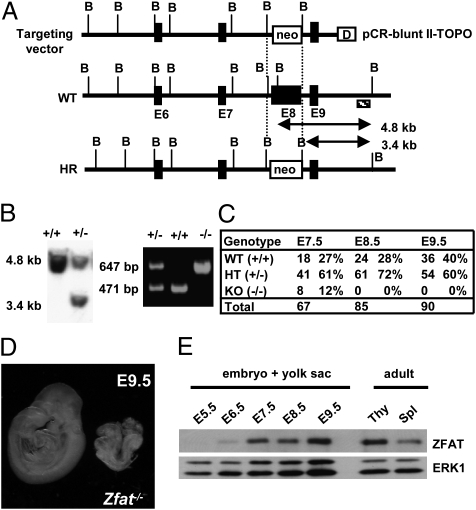
Fig. 1.
Zfat is indispensable for mouse embryonic development. (A) Targeting disruption of the Zfat gene. WT, wild-type; HR, homologous recombinant; B, BglII site; closed box: E, exon; neo, neomycin resistance cassette; D, DTA (diphtheria-toxin A fragment); shaded box: external probe. (B) Southern blotting of BglII-digested DNA using 3′ external probe (Left). PCR-based genotyping of Zfat+/− progeny (Right). (C) Genotyping statistics of progeny from Zfat+/− mice. The number and ratio of embryos showing normal development are shown. (D) Typical phenotype of Zfat−/− embryos at E9.5. (E) ZFAT expression during early developmental stage. Thy, thymocyte; Spl, splenocyte; ERK1, loading control.
Intercrosses between Zfat+/− mice failed to produce Zfat−/− mice, indicating that Zfat−/− mice died either in utero or shortly after birth. Developmental abnormalities in Zfat−/− embryos did occur by E8.5 (Fig. 1C) and no Zfat−/− embryos showed the embryonic turning at E9.5 (Fig. 1D), suggesting that embryonic development in Zfat−/− mice was severely impaired before the stage of embryonic turning. ZFAT protein expression in embryos with yolk sacs was observed from E6.5 and was gradually increased to the expression level of thymocytes or splenocytes in adult tissues, and was kept high at least by E9.5 (Fig. 1E). All these results indicated that ZFAT is a critical molecule during midgestation and its deficiency results in early embryonic lethality, demonstrating that ZFAT is essential for mouse embryonic development.
Impaired Differentiation of Hematopoietic Progenitor Cells in Blood Islands of Zfat-Deficient Mice.
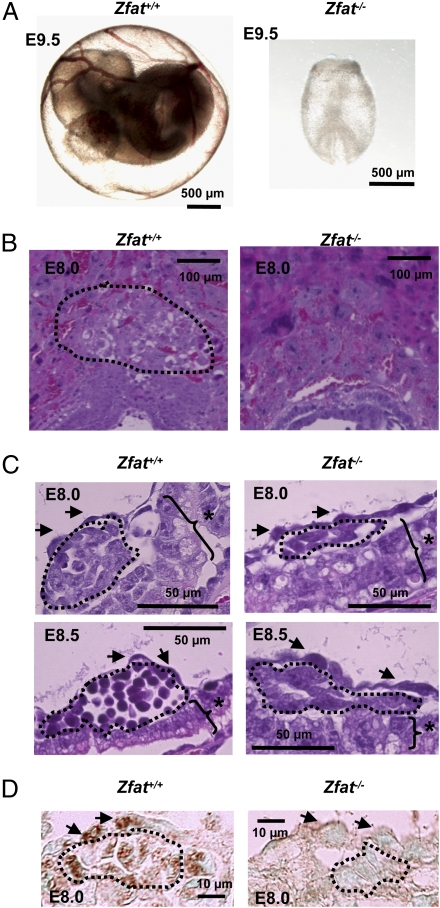
Dysfunction of the vascular system is a common cause of early embryonic lethality during midgestation (24). Initial inspection using a microscope indicated that Zfat−/− yolk sacs were bloodless at E9.5 (Fig. 2A), whereas the vascular system in Zfat+/− yolk sacs seemed to be normally developed (Fig. S2). Histological analyses of placentas revealed that the spongiotrophoblast layer was not well developed in Zfat−/− placentas at E8.0, the abnormality of which was consistently detected in Zfat−/− placentas (Fig. 2B). The phenotype observed in the spongiotrophoblast layer was utilizable as a marker for Zfat−/− yolk sacs. Histological analyses revealed that hematopoietic progenitor cells in Zfat+/+ blood islands differentiated into the more developed cells from E8.0 to E8.5, whereas those in Zfat−/− blood islands were spindle-shaped at both E8.0 and E8.5 (Fig. 2C), suggesting that differentiation of hematopoietic progenitor cells in Zfat−/− blood islands was impaired.
Fig. 2.
Impaired differentiation of hematopoietic progenitor cells in Zfat−/− blood islands. (A) Embryos with yolk sacs from Zfat+/+ or Zfat−/− mice at E9.5. (Scale bars, 500 μm.) (B) H&E-stained sections of Zfat+/+ and Zfat−/− placentas at E8.0. Region surrounded by the dotted line represents spongiotrophoblast layer. (Scale bars, 100 μm.) (C) H&E-stained sections of blood islands of Zfat+/+ and Zfat−/− yolk sacs at E8.0 (Upper) and E8.5 (Lower). Region surrounded by the dotted line represents hematopoietic progenitor cells. Arrows, endothelial cells; asterisks, visceral endodermal cells. (Scale bars, 50 μm.) (D) ZFAT protein expression in endothelial and hematopoietic progenitor cells in Zfat+/+ blood islands at E8.0. The region surrounded by the dotted line represents hematopoietic progenitor cells. Arrows, endothelial cells. (Scale bars, 10 μm.)
Reduction in the Number of Blood Islands and Hematopoietic Progenitor Cells in Zfat−/− Yolk Sacs.
To further characterize the abnormalities in Zfat−/− blood islands, the number of endothelial, hematopoietic progenitor, and visceral endodermal cells in blood islands were examined at E8.0 based on the morphological assessment described (25). The number of endothelial and visceral endodermal cells between Zfat+/+ and Zfat−/− blood islands were not significantly different (P > 0.1) (Table 1). Of interest was that the number of blood islands in Zfat−/− yolk sacs and hematopoietic progenitor cells in Zfat−/− blood islands were significantly decreased by 2.3-fold (*P = 0.014; Table 1) and 2.9-fold (**P = 0.004; Table 1), respectively, compared with those of Zfat+/+ mice. Furthermore, the ratios of hematopoietic progenitor cells per endothelial cells in Zfat+/+ and Zfat−/− blood islands were 1.43 and 0.71, respectively, with a statistically significant difference (P = 0.0037). Taken together, these results suggested that proper differentiation in the hematopoietic lineage was impaired in Zfat−/− blood islands.
Table 1.
Reduction in the number of blood islands and hematopoietic progenitor cells in Zfat-deficient yolk sac at E8.0
| Mean ± SD |
|||||||||
| WT-1 | WT-2 | WT-3 | KO-1 | KO-2 | KO-3 | WT | KO | t test (P value) | |
| Number of slides analyzed | 21 | 19 | 13 | 16 | 14 | 12 | |||
| Number of blood islands | 44 | 42 | 32 | 25 | 14 | 13 | 39 ± 6 | 17 ± 6 | 0.014* |
| Number of endothelial cells | 328 | 433 | 350 | 383 | 206 | 183 | 370 ± 55 | 257 ± 109 | 0.19 |
| Number of hematopoietic progenitor cells | 439 | 632 | 526 | 232 | 148 | 173 | 532 ± 96 | 184 ± 43 | 0.004** |
| Number of visceral endodermal cells | 376 | 414 | 395 | 412 | 205 | 207 | 395 ± 19 | 275 ± 118 | 0.16 |
*P <0.05; **P < 0.01.
ZFAT Expression Does Not Affect Apoptosis or Proliferation in Yolk Sac Blood Islands.
In immunohistochemical analysis using anti-ZFAT monoclonal antibody M16 (Fig. S3), ZFAT signals were evidently detected in endothelial and hematopoietic progenitor cells of Zfat+/+ blood islands at E8.0, whereas ZFAT signals were not observed in endothelial cells or hematopoietic progenitor cells of Zfat−/− blood islands (Fig. 2D), indicating that ZFAT was exactly expressed in endothelial and hematopoietic progenitor cells in blood islands at E8.0. Furthermore, signals of Ki-67 as a proliferation marker were evenly detected in endothelial and hematopoietic progenitor cells in both Zfat+/+ and Zfat−/− blood islands at E8.0, and signals of activated caspase-3 as an apoptosis marker were rarely detected in Zfat+/+ or Zfat−/− blood islands at E8.0 (Fig. S4). Taken together, these results indicate that ZFAT expression in blood islands does not function by inhibiting apoptosis or promoting progenitor cell proliferation, suggesting that ZFAT may instead be involved in promoting hematopoietic progenitor differentiation.
ZFAT Regulates the Genes Involved in Hematopoietic Differentiation in Blood Islands.
To address a possibility whether ZFAT regulates the genes essential for development of hematopoietic progenitor cells in blood islands, we performed real-time quantitative RT-PCR (qRT-PCR) assay for the hematopoiesis-related genes, including Tal1, Lmo2, Gata1, Gata2 (26) and Kit (1, 27), and Gapdh as a control gene in yolk sacs at E7.5. Expression levels of Tal1, Lmo2, and Gata1 in Zfat−/− yolk sacs were decreased by 50-, 20-, and 200-fold, respectively, compared with those of Zfat+/+ yolk sacs (Fig. 3A; *, P < 0.001), whereas the expressions of Gata2 and Kit were not different between Zfat+/+ and Zfat−/− yolk sacs (Fig. 3A; P > 0.05). Reduced expressions of Tal1, Lmo2, and Gata1 were consistent with the histological features in blood islands in Zfat−/− mice (Fig. 2), suggesting that ZFAT is an essential regulator for the expression of the hematopoiesis-related genes, including Tal1, Lmo2, and Gata1 in blood islands.
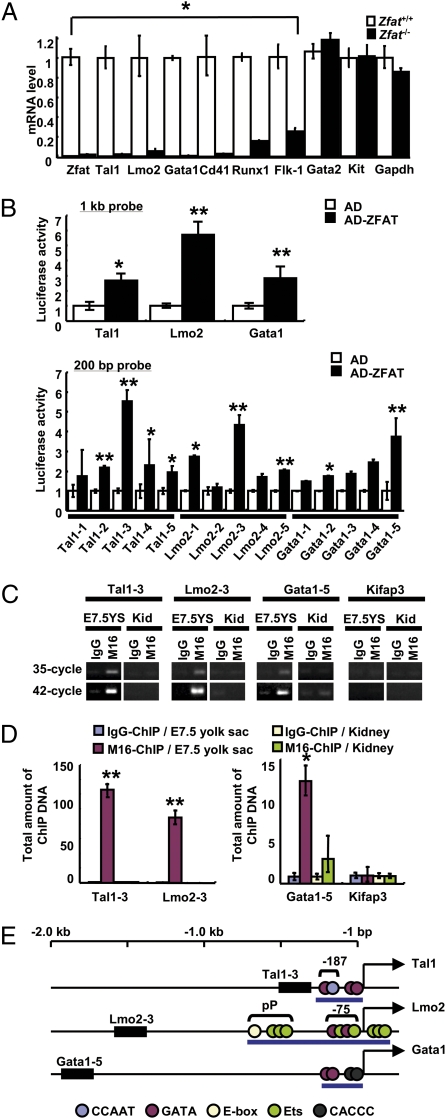
Fig. 3.
ZFAT directly regulates expressions of Tal1, Lmo2, and Gata1 genes in blood islands. (A) MicroRNA expression levels for Zfat, Tal1, Lmo2, Gata1, Cd41, Runx1, Flk-1, Gata2, Kit, and Gapdh genes in Zfat−/− (black bar) and Zfat+/+ (white bar) yolk sacs at E7.5. *P < 0.001. (B) Luciferase assay using 1-kb probes (Upper) and 200-bp probes (Lower) for detection of ZFAT-DNA binding. Activation domain (AD)-ZFAT-binding activity, black bar; AD-binding activity, white bar. *P < 0.05; **P < 0.01. (C) ChIP-PCR assay for detection of the bindings of ZFAT with the DNA elements (region for 200-bp probe in B) in yolk sacs at E7.5 and adult kidney as a control tissue. End-point PCR products at 35- and 42-cycled PCR. YS, yolk sac; Kid, kidney. (D) Quantification of ChIP DNA (region for 200-bp probe in B). Quantities of the ChIP DNA in yolk sacs at E7.5 with M16 anti-ZFAT antibody (red bar) and control IgG (blue bar). Quantities of the ChIP DNA in kidney with M16 (green bar) and control IgG (yellow bar). Bar indicates the total amount of ChIP DNA normalized by M16-ChIP DNA for Kifap3 promoter in kidney as 1.0 unit. *P < 0.05; **P < 0.01. (E) ZFAT-binding regions in the Tal1, Lmo2, and Gata1 promoters. Closed box, ZFAT binding regions; blue circle, CCAAT element; red circle, GATA binding site; yellow circle, E-box; green circle, Ets binding site; black circle, CACCC element; pP, proximal promoter; blue bar, known regulatory region for each gene expression; arrow, transcriptional start site for each gene.
Direct-Regulation of Tal1, Lmo2, and Gata1 by ZFAT.
We, next, determined whether or not ZFAT directly regulates Tal1, Lmo2, and Gata1 expressions. In luciferase reporter assay using 1-kb probes for the promoter regions of Tal1, Lmo2, and Gata1 genes, the luciferase activities by ZFAT fused with a transcriptional activator-domain (AD-ZFAT) were increased by 2.6-, 5.7-, and 2.8-fold, compared with those by a transcriptional activator-domain construct (AD), respectively (Fig. 3B, P < 0.05). ZFAT binding regions were further narrowed down with 200-bp probes from the 1-kb probes showing the activities. The luciferase activities for the 200-bp probes for Tal1, Lmo2, and Gata1 were increased to 5.5-fold (Tal1-3), 4.3-fold (Lmo2-3), and 3.7-fold (Gata1-5), respectively (Fig. 3B; **, P < 0.01).
To address the bindings of ZFAT with these DNA sequences in vivo, ChIP-PCR assays on yolk sacs at E7.5 and on adult kidney as a control tissue, where ZFAT is rarely expressed (21), using anti-ZFAT M16 antibody (Fig. S3) and control IgG, were done for the 200-bp regions with the highest luciferase activity (Tal1-3, Lmo2-3, and Gata1-5) and the promoter region of Kifap3 as a hematopoiesis-unrelated control gene. Differences of ChIP DNA concentrations were semiquantified by 35- and 42-cycle end-point PCR products. Promoter regions for Tal1, Lmo2, and Gata1 in the M16-ChIP DNA from E7.5 yolk sacs were enriched and compared with those of control IgG-ChIP DNA, whereas M16-ChIP DNA for Tal1, Lmo2, and Gata1 in kidney as a control tissue were not enriched (Fig. 3C); taken together, these data are suggestive of the specificity of anti-ZFAT M16 antibody and the bindings of ZFAT with these promoter regions. Furthermore, quantification by real-time qPCR assay for ChIP DNA showed that total amount of promoter regions for Tal1, Lmo2, and Gata1 in the M16-ChIP DNA were 126.4 units, 88.5 units, and 13.2 units, respectively (Fig. 3D, P < 0.05), whereas M16-ChIP DNA on the promoter regions for Cd41, Runx1, and Flk-1—the expressions of which are reported to be regulated by a TAL1-LMO2-GATA1 transcriptional complex (4, 5, 14, 28–32)—were not enriched in the end-point PCR or ChIP-qPCR assays (Fig. S5), suggesting that ZFAT specifically binds to the promoter regions for Tal1, Lmo2, and Gata1 in yolk sacs at E7.5.
The ZFAT binding regions detected in the Tal1, Lmo2, and Gata1 genes are mapped in the genome, showing that ZFAT binds to the distinct regions from the known regulatory regions including the −187 element in Tal1 (33), the proximal promoter and the −75 enhancer element in Lmo2 (34, 35), and the CACCC motif in Gata1 (36, 37) (Fig. 3E).
Reduction in Protein Expressions of TAL1, LMO2, and GATA1 and TAL1-Downstream Genes in Zfat−/− Blood Islands.
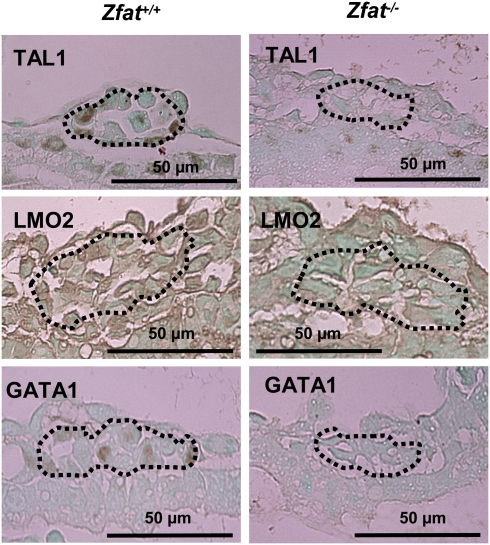
Immunohistochemical analysis on Zfat+/+ and Zfat−/− blood islands at E8.0 was performed to confirm the expression levels of TAL1, LMO2, and GATA1. The signals for TAL1, LMO2, and GATA1 were observed in Zfat+/+ blood islands, especially in hematopoietic progenitor cells, whereas all these expressions were much reduced in Zfat−/− blood islands (Fig. 4), suggesting that ZFAT is indispensable for the proper expressions of TAL1, LMO2, and GATA1 in hematopoietic progenitor cells in blood islands at E8.0.
Fig. 4.
Reduced expressions of TAL1, LMO2, and GATA1 in Zfat−/− blood islands at E8.0. Expressions were detected by immunohistochemical staining using each antibody. The region surrounded by the dotted line represents hematopoietic progenitor cells. Arrows, endothelial cells. (Scale bars, 50 μm.)
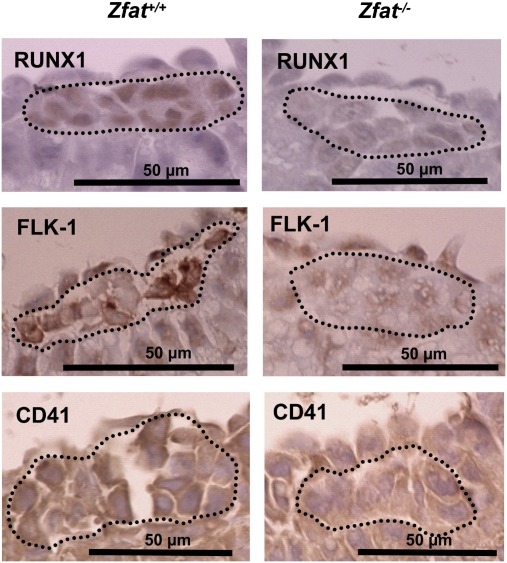
Real-time qRT-PCR assay at E7.5 showed that expression levels of Cd41, Runx1, and Flk-1 in Zfat−/− yolk sacs were decreased by 50-, 6.6-, and 4-fold, respectively, compared with Zfat+/+ yolk sacs (Fig. 3A; *, P < 0.001), although these genes were not directly regulated by ZFAT (Fig. S5). Protein expression of RUNX1 (38) in Zfat−/− hematopoietic progenitor cells at E8.0 was much reduced, compared with that of Zfat+/+ hematopoietic progenitor cells (Fig. 5). Intriguingly, FLK-1 expressions (39) in endothelial cells of blood islands at E8.0 were similarly observed between Zfat+/+ and Zfat−/− mice; however, FLK-1 expression in Zfat−/− hematopoietic progenitor cells was much reduced compared with that of Zfat+/+ hematopoietic progenitor cells (Fig. 5). On the other hand, CD41 expression in Zfat+/+ hematopoietic progenitor cells was not evidently detected at E8.0 and its expression was gradually increased from E8.0 to E8.5 (28, 40) (Fig. S6). CD41 expression at E8.25 in Zfat+/+ hematopoietic progenitor cells was evidently detected at the cellular region lining the plasma membrane (Fig. 5). In contrast, CD41 expression in Zfat−/− hematopoietic cells at E8.25 was not evidently detected (Fig. 5). All of these results suggested that ZFAT is essential for the proper expressions of RUNX1, FLK-1, and CD41 thorough the direct regulation of Tal1, Lmo2, and Gata1 genes in hematopoietic progenitor cells in blood islands, although a possibility of the involvement of unidentified factors regulated by ZFAT is not excluded.
Fig. 5.
Reduced expression of RUNX1, FLK-1, and CD41 in Zfat−/− blood islands. Expressions of RUNX1 at E8.0 (Top), FLK-1 at E8.0 (Middle), and CD41 at E8.25 (Bottom) in the blood islands. Expressions were detected by immunohistochemical staining using each antibody. The region surrounded by the dotted line represents hematopoietic progenitor cells. (Scale bars, 50 μm.)
Discussion
In this study, we generated Zfat−/− mice and demonstrated that Zfat-deficiency results in early embryonic lethality with the reduction in the number of blood islands and impaired differentiation of hematopoietic progenitor cells in blood islands (Fig. 1 C and D, Fig. 2, and Table 1) and that ZFAT is a critical transcription factor directly regulating Tal1, Lmo2, and Gata1 expressions in blood islands (Fig. 3). In hematopoietic differentiation, TAL1 is thought to function through a complex with GATA1 and LMO2 in the process of differentiation from hemangioblasts to hemogenic endothelium, and the development of extraembryonic vasculature is cooperatively regulated by the limited members of transcriptional factors (1, 14, 16, 32, 41). The findings of direct regulations of Tal1, Lmo2, and Gata1 by ZFAT in hematopoietic progenitor cells and ZFAT-mediated expressions of Flk-1, Runx1, and Cd41 through the direct regulation of Tal1, Lmo2, and Gata1 expressions may shed light on the transcriptional network in the developmental program of blood islands. However, this study only suggests a role for ZFAT in the differentiation of primitive hematopoietic progenitor cells; thus, the in vitro yolk sac progenitor culture system (42) would be useful to demonstrate the precise roles for ZFAT in the differentiation of primitive hematopoietic cells.
FLI-1 was reported to be an upstream regulator for Tal1 and Lmo2 (43–46) and is also involved in immunological disease (47, 48). Thus, elucidation of the relation between ZFAT and FLI-1 might lead to a better understanding of the transcriptional network not only in hematopoietic differentiation, but also in immune regulation. Genetic variants in ZFAT were recently reported to be associated with height in the Japanese and Korean populations (49, 50), and development of Zfat−/− embryos were impaired by E8.5, suggesting that ZFAT might be involved in development of mesodermal cells; however, molecular functions of ZFAT in embryonic development and mesoderm lineage should await future studies.
Elucidation of ZFAT functions in hematopoiesis will lead to a better understanding of transcriptional networks in differentiation and cellular programs of hematopoietic lineage, and provide useful information for applied medicine in stem cell therapy.
Materials and Methods
Cells and Animals.
HEK293 cells were cultured as described previously (22). Mice were maintained according to the National Institute Health standards, Guidelines for the Care and Use of Experimental Animals. All of the experimental protocols were approved by the Animal Investigation Committee of Fukuoka University.
Generation of Zfat-Deficient Mice.
We isolated a genomic DNA of the Zfat gene from a 129/SV mouse genomic library (Stratagene) and constructed the targeting vector by replacing a 1.4-kb fragment containing exon 8 of the Zfat gene (GenBank accession no. NT_078782) with a neomycin resistance gene cassette in the opposite transcriptional orientation. The 5′ and 3′ arms of the targeting construct were composed of 10.4 and 2.0 kb, respectively. Diphtheria-toxin A fragment cassette (DTA) flanked the 3′ short arm. The targeting vector was linearized with SalI and electroporated into ES cells and targeted ES clones were obtained. The mutant ES cells were microinjected into C57BL/6 blastocysts, as described previously (51), and the resultant male chimeras were mated with C57BL/6 mice. Zfat+/− mice were backcrossed six times and maintained in the genetic background of C57BL/6 mice. Heterozygous offspring were intercrossed to obtain Zfat−/− mice.
Genotyping.
Genotyping was performed by standard PCR using the specific primer set (Dataset S1). PCR was done by GeneAmp PCR System 9700 (Applied Biosystems).
Histopathological Examination.
Embryos with yolk sacs were fixed in 3.7% paraformaldehyde and embedded in paraffin. Sections (3 μm) were prepared at every 12-μm intervals throughout the tissues and were stained with H&E. Sections were analyzed using Biorevo BZ-9000 inverted-phase microscope at high-power magnification (×600) (Keyence).
Anti-ZFAT Monoclonal Antibody.
Recombinant mouse ZFAT protein (amino acid residues 513–699) was expressed as a GST fusion protein using the pGEX6P-1 vector (GE Healthcare). The fusion protein was soluble in nondenaturing buffer and was purified with glutathione-Sepharose 4 fast flow (GE Healthcare). Clone M16, rat monoclonal antibody against the fusion protein, was established following a general protocol.
Real-Time qRT-PCR.
Real-time qRT-PCR was performed as previously described (52). The primer set ID for the assay is listed in Dataset S1. Data were analyzed by the ΔΔCt method as previously described (53, 54).
Luciferase Assay.
The pGL3 firefly reporter plasmid and Dual-Luciferase Reporter Assay System (Promega) were used according to the manufacturers’ instructions. The primer sets used are listed in Dataset S1. The probes (1-kb length) used for the assay were selected from the 5-kb upstream or 2-kb downstream region from a transcriptional start site for each gene. ZFAT protein fused with VP16-transcriptional activator-domain (AD-ZFAT) or the VP16-transcriptional activator-domain (AD) as a control was expressed in HEK293 cells in 96-well plates (Thermo Scientific). Luminescence was measured using GloMax 96 Microplate Luminometer (Promega).
ChIP-PCR Assay.
ChIP-PCR assays for Tal1, Lmo2, and Gata1 were performed on yolk sacs at E7.5 and adult kidney as a control tissue, where ZFAT is rarely expressed (21). Kifap3 was used as a hematopoiesis-unrelated control gene. As for immunoprecipitation, 100 μg of anti-ZFAT monoclonal antibody M16 or Rat IgG (SM14LE, Acris) as a control were used. End-point PCR assays were performed at 35- and 42-cycled PCR. In ChIP-qPCR assay, the total amount of ChIP DNA was normalized by M16-ChIP DNA for Kifap3 in kidney as 1.0 unit. Primer sets for the assay are listed in Dataset S1.
Immunohistochemical Examination.
Tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections (3 μm) for staining of ZFAT, TAL1, LMO2, GATA1, and CD41 were treated with 0.3% hydrogen peroxidase in methanol for 30 min. Additionally, sections for CD41-staining were antigen-retrieved by TE buffer (10 mM Tris, 1 mM EDTA, pH8.0). Sections for staining of RUNX1 or FLK-1 were treated by citrate buffer (10 mM citric acid, pH 6.0) for the inactivation of endogenous peroxidase and antigen-retrieval. Sections were applied to immunohistochemical analysis using anti-ZFAT antibody (M16), anti-TAL1 antibody (ab75739, Abcam), anti-LMO2 antibody (G-16, Santa Cruz Biotechnology), anti-GATA1 antibody (N6, Santa Cruz Biotechnology), anti-FLK-1 antibody (55B11, Cell Signaling Technology), anti-RUNX1 antibody (ab35962, Abcam), and anti-CD41 antibody (MWReg30, BD Pharmingen). Signals were detected using HISTOFINE simple stain MAX PO (Nichirei) and DAB substrate (Nichirei). Sections were counterstained with 1% methyl green (Muto Pure Chemicals) for staining of ZFAT, TAL1, LMO2, and GATA1 and with hematoxylin for staining of RUNX1, FLK-1, and CD41. Sections were examined using Biorevo BZ-9000 inverted-phase microscope (Keyence).
Statistical Analysis.
Data are presented as means ± SDs of means of triplicate samples. Statistical analyses were performed with an unpaired Student's t test. Differences at P < 0.05 are considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank T. Danno and T. Umezu for their technical assistance. This work was supported by a grant from the Genome Network Project; a Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology; and a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of the Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002494107/-/DCSupplemental.
References
- 1.Lancrin C, et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gläsker S, et al. Hemangioblastomas share protein expression with embryonal hemangioblast progenitor cell. Cancer Res. 2006;66:4167–4172. doi: 10.1158/0008-5472.CAN-05-3505. [DOI] [PubMed] [Google Scholar]
- 3.Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp Hematol. 2001;29:927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 4.Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 5.Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 6.Oberlin E, Tavian M, Blazsek I, Péault B. Blood-forming potential of vascular endothelium in the human embryo. Development. 2002;129:4147–4157. doi: 10.1242/dev.129.17.4147. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson JE, 3rd, Kelley RW, Patterson C. Mechanisms of endothelial differentiation in embryonic vasculogenesis. Arterioscler Thromb Vasc Biol. 2005;25:2246–2254. doi: 10.1161/01.ATV.0000183609.55154.44. [DOI] [PubMed] [Google Scholar]
- 8.Begley CG, et al. Molecular cloning and chromosomal localization of the murine homolog of the human helix-loop-helix gene SCL. Proc Natl Acad Sci USA. 1991;88:869–873. doi: 10.1073/pnas.88.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robb L, et al. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson NK, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 13.Warren AJ, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 14.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada Y, Pannell R, Forster A, Rabbitts TH. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci USA. 2000;97:320–324. doi: 10.1073/pnas.97.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson LJ, et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 2007;109:2389–2398. doi: 10.1182/blood-2006-02-003087. [DOI] [PubMed] [Google Scholar]
- 17.Lécuyer E, et al. Protein stability and transcription factor complex assembly determined by the SCL-LMO2 interaction. J Biol Chem. 2007;282:33649–33658. doi: 10.1074/jbc.M703939200. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez P, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokomizo T, et al. Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo. EMBO J. 2007;26:184–196. doi: 10.1038/sj.emboj.7601480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirasawa S, et al. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum Mol Genet. 2004;13:2221–2231. doi: 10.1093/hmg/ddh245. [DOI] [PubMed] [Google Scholar]
- 21.Koyanagi M, et al. ZFAT expression in B and T lymphocytes and identification of ZFAT-regulated genes. Genomics. 2008;91:451–457. doi: 10.1016/j.ygeno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto T, et al. ZFAT is an antiapoptotic molecule and critical for cell survival in MOLT-4 cells. FEBS Lett. 2009;583:568–572. doi: 10.1016/j.febslet.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 23.Comabella M, et al. Genome-wide scan of 500,000 single-nucleotide polymorphisms among responders and nonresponders to interferon beta therapy in multiple sclerosis. Arch Neurol. 2009;66:972–978. doi: 10.1001/archneurol.2009.150. [DOI] [PubMed] [Google Scholar]
- 24.Copp AJ. Death before birth: Clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 25.Carmeliet P, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 26.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 27.Lécuyer E, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 28.Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 29.Landry JR, et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood. 2008;111:3005–3014. doi: 10.1182/blood-2007-07-098830. [DOI] [PubMed] [Google Scholar]
- 30.Yokomizo T, et al. Requirement of Runx1/AML1/PEBP2alphaB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- 31.Ribatti D. Hemangioblast does exist. Leuk Res. 2008;32:850–854. doi: 10.1016/j.leukres.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Kappel A, et al. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood. 2000;96:3078–3085. [PubMed] [Google Scholar]
- 33.Bockamp EO, et al. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood. 1995;86:1502–1514. [PubMed] [Google Scholar]
- 34.Landry JR, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- 35.Landry JR, et al. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
- 36.Ohneda K, Ohmori S, Ishijima Y, Nakano M, Yamamoto M. Characterization of a functional ZBP-89 binding site that mediates Gata1 gene expression during hematopoietic development. J Biol Chem. 2009;284:30187–30199. doi: 10.1074/jbc.M109.026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zon LI, Orkin SH. Sequence of the human GATA-1 promoter. Nucleic Acids Res. 1992;20:1812. doi: 10.1093/nar/20.7.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.North TE, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 39.Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 40.Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- 41.Dzierzak E, Speck NA. Of lineage and legacy: The development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 43.Watson DK, et al. The ERGB/Fli-1 gene: Isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3:705–713. [PubMed] [Google Scholar]
- 44.Seth A, Robinson L, Thompson DM, Watson DK, Papas TS. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: Stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8:1783–1790. [PubMed] [Google Scholar]
- 45.Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 46.Pimanda JE, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, et al. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowling TK, Gilkeson GS. Regulation of Fli1 gene expression and lupus. Autoimmun Rev. 2006;5:377–382. doi: 10.1016/j.autrev.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Takeuchi F, et al. Evaluation of genetic loci influencing adult height in the Japanese population. J Hum Genet. 2009;54:749–752. doi: 10.1038/jhg.2009.99. [DOI] [PubMed] [Google Scholar]
- 50.Cho YS, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 51.Shirasawa S, et al. Rnx deficiency results in congenital central hypoventilation. Nat Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- 52.Tsunoda T, et al. Three-dimensionally specific inhibition of DNA repair-related genes by activated KRAS in colon crypt model. Neoplasia. 2010;12:397–404. doi: 10.1593/neo.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bubner B, Gase K, Baldwin IT. Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnol. 2004;4:14. doi: 10.1186/1472-6750-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.