Abstract
Dysregulation of autophagy, a cellular catabolic mechanism essential for degradation of misfolded proteins, has been implicated in multiple neurodegenerative diseases. However, the mechanisms that lead to the autophagy dysfunction are still not clear. Based on the results of a genome-wide screen, we show that reactive oxygen species (ROS) serve as common mediators upstream of the activation of the type III PI3 kinase, which is critical for the initiation of autophagy. Furthermore, ROS play an essential function in the induction of the type III PI3 kinase and autophagy in response to amyloid β peptide, the main pathogenic mediator of Alzheimer's disease (AD). However, lysosomal blockage also caused by Aβ is independent of ROS. In addition, we demonstrate that autophagy is transcriptionally down-regulated during normal aging in the human brain. Strikingly, in contrast to normal aging, we observe transcriptional up-regulation of autophagy in the brains of AD patients, suggesting that there might be a compensatory regulation of autophagy. Interestingly, we show that an AD drug and an AD drug candidate have inhibitory effects on autophagy, raising the possibility that decreasing input into the lysosomal system may help to reduce cellular stress in AD. Finally, we provide a list of candidate drug targets that can be used to safely modulate levels of autophagy without causing cell death.
Keywords: reactive oxygen species, type III PI3 kinase, neurodegeneration, signaling, transcriptional regulation
Autophagy, a lysosome-dependent catabolic process mediating turnover of cellular components, plays an important role in regulating cellular homeostasis in the nervous system (1). Even in the absence of any other risk factors, autophagy deficiency in the CNS has been shown to lead to the accumulation of protein aggregates and progressive neurodegeneration (2). Thus, autophagy has been established as an important mechanism mediating degradation of misfolded proteins in the CNS. Because accumulation of misfolded proteins is a common feature in multiple human neurodegenerative diseases, activation of autophagy has been proposed as a strategy for combating neurodegeneration (3). However, little is currently known about how defects in autophagy might be involved in specific neurodegenerative diseases. Furthermore, as induction of autophagy is frequently associated with cell death, it remains a challenge to identify molecular targets whose inhibition can specifically activate autophagy without compromising cell viability.
Pathological evidence supports the involvement of autophagy dysfunction in neurodegenerative diseases in humans. In Alzheimer's disease (AD), one of the earliest pathological changes include accumulation of autophagic vesicles (AVs) specifically within damaged neuritic processes and synaptic terminals (4). This phenotype is also observed in AD animal models and in cell-based models upon exposure to amyloid β peptide (Aβ). However, the mechanisms leading to the accumulation of AVs and the causal relationship to neurodegeneration are not yet established.
We have recently conducted a genome-wide screen using siRNA library to identify genes regulating autophagy in human cells under normal nutritional conditions (5). In this image-based screen we took advantage of the autophagy specific GFP-LC3 reporter whose translocation from the cytosol to autophagosomes can serve as a quantitative measure of autophagy. In this study, we specifically explore the mechanisms that regulate autophagy in neural cells using the hits identified in our screen. We demonstrate that reactive oxygen species (ROS) play a general function in mediation of autophagy upstream of the type III PI3 kinase and that this pathway is essential for the up-regulation of autophagy by Aβ. Interestingly, our data show that genes regulating autophagy are differentially expressed in normal aging and in AD patient brains. Finally, we identify candidate molecular targets that may be safely manipulated to modulate autophagy to treat neurodegenerative diseases.
Results
ROS Functions Upstream of the Type III PI3 Kinase to Induce Autophagy.
Genes whose knock-down induced autophagy in our screen included components of the ROS detoxification pathway GPx2 and SOD1, whose mutations are known to cause familial ALS. The screen hits also included several mitochondrial proteins, some of which are involved in oxidative respiration and electron transport (Table S1) as well as genes reported to be involved in ROS homeostasis based on a literature co-citation analysis (Table S2). This suggests a role for ROS in the induction of autophagy and a possible function for this pathway in neurodegeneration.
To determine whether ROS are sufficient to induce autophagy, we confirmed that transfection of SOD1 siRNA led to both an induction of autophagy in H4 human neuroblastoma cells, as well as to elevated levels of ROS (Fig. S1 A–C). Consistent with a causal role of ROS, treatment with an antioxidant N-acetyl-L-cysteine (NAC) significantly attenuated the induction of autophagy (Fig. S1D). Therefore, interference with normal cellular ROS homeostasis by inactivation of a gene involved in neurodegeneration is sufficient to induce autophagy in the absence of any other noxious stimuli.
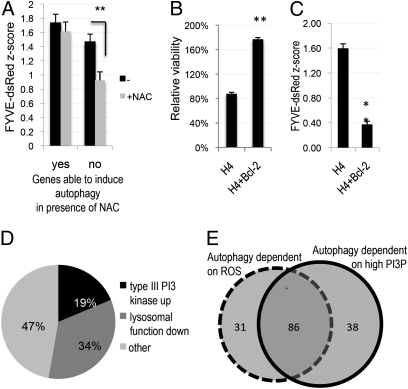
To determine whether ROS may have a general signaling function in autophagy, we compared the levels of autophagy induced by knock-down of our screen hit genes in the presence and absence of NAC. We uncovered a large group of genes (117, or 54% of all genes tested, Fig. S2A and Table S3) that when knocked-down led to translocation of GFP-LC3 to autophagosomes in the absence but not the presence of NAC, suggesting that ROS were required for the induction of autophagy. Interestingly, the presence of NAC also reduced the vesicular accumulation of FYVE-dsRed, a reporter protein for PtdIns3P, induced by the knock-down of these genes (Fig. 1A). As PtdIns3P is the product of type III PI3 kinase, this result suggests that ROS may serve an important general signaling function in the early steps of autophagic pathway mediated by the type III PI3 kinase.
Fig. 1.
Suppression of ROS and expression of Bcl-2 lead to decrease in the levels of autophagy and the type III PI3 kinase activity. (A) Quantification of average type III PI3 kinase activity following knock-down of genes able (yes) and unable (no) to induce autophagy in the presence of NAC. H4 FYVE-dsRed cells were transfected with hit siRNA for 72 h, followed by fixation and imaging on a high-throughput fluorescent microscope at 10× magnification. Average z-scores as compared with nontargeting siRNA are shown. (B) Comparison of the relative average viability of WT or pBabe-Bcl-2–expressing H4 cells transfected with hit gene siRNAs for 72 h. (C) Comparison of average type III PI3 kinase activity in WT or pBabe-Bcl-2–expressing H4 FYVE-dsRed cells following hit siRNA transfection for 72 h. (D) Subdivision of hits whose knock-down was able to induce autophagy under conditions of low PtdIns3P into functional categories based on their ability to up-regulate type III PI3 kinase activity or to alter lysosomal processing. (E) Subdivision of genes whose knock-down led to the induction of autophagy into functional categories based on their dependence on ROS and elevated levels of PtdIns3P (PI3P). *P < 0.05, **P < 0.01 based on two-tailed t test with equal variance. All error bars indicate SEM.
Knock-down any of the remaining 98 (46%, Table S4) genes was able to induce accumulation of GFP-LC3 in the presence of NAC, suggesting that, in these cases, autophagy can be induced independently of ROS. Knock-down of these genes was also able to induce comparable average levels of vesicular FYVE-dsRed in the presence and absence of NAC (Fig. 1A). Thus, knock-down of this group of genes led to induction of the type III PI3 kinase through a mechanism independent of ROS.
Because our data indicated that the type III PI3 kinase plays an especially crucial role in the mediation and regulation of autophagy downstream of ROS, we performed an additional screen to determine how our hit genes are influenced by the type III PI3 kinase activity. In addition to its prominent function in regulation of apoptotic cell death, Bcl-2 may negatively regulate autophagy through its interaction with beclin 1 and consequent inhibition of the type III PI3 kinase activity (6). Consistently, we observed a significant increase in cell viability and a decrease in levels of PtdIns3P following knock-down of the hit genes in H4 cells expressing Bcl-2 as compared with WT controls (Fig. 1 B and C and Fig. S2B). Knock-down of 124 (58%) of the 215 tested hit genes was unable to induce accumulation of vesicular GFP-LC3 in cells overexpressing Bcl-2, confirming that, in the majority of cases, up-regulation of the type III PI3 kinase activity is necessary for the induction of autophagy (Fig. S2C and Table S5). On the other hand, knock-down of the remaining 91 (42%) genes was able to induce translocation of GFP-LC3 to autophagosomes in the presence of Bcl-2 (Table S6). For 17 (19%) of these genes, induction of autophagy was accompanied by an increase in type III PI3 kinase activity, suggesting additional mechanisms that regulate production of PtdIns3P downstream of Bcl-2 (Fig. 1D). However, knock-down of the remaining 74 genes was able to induce autophagy without additional activation of the type III PI3 kinase. Knock-down of 31 (34%) of these genes led to the expansion of the lysosomal compartment as assessed by the accumulation of Lamp-1-RFP marker, indicating that, in these cases, a block in lysosomal degradation may contribute to the apparent increase in autophagy (Fig. 1D). No changes in the lysosomal function were observed for the remaining 43 (47%) genes. These genes may function either downstream or independently of the type III PI3 kinase, suggesting that the apparent inhibitory effect of Bcl-2 on the type III PI3 kinase is not always incompatible with the induction of autophagy, which, in these cases, can occur without an increase in PtdIns3P levels.
Interestingly, we observed a substantial overlap between genes whose knock-down was unable to induce autophagy in the presence of NAC or Bcl-2 (86, 40% of all genes tested; Fig. 1E). The overlapping genes include the majority (11 of 18) of the mitochondrial genes, confirming that mitochondria-generated ROS induce autophagy by positively regulating the type III PI3 kinase. Because overexpression of Bcl-2 has been shown to reduce stress induced generation of ROS (7), our data imply that, in addition to directly binding and suppressing the activity of Beclin1 (6), Bcl-2 may be able to contribute to the regulation of the type III PI3 kinase and autophagy through its ability to attenuate generation of ROS.
Differential Expression of Autophagy Regulators in AD Brain Samples.
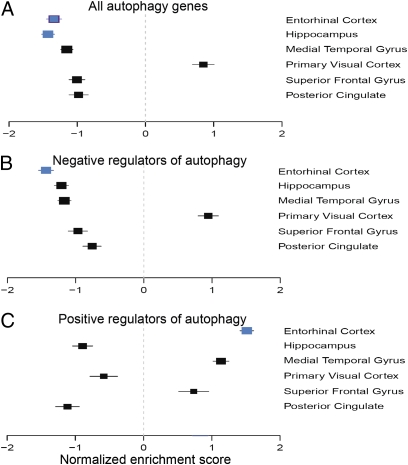
Accumulation of both ROS and AVs are early features in AD (4). To determine whether we could detect changes in the expression of genes involved in regulation of autophagy in this disease, we analyzed expression of the autophagy screen hit genes in six brain regions of 34 patients with AD and 14 age-matched normal controls (8). We observed an overall significant underexpression of the hit genes in AD patient samples compared with controls specifically in the hippocampus and entorhinal cortex, the brain regions most affected by the disease (Fig. 2A). Consistent trends were observed in other brain regions affected by AD (superior frontal gyrus, posterior cingulate, and medial temporal gyrus), but did not reach statistical significance. Notably, in the visual cortex, a brain region relatively resistant to AD pathology, these changes were absent. Further subdivision of the hit genes revealed that in the entorhinal cortex negative regulators of autophagy flux were specifically negatively enriched (Fig. 2B). A similar trend was also observed in other brain areas affected by AD. Conversely, positive regulators of autophagy were positively enriched in the entorhinal cortex (Fig. 2C). Such differential expression patterns of autophagy regulators suggest transcriptional up-regulation of autophagy in AD brains.
Fig. 2.
Differential gene expression leads to transcriptional up-regulation of autophagy in Alzheimer's disease. Forrest plots of NES estimates with SD for the screen hit gene sets are shown. (A) GSEA analysis of overall screen hit gene expression in different regions of AD brain as compared with unaffected age-matched controls. (B and C) GSEA analysis of hit genes determined to function as negative (B) or positive (C) regulators of autophagy flux. Blue squares indicate enrichment signals with P ≤ 0.05 in an individual comparison. The size of a square is inversely proportional to the respective SD.
KD of Mitochondrial Complex IV Gene Cox5a Leads to ROS-Induced Autophagy.
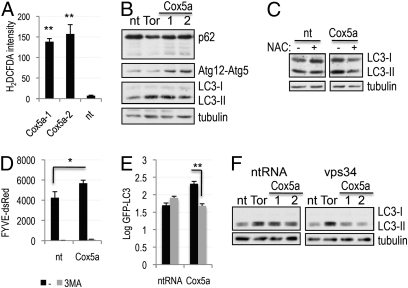
The screen hits included Cox5a, a nuclear-encoded component of the mitochondrial electron transport complex IV (Table S1). Inactivation of this complex has been suggested to occur in AD and to contribute to accumulation of ROS (9). We determined that inhibition of complex IV by knock-down of Cox5a led to the accumulation of ROS (Fig. 3A and Fig. S3 A and B) and induction of autophagy (Fig. 3B and Fig. S3C). Consistent with the requirement for ROS, induction of autophagy following knock-down of Cox5a was attenuated in the presence of NAC (Fig. 3C).
Fig. 3.
Knock-down of the mitochondrial gene Cox5a leads to generation of ROS and induction of autophagy. (A) Induction of ROS in H4 cells transfected with two independent siRNAs against Cox5a or nontargeting siRNA for 72 h. Cells were stained in 25 mM carboxy-H2DCFDA and Hoechst and imaged at 40×. (B) Induction of autophagy in cells transfected with siRNAs against Cox5a or controls for 72 h; autophagy levels were assessed with antibodies against p62, Atg5 (Atg12-Atg5 complex is shown), and LC3. (C) Induction of autophagy by Cox5a knock-down is suppressed in the presence of the antioxidant NAC. Cells were prepared as in B, except that, where indicated, 10 mM NAC was added to the culture media 24 h after transfection. (D) Knock-down of Cox5a leads to the accumulation of PtdIns3P in a manner dependent on the function of the type III PI3 kinase. Quantification of FYVE-dsRed reporter is shown. Cells were transfected as in B; where indicated, the type III PI3 kinase inhibitor 3MA (10 mM) was added for 8 h before fixation and imaging. (E) Induction of autophagy following Cox5a knock-down depends on the function of the type III PI3 kinase. GFP-LC3 H4 cells were prepared and imaged as in D. (F) Levels of autophagy induced following knock-down of Cox5a were assessed in H4 cells transfected with control siRNA (nt, nontargeting; Tor, mTOR) or siRNA against Vps34 for 72 h by Western blot. *P < 0.05, **P < 0.01 based on two-tailed t test with equal variance. All error bars indicate SEM.
Our screen data indicated that ROS is regulating autophagy by activating the type III PI3 kinase. In agreement, knock-down of Cox5a led to the increase in the levels of PtdIns3P (Fig. 3D). Levels of PtdIns3P were suppressed in the presence of the type III PI3 kinase inhibitor, 3MA, confirming the involvement of this kinase. In addition, 3MA, as well as the knock-down of the catalytic subunit of the kinase, Vps34, attenuated the induction of autophagy in response to the loss of Cox5a (Fig. 3 E and F and Fig. S3D). Together, these data indicate that knock-down of Cox5a leads to the induction of autophagy by increasing ROS dependent type III PI3 kinase activity.
ROS Mediate Autophagy in Response to Amyloid β.
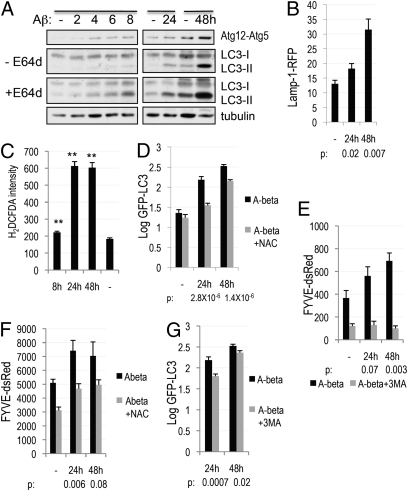
Aβ, the main pathogenic factor in AD, has been proposed to cause mitochondrial damage leading to the generation of ROS (9). We hypothesized that induction of autophagy by Aβ may also be mediated by ROS. We observed increased levels of autophagy following treatment of H4 cells with Aβ (Fig. 4A and Fig. S4A). To determine whether this was due to an increase in the initiation of autophagy or to a block in lysosomal degradation, we compared the accumulation of LC3-II following Aβ treatment in the absence and presence of lysosomal protease inhibitor, E64d (Fig. 4A). Up to 8 h after treatment the accumulation of LC3-II could be observed only in the presence of E64d. At 48 h after the addition of Aβ, the increased levels of LC3-II were observed even without E64d but were further increased in the presence of E64d. In addition, we observed increased conjugation of Atg12-Atg5 starting 4 h after Aβ treatment. Together these data suggest increased initiation of autophagy in response to Aβ. However, as previously reported (4), lysosomal proteolysis also appeared to be inhibited, as demonstrated by the expansion of the lysosomal compartment (Fig. 4B) and altered mobility of lysosomal proteins, cathepsin D (CtsD) and Lamp-2 on a Western blot, starting at 24 h after Aβ treatment (Fig. S4B, Left). Therefore, Aβ regulates the apparent levels of cellular autophagy by both enhancing initiation of autophagy and decreasing the rate of autophagosome clearance due to lysosomal inhibition.
Fig. 4.
Amyloid β up-regulates autophagy by inducing accumulation or ROS and the type III PI3 kinase activity. (A) Comparison of levels of LC3-II accumulation in the presence or absence of 10 μM E64d following treatment of H4 cells with 5 μM Aβ. (B) Expansion of the lysosomal compartment following Aβ treatment based on the Lamp-1-RFP lysosomal reporter. Cells were treated as in A, then fixed and imaged at 10×. (C) Treatment with Aβ leads to generation of ROS. Cells were stained in 25 mM carboxy-H2DCFDA and imaged at 40× magnification. (D) Induction of autophagy by Aβ is suppressed in the presence of the antioxidant NAC. GFP-LC3 cells were prepared as in B except that, where indicated, 10 mM NAC was added. (E) Aβ induces accumulation of PtdIns3P. FYVE-dsRed cells were prepared and imaged as in B; where indicated, the type III PI3 kinase inhibitor 3MA (10mM) was added for 8 h before fixation. (F) Induction of the type III PI3 kinase activity by Aβ is suppressed in the presence of antioxidant. Cells were prepared as in D. (G) Induction of autophagy by Aβ is dependent on the type III PI3 kinase activity. H4 GFP-LC3 cells were treated and imaged as in E. **P < 0.01 based on two-tailed t test with equal variance. All error bars represent SEM.
To determine whether ROS are involved in the induction of autophagy by Aβ, we first checked that Aβ induced ROS (Figs. 4C and S4C). Confirming the essential function of ROS, NAC attenuated induction of autophagy in response to Aβ (Fig. 4D). Interestingly, NAC was unable to suppress the Aβ-induced changes in lysosomal proteins (Fig. S4B, Right), suggesting that lysosomal damage may be mediated by a different mechanism.
We then investigated the involvement of the type III PI3 kinase in the induction of autophagy by Aβ. We observed increased accumulation of PtdIns3P (Fig. 4E), which was suppressed in the presence of 3MA (Fig. 4E), confirming the involvement of the type III PI3 kinase. In agreement with a causal role of ROS, accumulation of PtdIns3P was suppressed in the presence of NAC (Fig. 4F). Finally, treatment with 3MA (Fig. 4G) or knock-down of Vps34 (Fig. S4D) was able to attenuate induction of autophagy in response to Aβ. On the other hand, an Aβ induced change in Lamp-2 mobility was not affected (Fig. S4D). Therefore, Aβ influences autophagy by two distinct mechanisms: by increasing initiation, and by blocking lysosomal degradation. The initiation of autophagy is an earlier event and is dependent on the accumulation of ROS and up-regulation of the type III PI3 kinase activity. The blockade of lysosomal degradation is mediated by a ROS and type III PI3 kinase independent mechanism.
Modulation of Autophagy as Treatment Against Neurodegeneration.
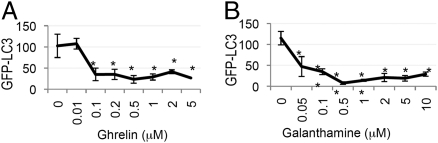
To explore the feasibility of modulating autophagy as treatment for neurodegenerative diseases, we compared our screen hits with the known neurodegenerative disease drug targets. This revealed that several of the genes identified in the screen are targets of drugs currently in use or in clinical trials against neurological and neurodegenerative diseases, including AD (Table S7). Consistent with the predicted negative effect of the growth hormone secretagogue receptor activity on the levels of autophagy, treatment of H4 cells with its ligand, Ghrelin, a candidate drug for AD, led to a dose-dependent suppression of autophagy (Fig. 5A). Similarly, galanthamine hydrochloride, an agonist of the nicotinic acetylcholine receptor (CHRND) and an AD drug, led to the suppression of autophagy levels (Fig. 5B). Neither drug significantly affected cell viability (Fig. S5A and B). Therefore, these drugs may be able to partially suppress up-regulation of autophagy.
Fig. 5.
Drugs against AD suppress autophagy. H4 GFP-LC3 cells were treated with indicated concentrations of GSHR ligand Ghrelin (A) or CHRND agonist Galanthamine (B) for 24 h and imaged at 10×. *P < 0.05, ** P < 0.01 based on two-tailed t test with equal variance. All error bars represent SEM.
Candidate Autophagy Drug Targets Against Neurodegeneration.
To modulate autophagy as a treatment against neurodegenerative diseases, we need to identify novel molecular drug targets that can up-regulate autophagy flux without affecting cell viability. As ROS can be the cause of cellular damage, these genes should up-regulate autophagy in a ROS-independent manner. To this end, we analyzed our screen data and identified 26 such candidate genes (Table 1 and Table S8). We propose these genes as candidate inhibitory drug targets against neurodegenerative diseases where up-regulation of autophagy is beneficial.
Table 1.
Candidate inhibitory drug targets for modulation of autophagy in neurodegenerative disease
| Gene symbol | Gene ID | Gene name |
| Drug targets for up-regulation of autophagy | ||
| PPFIA4 | 8497 | Protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| ADMR | 11318 | Adrenomedullin receptor |
| PTGER2 | 5732 | Prostaglandin E receptor 2 (subtype EP2), 53kDa |
| EP300 | 2033 | E1A binding protein p300 |
| CXCL12 | 6387 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) |
| NCR3 | 259197 | Natural cytotoxicity triggering receptor 3 |
| GABBR2 | 9568 | Gamma-aminobutyric acid (GABA) B receptor, 2 |
| GJA4 | 2701 | Gap junction protein, alpha 4, 37kDa |
| KRT18 | 3875 | Keratin 18 |
| SDHB | 6390 | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| GTF2IRD2 | 84163 | GTF2I repeat domain containing 2 |
| GNRH2 | 2797 | Gonadotropin-releasing hormone 2 |
| HIVEP2 | 3097 | HIV type I enhancer binding protein 2 |
| USP19 | 10869 | Ubiquitin specific peptidase 19 |
| EPHA6 | 203806 | EPH receptor A6 |
| TACR2 | 6865 | Tachykinin receptor 2 |
| MYL3 | 4634 | Myosin, light chain 3, alkali; ventricular, skeletal, slow |
| CAPN1 | 823 | Calpain 1, (mu/I) large subunit |
| SORCS2 | 57537 | Sortilin-related VPS10 domain containing receptor 2 |
| BAIAP2 | 10458 | BAI1-associated protein 2 |
| THBS2 | 7058 | Thrombospondin 2 |
| PCGF1 | 84759 | Polycomb group ring finger 1 |
| MMP10 | 4319 | Matrix metallopeptidase 10 (stromelysin 2) |
| CHRND | 1144 | Cholinergic receptor, nicotinic, delta |
| CDKN2D | 1032 | Cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| FBXL20 | 84961 | F-box and leucine-rich repeat protein 20 |
| Drug targets for down-regulation of autophagy | ||
| ATG7 | 10533 | ATG7 autophagy related 7 homolog (S. cerevisiae) |
| MEGF10 | 84466 | Multiple EGF-like-domains 10 |
| KIF5C | 3800 | Kinesin family member 5C |
| SMYD3 | 64754 | SET and MYND domain containing 3 |
| LOC285647 | 285647 | Suppressor of defective silencing 3 pseudogene |
| STIM1 | 6786 | Stromal interaction molecule 1 |
| GAB1 | 2549 | GRB2-associated binding protein 1 |
| GPR18 | 2841 | G-protein–coupled receptor 18 |
| CATSPER4 | 378807 | Cation channel, sperm associated 4 |
In addition, our screen included nine genes whose inhibition down-regulated autophagy without affecting cell viability and which are not known to affect ROS homeostasis (Table 1 and Table S8). These genes represent potential drug candidates against diseases such as late-stage AD, in which reducing autophagic input might be advantageous.
Transcriptional Regulation of Autophagy in Normal Brain Aging.
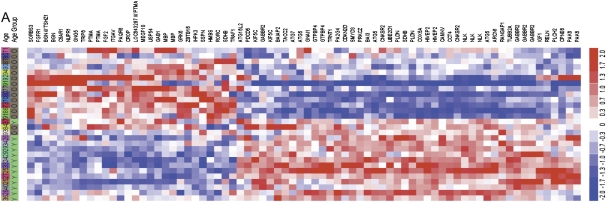
To determine whether the regulation of autophagy may have wider implications in normal aging of the human brain, we analyzed expression of the autophagy screen hit genes in a set of younger versus older human brain samples (10). We observed differential expression of a large subset of genes, including a group of 32 genes significantly (P < 0.05) up-regulated and 46 down-regulated with age (Fig. 6A and Fig. S6 A and B and Table S9). Gene ontology biological process analysis revealed that the age up-regulated group was highly enriched in genes involved in mediation and regulation of the MAP kinase pathway (P = 1.6 × 10−4). An increase in the activity of MAP kinase pathway was predicted by our previous analysis to lead to the suppression of autophagy (5). Conversely, expression of the key autophagy genes, such as Atg5 and Atg7, was down-regulated in aging. This is consistent with our previous data demonstrating transcriptional down-regulation of beclin 1, in normal human brain aging (11). Together, this suggests, that unlike AD, the normal aging process may lead to transcriptional down-regulation of autophagy.
Fig. 6.
Expression of autophagy screen hit genes in normal human aging. Clustering analysis (dChip) of mRNA expression levels of select autophagy hit genes in younger (≤40 y old) versus older (≥70 y old) human brain samples, based on (i) minimum 1.2-fold change between the average expression, and (ii) P value <0.05 using unpaired t test.
To further define the biological processes affected by down-regulation of autophagy in aging, we used gene ontology canonical pathway analysis. It revealed a significant enrichment in the “Axon guidance” (P = 0.0009) and “Regulation of actin cytoskeleton” (P = 0.038) pathways, suggesting a connection between regulation of autophagy, axon guidance and actin dynamics. Construction of protein–protein interaction networks anchored by the hit genes belonging to these pathways (12, 13) revealed two related networks encompassing, respectively, 27 (11%) and 61 (26%) of the hit genes (Fig. S6 C and D). Importantly, both networks directly connect to the known autophagy machinery through the interaction of the RIP kinase (RIPK1) and PKCζ (PRKCZ) with p62/sequestrosome (SQSTM1). In addition, syndecan 2 (SDC2), a part of the “Regulation of actin cytoskeleton” network, interacts with syntenin, a binding partner of ULK1, the human ortholog of yeast Atg1 (14). ULK1 is known to play a role in the regulation of endocytic processes involved in axon guidance (15) and to promote synapse formation in Drosophila (16). These data suggest that some of the molecular networks involved in the regulation of autophagy are closely connected to those regulating endocytosis, actin dynamics, and neuronal axon guidance, and that autophagy may play a wider role in the development and maintenance of neuronal function.
Discussion
In this study, we demonstrate that the type III PI3 kinase plays a fundamental role in the regulation of autophagy and that ROS function as general mediators of autophagy induction upstream of this kinase. This pathway has an essential function in the initiation of autophagy in response to mitochondrial damage following exposure to Aβ, the main pathogen of AD. At the same time, Aβ is able to slow down autophagic processing through ROS independent inhibition of lysosomal degradation. In addition, our analysis of expression of the autophagy screen hits suggests that autophagy is differentially regulated at the transcriptional level in normal human aging and in AD, with overall levels decreased in normal aging but elevated in AD.
ROS Are General Mediators of Autophagy Upstream of Type III PI3 Kinase.
Although ROS have been implicated in the regulation of starvation-induced autophagy (17), their general significance in mediation of autophagy has not been appreciated. Furthermore, the biochemical step at which ROS intersect with the autophagic pathway has remained unknown. Our data demonstrate that up-regulation of autophagy and the type III PI3 kinase activity by the knock-down of a large fraction of our autophagy screen hit genes can be inhibited in the presence of the antioxidant NAC. Thus, ROS might serve a previously unappreciated role as a common signaling molecule essential for the regulation of the basal levels of autophagy upstream of the type III PI3 kinase. Although ROS have been implicated in regulation of different cellular events, such as cell death, such extensive involvement in a specific signaling pathway has not been appreciated before. Because an increase in the levels of ROS is a frequent consequence of the accumulation of misfolded proteins and expired organelles such as old mitochondria, we propose that ROS may serve as an important intracellular signal for the homeostatic activation of autophagy under basal physiological conditions as well as in neurodegenerative diseases including AD.
Both excessive generation of ROS and accumulation of AVs are some of the earliest hallmarks of AD (4, 18, 19). Until now, however, these phenotypes were not considered to be related. Although further confirmation in primary neurons will be necessary, our data suggest a mechanistic link between the generation of ROS and the early accumulation of AVs observed in AD. In addition, they point to the previously unappreciated involvement of the type III PI3 kinase and the accumulation of PtdIns3P in the etiology of AD.
Differential Expression of Autophagy Regulators in Normal Aging and in AD.
Our gene expression data suggest that autophagy is also differentially regulated at the transcriptional level in normal human brain aging versus in AD. Because autophagy is known to play a protective role against onset of neurodegeneration in animal models (2, 3, 20, 21), its down-regulation in normal aging could contribute to the observed age-dependent predisposition to development of chronic neurodegenerative diseases. In addition, the extensive overlap of the autophagy screen hits with regulatory networks mediating actin dynamics and endocytic processes involved in axonal guidance suggests that autophagy may directly contribute to the regulation of growth and maintenance of neuronal processes. Therefore, progressive transcriptional down-regulation of autophagy in normal human brain aging may also contribute to age related decline in synaptic plasticity, memory, and cognitive function.
Conversely, our data indicate that autophagy is specifically up-regulated in AD, due both to ROS-dependent activation of the type III PI3 kinase as well as at the transcriptional level. We hypothesize that this may represent, respectively, an acute and a long-term attempt by the affected neuronal cells to rid themselves of the harmful effects of Aβ exposure, such as accumulation of defective mitochondria and protein aggregates.
Surprisingly, our study also demonstrated that drugs in clinical trials or used against AD lead to the inhibition of autophagy. Because increasing autophagy is predicted to be beneficial for the treatment of neurodegenerative diseases, one may wonder if this inhibition may be an unintended side effect, as autophagy was not a parameter tested when developing these drugs. Alternatively, inhibition of autophagy could contribute to the efficacy of those drugs. Our data indicate that treatment with Aβ leads to both an increase in the initiation of autophagy as well as a decrease in the lysosomal degradation. We hypothesize that as the downstream degradative pathway is partially blocked, increasing the autophagic input may further exacerbate the stress on the system. In this case, reducing the initiation of autophagy might be beneficial by decreasing input into the lysosomal system, thus allowing for gradual turnover of the accumulated material. Therefore, although up-regulation of autophagy might be beneficial as a preventive measure during normal aging, as well as to combat some neurodegenerative diseases including early stages of AD (21, 22), increasing autophagic input may backfire if applied to later stages of AD, in which the lysosomal blockage is preeminent.
Dissociation of Autophagy from Cell Death for Clinical Benefit.
Another important consideration in developing autophagy-based therapies is the ability to change levels of autophagy without inducing neuronal cell death. As a first important step toward this goal, we provide a list of potential molecular drug targets for the modulation of autophagy in neurodegenerative diseases. Interestingly, this list includes calpain 1 which, as we have previously shown, can be inhibited by fluspirilene, an inhibitor of intracellular Ca2+ flux, to induce autophagy by reducing cleavage of full-length Atg5 and increasing conjugated Atg12-Atg5 without inducing cell death (23). We propose that by developing inhibitors against these gene targets, autophagy can be safely manipulated without harming neuronal cells, thus providing potential novel therapies against neurodegenerative diseases.
Materials and Methods
Cell Lines and Culture Conditions.
H4 human neuroblastoma cells were cultured and treated as described elsewhere (5). GFP-LC3, FYVE-dsRed, GFP-LC3 pSRP-Beclin1 knock-down (11) and Lamp-1-RFP (5) H4 cells have been previously described.
For antioxidant assay, cells were treated with 2.5 mM NAC (Sigma). Aβ was prepared using a modified method from ref. 24.
Imaging and Image Quantification.
Cells were imaged as previously described (5) on a CellWoRx microscope (Applied Precision) at ×10 magnification. Images were quantified using VHSscan and VHSview software (Cellomics).
Quantification of Cellular ROS Levels.
ROS were quantified using Image-iT LIVE Green ROS Detection Kit for microscopy (Molecular Probes) according to manufacturers instructions. Images were acquired on a Nikon Eclipse E800 microscope at ×40 magnification and quantified using CellProfiler software (25).
Bioinformatics Analysis.
Gene set enrichment analysis.
Associations of autophagy gene sets with AD were evaluated using GSEA (26) and gene expression data from laser-capture microdissected non-tangle-bearing neurons of 34 late-onset AD-afflicted individuals with a mean age at death of 79.9 ± 6.9 y and 14 neurologically normal healthy elderly controls (27) (GEO accession number GSE5281).
Analysis of hit gene expression in aging.
Analysis was based on Affymetrix HG-U133_Plus_2 microarray data of young (≤40 y old) and old (≥70 y old) human brain samples (10). Array normalization, expression value calculation and clustering analysis were performed using the dChip software (28) (www.dchip.org).
Supplementary Material
Acknowledgments
We thank the members of the Yuan laboratory and Institute of Chemistry and Cell Biology (ICCB)-Longwood for help during this work. This work was supported in part by National Institutes of Health Grants R37 AG012859 (to J.Y.), PO1 AG027916 (to B.A.Y. and J.Y.), AI062773 and DK043351 (to R.J.X.), as well as R01 NS064155, R21 NS060227, P01 NS058793, and an RJG Foundation grant (to C.R.S.). A.N. is supported by the Crohn's and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009485107/-/DCSupplemental.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 3.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 4.Nixon RA, et al. Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 5.Lipinski MM, et al. Multiple mTORC1 independent signaling pathways regulate autophagy through type III PI3 kinase under normal nutritional conditions. Dev Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Kane DJ, et al. Bcl-2 inhibition of neural death: Decreased generation of reactive oxygen species. Science. 1993;262:1274–1277. doi: 10.1126/science.8235659. [DOI] [PubMed] [Google Scholar]
- 8.Liang WS, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takuma K, et al. ABAD enhances Abeta-induced cell stress via mitochondrial dysfunction. FASEB J. 2005;19:597–598. doi: 10.1096/fj.04-2582fje. [DOI] [PubMed] [Google Scholar]
- 10.Loerch PM, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 12.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lustbader JW, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 19.Reddy PH, Beal MF. Amyloid beta, mitochondrial dysfunction and synaptic damage: Implications for cognitive decline in aging and Alzheimer's disease. Trends Mol Med. 2008;14:45–53. doi: 10.1016/j.molmed.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 21.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy. 2009;5:502–510. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 23.Xia HG, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–66. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh DM, et al. Amyloid beta-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 25.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: A reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.