Abstract
Mitochondria are key regulators of cell viability and provide essential functions that protect against neurodegenerative disease. To develop a model for mitochondrial-dependent neurodegeneration in Caenorhabditis elegans, we used RNA interference (RNAi) and genetic ablation to knock down expression of enzymes in the Coenzyme Q (CoQ) biosynthetic pathway. CoQ is a required component of the ATP-producing electron transport chain in mitochondria. We found that reduced levels of CoQ result in a progressive uncoordinated (Unc) phenotype that is correlated with the appearance of degenerating GABA neurons. Both the Unc and degenerative phenotypes emerge during late larval development and progress in adults. Neuron classes in motor and sensory circuits that use other neurotransmitters (dopamine, acetylcholine, glutamate, serotonin) and body muscle cells were less sensitive to CoQ depletion. Our results indicate that the mechanism of GABA neuron degeneration is calcium-dependent and requires activation of the apoptotic gene, ced-4 (Apaf-1). A molecular cascade involving mitochondrial-initiated cell death is also consistent with our finding that GABA neuron degeneration requires the mitochondrial fission gene, drp-1. We conclude that the cell selectivity and developmental progression of CoQ deficiency in C. elegans indicate that this model may be useful for delineating the role of mitochondrial dysfunction in neurodegenerative disease.
Keywords: cell death, disease, mitochondria, neurodegeneration, necrosis
Mitochondrial dysfunction, selective neuronal vulnerability, and progressive onset are unifying features of major human neurodegenerative diseases including amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD) (1). Genetic mutations linked to these diseases have been shown to disrupt the electron transport chain (ETC) (2), a mitochondrial pathway that accounts for the majority of cellular energy production (3). Chemical inhibitors of the ETC can also produce symptoms reminiscent of neurodegenerative diseases (4). Despite the broad disruption of mitochondrial function in these cases, specific neuron classes are preferentially targeted. The molecular basis for this selectivity is largely unknown (5).
The key role of mitochondrial fragmentation in both necrosis and apoptosis (6) parallels emerging evidence that excessive mitochondrial fission may contribute to neurodegenerative disease (7). Inhibition of the fission-promoting dynamin GTPase, Drp-1, for example, blocks mitochondrial fragmentation and death induced by overexpression of the mutant Huntingtin protein (mtHtt) in cultured cells (8). The progressive onset of these diseases is correlated with an age-dependent decline in mitochondrial function, but the mechanistic link between diminished mitochondrial activity and neurodegeneration is poorly understood.
Although the cell death pathways of necrosis and apoptosis feature distinct biochemical and morphological components, recent studies have revealed “aponecrotic” cell killing pathways that include elements of both mechanisms (9). Additionally, apoptosis and necrosis share common molecular triggers, such as oxidative stress and energy depletion, two hallmarks of mitochondrial dysfunction (10, 11). These findings suggest that apoptotic and necrotic pathways may represent related cell death responses and can be activated by mitochondrial impairment.
Coenzyme Q (CoQ) transfers electrons from complexes I and II to complex III in the mitochondrial ETC and fulfills a critical role in mitochondrial ATP production (12). CoQ also limits the production of reactive oxygen species that can damage cellular processes. Human patients with CoQ deficiency exhibit diverse symptoms ranging in severity from infantile multiorgan disease (13, 14) to discrete late onset cerebellar ataxia (15, 16). Although the essential role of CoQ in mitochondrial energy production and cell survival are likely to account for these deficits, the molecular basis for this heterogeneity is unclear, as are the specific cellular pathologies arising from CoQ deficiency (17).
We developed a model of CoQ deficiency for the purpose of delineating the molecular events that contribute to cell death arising from mitochondrial dysfunction. Because of its short life span, anatomic simplicity, and genetic tractability, the nematode Caenorhabditis elegans is particularly amenable to this approach. We used RNA interference (RNAi) and genetic ablation to knock down genes in the CoQ synthesis pathway (coq-1, coq-2, coq-3) to mimic CoQ deficiency. These treatments result in age-dependent loss of motor coordination that is correlated with progressive degeneration of GABA neurons. Our results show that CoQ deficiency in C. elegans results in a selective cell death pathway that includes features of both apoptotic and necrotic processes and, therefore, suggest that this experimental model may be useful for delineating the mechanism of neuron cell death in human CoQ deficiency and in related neurodegenerative diseases that are also linked to defective mitochondrial function.
Results
Knockdown of coq-1 Results in Age-Dependent Loss of Coordinated Movement.
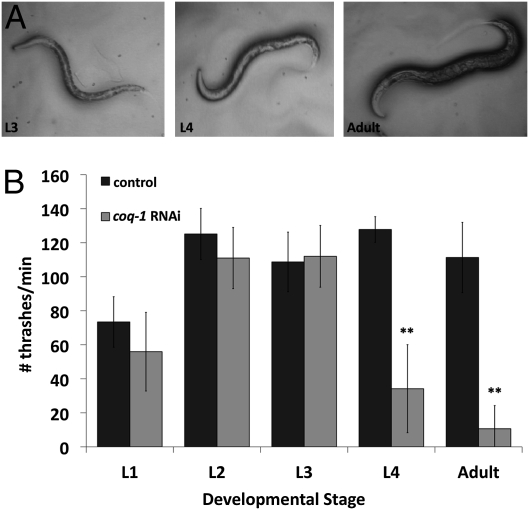
COQ-1 catalyzes the first step in Coenzyme Q synthesis, the assembly of the lipophillic polyisoprenoid tail (18). RNAi knockdown of coq-1 is reported to induce uncoordinated (Unc) and Egg-laying defective (Egl) phenotypes, but the mechanism of these effects has not been studied (19). We replicated this experiment, using the RNAi “feeding” method to expose an RNAi-hypersensitive strain, eri-1(mg366) (20), to bacteria expressing coq-1 double-stranded RNA (dsRNA). Loss of motor coordination first appeared at the L4 larval stage as a kink in the normal sinusoidal wave that drives locomotion. Movement then gradually declined in adults, often culminating in paralysis (Fig. 1A and Movie S1). Quantification of movement loss with a swimming assay (21) verified the developmental progression of the Unc phenotype (Fig. 1B).
Fig. 1.
RNAi knockdown of coq-1 results in progressive loss of motor coordination. (A) Examples of the abnormal postures adopted by L4 larvae and adults after exposure to coq-1 RNAi. (B) Swimming assay quantifying movement defects (bars represent avg ± SD, **P < 0.005, n = 10).
RNAi or Genetic Depletion of CoQ Induces Age-Dependent Degeneration of GABA Neurons.
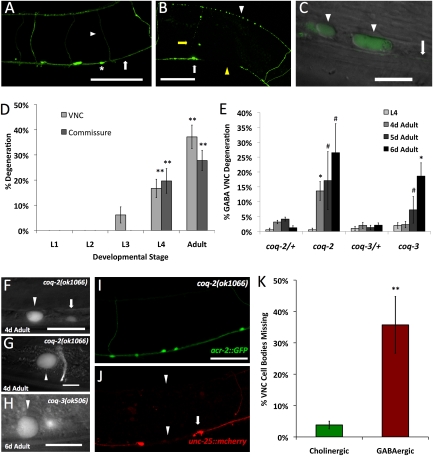
Observation of GABA motor neurons in the ventral nerve cord, labeled with the unc-25::GFP reporter (Fig. 2A), suggested a possible explanation for the loss in motor coordination. These GABA neurons degenerate in coq-1 knockdown animals, and cell bodies, viewed under differential interference contrast (DIC), demonstrated the swollen appearance characteristic of necrotic cell death (Fig. 2C). GABAergic processes in the ventral nerve cord, circumferential commissures and GABAergic processes in the dorsal cord appear discontinuous with apparent breaks (Fig. 2B). These morphological defects first appeared in late larval development and progressed as animals aged (Fig. 2D), thereby mirroring the age-dependent pattern of the Unc phenotype.
Fig. 2.
CoQ depletion results in GABA neuron degeneration. (A) Wild-type adult GABA neuron cell bodies (asterisks), processes in the ventral nerve cord (VNC) (arrow), and dorsally projecting commissures (arrowhead) labeled with unc-25::GFP. (B) Adult coq-1 RNAi-treated worm showing signs of degeneration (white arrowhead, dorsal process beading; white arrow, degenerating GABA cell body; yellow arrow, degenerating commissure; yellow arrowhead, VNC process break). (Scale bars: 50 μm.) (C) unc-25::GFP-labeled GABA neurons appear swollen in coq-1 RNAi-treated adults. Shown is the ventral view of GABAergic cell bodies (arrowheads) and VNC (arrow). (Scale bar: 10 μm.) (D) Quantification of progressive degeneration of GABA neuron VNC processes and commissures (n ≥ 3, error bars = SEM; **P < 0.001 vs. empty vector-treated animals). No degeneration was observed in empty vector-treated animals. (E) Progressive GABA neuron degeneration in coq-2(ok1066) and coq-3(ok506) mutants. GABA neuron degeneration was not observed in L1–L3 larval stages but was detectable in L4 larvae and pronounced in adults (day, d). #P < 0.04 and *P < 0.001 vs. heterozygous controls, n ≥ 20 for each developmental stage. (F) Enlarged cell size of degenerating GABA neuron (arrowhead) compared with an unaffected GABA neuron cell body (arrow). (F–H) Swollen D-class (F and H) and RME GABA neurons (G) in coq-2(ok1066) (F and G) and coq-3(ok506) (H) mutants visualized with unc-25::GFP. (Scale bars: 10 μm.) Cholinergic neurons (acr-2::GFP) (I) are unaffected, whereas GABA neurons (unc-25::mcherry) (J) show degenerating processes (arrowheads) and cell bodies (arrows) in a single coq-2(ok1066) animal. (Scale bar: 30 μm.) (K) VNC acr-2::GFP-cholinergic neurons were counted from VA2–VA11 (30 total) and VNC unc-25::mcherry-GABA neurons from VD3–VD11 (13 total). Fraction of missing cholinergic vs. GABA neurons in coq-2(ok1066). **P < 0.001, n = 16. Anterior left, ventral down in A–C and F–J.
We also validated the coq-1 RNAi results by examining coq-1(ok749) mutant animals (Fig. S1). The ok749 deletion removes a C-terminal region comprising ≈70% of the coding sequence and is therefore a likely null allele. Prior studies of coq-1(ok749) reported paralysis and early larval lethality (22). We observed that homozygous coq-1 animals develop to at least the third larval stage with a few adult escapers. This finding is likely to mean that the coq-1(ok749) mutation results in a maternal effect lethal phenotype in which the first generation of viable offspring progress through larval development with maternally provided CoQ (23). Homozygous coq-1 mutants appear Unc as L3 larvae. Degeneration of GABA neurons is initially observed in young adults (Fig. S1B) (8/13 adult animals; Fig. S1C); a swollen ventral cord cholinergic motor neuron was observed in a single animal at this stage (n = 20). Older coq-1(ok749) animals show extensive swelling and degeneration of other tissues as reported (22).
We tested deletion mutants for coq-2 and coq-3 to ask whether genetic ablation of other CoQ biosynthetic genes phenocopies coq-1 knockdown (24). GABA neurons of coq-2(ok1066) and coq-3(ok506) animals also show the swollen morphology (Fig. 2 F–H) and progressive degeneration of coq-1-deficient animals (Fig. 2E). The similar phenotypes displayed by coq-2 and coq-3 mutants substantiate the idea that the movement and degenerative defects result from reduced CoQ synthesis, rather than off-target effects or additional roles of the COQ-1 enzyme.
Treatment with exogenous CoQ10 has successfully slowed disease progression of some cases of human CoQ deficiency (25, 26). To verify that CoQ deficiency was the cause of the degenerative phenotype in C. elegans, we incubated coq-1 RNAi-treated animals with CoQ10. We found that supplemental Coenzyme Q10 rescued degeneration in a dose-dependent manner, with an EC50 of 72 mg/mL (Fig. S2). A requirement for high CoQ doses for efficacy has also been observed in human patients and may reflect poor uptake of the drug by neurons (27).
The similar degenerative pattern and age-dependence shown by the coq-1, coq-2, and coq-3 knockout animals validates the specificity of the coq-1 RNAi treatment. Hence, for most subsequent experiments, we used RNAi knockdown of coq-1 to bypass the developmental delay of the CoQ pathway mutants and to produce large numbers of viable adult animals with CoQ-deficient phenotype.
GABA Neurons Are Preferentially Sensitive to CoQ Depletion-Induced Degeneration.
Having observed that CoQ deficiency results in GABA neuron degeneration, we asked whether nearby cholinergic motor neurons in the ventral nerve cord were similarly affected. The results of this analysis revealed that GABA neurons are significantly more sensitive to CoQ depletion (Fig. 2 I and J and Fig. S3). For example, in coq-2(ok1066) adults in which >35% of GABA neurons have degenerated, <5% of ventral cord cholinergic motor neurons are affected (Fig. 2K).
We next used cell-specific GFP reporters to evaluate the sensitivity of additional neuron subtypes to CoQ depletion by coq-1 RNAi. Populations tested were cholinergic (acr-2::GFP), serotonergic (tph-1::GFP), glutamatergic (eat-4::GFP), and dopaminergic (dat-1::GFP) neurons. Although all animals displayed age-dependent Unc phenotype with coq-1 RNAi, none of these neuron classes showed signs of degeneration comparable with that observed for GABA neurons at a similar developmental stage (Fig. S4 A–E). We considered the possibility that this differential effect could be due to the relative insensitivity of these neuron classes to RNAi. This explanation is made less likely, however, by the finding that all of these neuronal types were equally vulnerable to RNAi against GFP (Fig. S4G).
Like neurons, muscle is also a highly metabolic tissue, and muscle degeneration is seen in some cases of CoQ deficiency (28). Therefore, we used a GFP-labeled myosin heavy chain protein (MYO-3::GFP) (29) to examine muscle structure in the coq-1 RNAi knockdown animals. We detected no morphological abnormalities in body wall muscle (Fig. S4F), verifying that the Unc phenotype associated with this RNAi construct is not the result of muscle degeneration. We also examined vulval muscle, reasoning that defects in these muscles might explain the Egl phenotype. However, we observed no structural differences between vulval muscles in control versus coq-1 RNAi-treated animals.
Although our results do not preclude the possibility that non-GABAergic neurons or muscle are functionally affected by coq-1 RNAi, the earliest degenerative phenotype was exclusively observed in GABA neurons. In fact, functional deficits in other neurons or muscle are likely because the movement defects of mutants in which GABA neurons are selectively disabled are less severe than the paralyzed adult phenotype that results from RNAi ablation of coq-1 (30). Ultimately, coq-1 RNAi-treated adults and become completely paralyzed and show widespread cell swelling and tissue necrosis after 5 d of RNAi treatment; non-GABA neuron cell types were similarly affected in older coq-2 and coq-3 adults.
coq-1 RNAi-Mediated Degeneration Is Ca2+-Dependent.
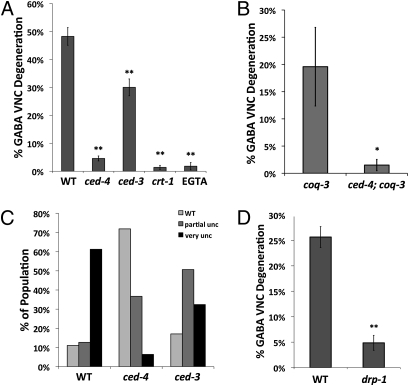
Calcium is a key effector of neurodegenerative diseases involving mitochondrial dysfunction (31). Calreticulin (crt-1) functions in the ER lumen to maintain Ca2+ levels for ready release on appropriate stimuli (32). In C. elegans, crt-1 mutants block necrotic degeneration of motor neurons (33). We tested a crt-1-null mutant (bz30) in our paradigm and found that it prevented the progressive degeneration of GABA neurons in coq-1 RNAi-treated animals (Fig. 3A). The calcium chelating agent EGTA (0.5 mM) was similarly protective (Fig. 3A). Taken together, these results demonstrate that Ca2+ signaling is important for coq-1 RNAi-mediated degeneration of GABA neurons.
Fig. 3.
Apoptotic genes and calcium and are required for coq-1 RNAi-induced GABA neuron degeneration. (A) ced-4(n1162), ced-3(n717), crt-1(bz30), and EGTA (0.5 mM)-treated animals were tested for coq-1 knockdown-induced GABA neuron degeneration. All results were obtained from coq-1 RNAi-treated animals containing eri-1(mg366); unc-25::GFP (Materials and Methods). GABA neurons were scored in adults as in Fig. 2. n ≥ 3 experiments for each genotype. Error bars = SEM, **P < 0.0001 vs. WT (wild-type). (B) ced-4(n1162) suppresses GABA neuron degeneration in coq-3(ok506) adults, *P < 0.02 vs. coq-3(ok506), n > 14. (C) Movement assay of ced-4 and ced-3 mutants treated with coq-1 RNAi. Adults were tapped on the head and the tail and scored for movement: “partial unc” worms are uncoordinated and “very unc” worms are unable to move, n > 100 worms. (D) drp-1 (tm1108) suppresses coq-1 knockdown-induced GABA degeneration, **P < 0.001 vs. WT, n > 60.
Specific Apoptotic Genes Are Required for GABA Neuron Cell Death in CoQ-Depleted Animals.
Ca2+ release from the endoplasmic reticulum can result in necrotic (34) or apoptotic (35) cell death. The cell swelling observed in coq-1 knockdown mutants is consistent with a necrotic mechanism. To investigate whether apoptotic genes are required for GABA cell death, we tested mutants of the canonical apoptotic pathway, egl-1, ced-9, ced-4, and ced-3 (Fig. 3 and Fig. S5). In apoptotic cells, the BH3-only protein, EGL-1, interacts with CED-9 (Bcl-2) to release CED-4 (Apaf-1) for activation of CED-3 (caspase) that, in turn dismembers sensitive target proteins that normally maintain cellular integrity (36). Loss-of-function mutations in egl-1 did not prevent GABA neuron degeneration after coq-1 knockdown. A gain-of-function allele of the anti-apoptotic protein CED-9 was similarly ineffective (Fig. S5). Surprisingly, mutations in ced-4 and ced-3 attenuated both the coq-1 RNAi-dependent GABA neuron degeneration and Unc phenotype (Fig. 3 A and C). These genetic results indicate that CED-4 is activated in coq-1–depleted GABA neurons by a pathway that is not regulated by either egl-1 or ced-9. In addition to strongly suppressing degeneration of GABA ventral nerve cord (VNC) processes, the ced-4 mutation also attenuates the elimination of GABA neuron cell bodies in coq-1 RNAi-treated animals (Fig. S6), which suggests that GABA neuron cell death is blocked. This result is supported by our finding of comparably strong ced-4 suppression of GABA neuron degeneration in coq-3(ok506) animals (Fig. 3B). Curiously, we note that a strong loss-of-function allele of ced-3 only partially rescued the uncoordinated and GABA neuron degeneration phenotypes, whereas the ced-4 mutation affords almost complete suppression of both defects in coq-1 RNAi-treated animals (Fig. 3). This observation is indicative of an additional ced-3-independent pathway functioning downstream of ced-4 to evoke GABA neuron degeneration. The cytoplasmic swelling of CoQ-depleted GABA neurons in conjunction with the essential roles of CED-4/Apaf-1 and CED-3/caspase suggest that loss of CoQ synthetic activity triggers selected components of both programmed cell death and necrotic pathways.
coq-1 Knockdown-Induced Cell Death Depends on the Mitochondrial Fission Gene drp-1.
Calcium has been shown to stimulate mitochondrial fission (37), which, in turn, has been linked to both apoptosis and necrosis in mammals (6, 38) and in C. elegans (39). We reasoned that CoQ deficiency might sensitize mitochondria to Ca2+-dependent fission-related cell death. To test this idea, we used RNAi knockdown of coq-1 in drp-1 mutant animals (Fig. 3D). DRP-1 (dynamin-related protein) has been shown to function in both mitochondrial fission (39) and in a ced-3–dependent and ced-9–independent cell death pathway (40). The drp-1 mutant blocked GABA neuron degeneration (Fig. 3D) and rescued the movement defect (Fig. S7), thereby implicating the fission machinery in the pathology associated with coq-1 knockdown in C. elegans. Similar results were obtained in coq-1+drp-1 double RNAi experiments (Fig. S8A). We also used double RNAi to test fzo-1, a gene required for mitochondrial fusion (41). In this case, fzo-1 knockdown actually enhances the GABA neuron degeneration phenotype of coq-1–deficient animals as expected for a treatment that impairs mitochondrial fusion (Fig. S8 B and C). The conclusion that mitochondrial function is defective in CoQ-depleted animals is also supported by our finding that mitochondrial ATP levels are significantly reduced by coq-1 RNAi treatment (Fig. S9). Together, these results indicate that CoQ depletion disrupts mitochondrial function and activates a cell death pathway that depends on the mitochondrial morphogenesis gene, drp-1 (Fig. 4).
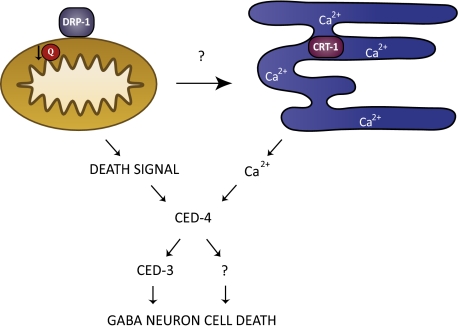
Fig. 4.
Model of GABA neuron death induced by CoQ depletion. Reduced CoQ (Q) function in mitochondria activates an unknown signal that may evoke CRT-1–dependent (calreticulin) Ca2+ release from the endoplasmic reticulum. GABA neuron degeneration requires CED-4 and at least one additional downstream pathway that functions in parallel to CED-3 (caspase). DRP-1, a dynamin-related protein that drives mitochondrial fission, is also required for degeneration in CoQ-depleted GABA neurons.
Discussion
We have shown that depletion of Coenzyme Q (CoQ) through RNAi and genetic ablation of CoQ biosynthetic genes, coq-1, coq-2, and coq-3, leads to the progressive loss of motor coordination and preferential degeneration of GABA neurons. The mechanism of cell death relies strongly on the function of ced-4 (Apaf-1) and partially depends on ced-3 (caspase) activity. The GABA neuron pathology that accompanies CoQ depletion also involves calcium signaling, possibly from ER stores, and the mitochondrial fission protein DRP-1 (Fig. 4). These results emphasize an important role for CoQ in neuron survival and link mitochondrial dysfunction to a calcium-dependent mechanism of selective neuron degeneration in C. elegans.
CoQ Depletion Triggers a Cell Death Pathway Featuring Elements of Both Apoptosis and Necrosis.
Our findings suggest that CoQ depletion in C. elegans GABA neurons triggers a cell death mechanism that includes features of both apoptotic and necrotic pathways. The affected GABA neurons morphologically resemble necrotic cells (i.e., swollen soma) yet require downstream components of apoptosis, ced-4, and ced-3. In C. elegans, necrosis and apoptosis have been reported to function as separate pathways (42); however, our results indicate that a subset of apoptotic genes is required for a pathology (cell swelling) normally restricted to a necrotic mechanism. In the canonical apoptotic pathway, CED-9 tethers CED-4 to the outer mitochondrial membrane. EGL-1 binding to CED-9 releases CED-4 for incorporation into the apoptosome and subsequent activation of CED-3/caspase (36). Our finding that neither EGL-1 nor CED-9 affects coq-1 RNAi-induced GABA neuron death indicates that CED-4 must be activated by an alternative mechanism in this case. Our results also suggest that CED-4 is likely to activate other downstream effectors in addition to CED-3. CED-4–dependent, CED-3–independent cell killing programs have been reported for yeast in which CED-4 is overexpressed (43) and in C. elegans with RNAi knockdown of the presumptive anti-apoptotic protein, ICD-1 (44). Mechanisms whereby CED-4 might activate these pathways are unknown. In the future, it will be interesting to determine whether the downstream CED-3-independent program that is triggered in CoQ-depleted GABA neurons includes other members of the C. elegans caspase family (45) or novel components (46).
Mitochondrial Morphogenesis Protein, DRP-1, Is Required for the Death of CoQ-Depleted GABA Neurons.
Mitochondrial fragmentation, which can trigger apoptosis, depends on the dynamin-related protein, DRP-1 (7). Previous studies have shown that coq-1 RNAi induces mitochondrial fragmentation in C. elegans (47). Thus, our finding that DRP-1 is required for the execution of GABA neurons in CoQ-deficient animals (Fig. 3 and Fig. S8) is consistent with a model in which the mitochondrial fragmentation or “excessive fission” accounts for the neurodegenerative effect of CoQ depletion. Enhancement of this effect by RNAi knockdown of the mitochondrial fusion gene, fzo-1, also supports this idea (Fig. S8). In addition to mediating mitochondrial fusion, the mammalian homolog of FZO-1, Mfn2, promotes expression of ETC proteins (48). This finding suggests that the loss of mitochondrial energy production in fzo-1–deficient animals could also contribute to cell death in CoQ-depleted GABA neurons. Although mitochondrial fission has been linked to apoptosis in mammals, conflicting results have been obtained in C. elegans (39, 40). However, our finding that DRP-1 is required for GABA neuron degeneration in C. elegans parallels results with mammalian cells showing that mitochondrial fission is a necessary step in experimental models of neurodegenerative disease resulting from mitochondrial injury. For example, exposure of cortical neurons to high concentrations of the neurotransmitter NO (nitric oxide) triggers a caspase-independent death pathway that requires mitochondrial fission (46). NO-induced degeneration is accompanied by ATP depletion and production of free radicals. Both of these traits are also induced by either environmental or genetic defects that block mitochondrial ETC (5, 49, 50). We therefore speculate that CoQ depletion in C. elegans disrupts mitochondrial ETC function, which in turn, triggers a degenerative pathway that shares common features such as downstream signals and effector molecules that also mediate neurodegeneration in mammalian neurons.
CoQ Deficiency in C. elegans as a Model for Human Disease.
The selective, age-dependent death of GABA neurons and loss of coordinated movement seen in CoQ-depleted worms are shared features of CoQ deficiency in humans. The most common outcome of this deficiency is cerebellar ataxia (51), although mutations that disrupt CoQ biosynthesis can also result in a more severe early-onset pathology involving multiple tissues (14, 25).
Despite the rarity of CoQ deficiency in humans, it shares important pathologies with prevalent glutamine repeat diseases. These include the autosomal dominant Huntington's Disease (HD) and spinocerebellar ataxias. The age-related death of medium spiny GABAergic neurons of the striatum occurs in HD (52) and GABAergic Purkinje cells are significant targets for degeneration in the cerebellar ataxias (53). Recent studies have linked neuronal degeneration in HD to disrupted mitochondrial function (54). For example, exogenous CoQ is neuroprotective in mouse models of HD (55). In addition, the pathology induced by overexpression of mtHtt in mammalian cells or in C. elegans depends on the mitochondrial fission machinery (8); ced-3 function is also required in C. elegans (56). Lastly, coq-1 RNAi in C. elegans enhances the toxicity of a mutant form of human tau that has been implicated in the pathology of AD (57). It is striking that salient features of these human genetic diseases including selective neuron sensitivity, mitochondrial dysfunction, Ca2+ dependence, and aberrant cell death (58, 59) are observed in C. elegans with the depletion of CoQ. These parallels suggest that neurodegenerative mechanisms arising from mitochondrial dysfunction are conserved and, thus, can be effectively delineated by studies in C. elegans.
Materials and Methods
Strains and Maintenance.
C. elegans strains (SI Materials and Methods) were maintained at 20 °C unless otherwise indicated (60). GFP reporters and the ced-3(n717), ced-4(n1162), crt-1(bz30), and drp-1(tm1108) mutant alleles were crossed into the RNAi-hypersensitive strain eri-1(mg366). eri-1(mg366) was verified by using single-worm PCR (SI Materials and Methods). unc-25::GFP(juIs76) was crossed into coq-2(ok1066)/hT2 and coq-3(ok506)/nT1 to mark GABA neurons, and the cholinergic neuron reporter acr-2::GFP(juIs14) and unc-25::mcherry(wdEx658) were crossed into coq-2(ok1066)/hT2. The unc-25::mcherry was generated by bombardment (61) with the plasmid pMLH41 [punc-25::mcherry, unc-119(+)] (SI Materials and Methods).
RNAi.
RNAi assays were performed by feeding (62). Briefly, 3 mL of LB/ampicillin (50 mg/mL) was inoculated with 30 μL of overnight culture and grown in a 37 °C shaker. At OD600 ≈ 0.800, the culture was diluted to 6 mL with LB/amp + IPTG (40 mM final concentration) and incubated at 37 °C for another 4 h. Bacteria were pelleted, dispersed in 250 mL of M9/IPTG, and spread onto 60-mm NGM plates. L4 larvae were added to plates and incubated at 20 °C for 5 d before scoring progeny. RNAi clones were sequenced to confirm inserts. EGTA (0.5 mM) was added to media before pouring plates.
Degeneration Assay.
Animals were anesthetized in 0.1% tricaine/tetramisole on 2% agar pads (63). Percent VNC degeneration was determined by dividing the number of degenerating VNC intervals between GABA motor neuron cell bodies by the total number of intervals between all GABA neuron cell soma per animal (for example, if 3 VNC intervals are scored as degenerating of a total of 12 total intervals in a given animal, the percent VNC degeneration would be 3/12 or 25%). Commissures were scored similarly, with the number of degenerating commissures divided by the total number of discrete GABA neuron commissures per animal (16 total). A 63× objective was used for scoring. The observer was blinded to experimental versus control samples to avoid bias. unc-25::GFP(juIs76) was used to mark GABA neurons for experiments with coq-1 RNAi, coq-1(ok749), coq-2(ok1066), and coq-3(ok506). unc-25::mcherry(wdEx658) was also used to mark GABA neurons in coq-2(ok1066) for results shown in Fig. 2. Swimming assays were performed as described (21). coq-3(ok506) and coq-3(ok506); ced-4(n1162) animals were scored as 7-d-old adults (Fig. 3B). The following mutants and treatments did not perturb GABA neuron morphology: ced-4(n1162), ced-3(n717); crt-1(bz30); drp-1(tm1108), CoQ10, 0.5 mM EGTA.
Microscopy.
Animals were imaged in a Zeiss Axioplan compound microscope with a CCD camera (ORCA I; Hamamatsu). Confocal images were obtained on a Zeiss LSM 510 confocal microscope and on a Leica TCS SP5 confocal microscope.
Supplementary Material
Acknowledgments
We thank D. Xue (University of Colorado, Boulder, CO) for drp-1(tm1108), Y. Jin (University of California, San Diego, CA) for plasmid pSC392, and C. Thorne (Vanderbilt University, Nashville, TN) for help with the ATP assays. Other strains were obtained from the C. elegans Genetics Center, which is supported by National Institutes of Health National Center for Research Resources. We thank A. Bowman (Vanderbilt University, Nashville, TN), B. McLaughlin (Vanderbilt University, Nashville, TN), and M. Driscoll (Rutgers University, Piscataway, NJ) for comments on the manuscript. This work was supported by National Institutes of Health Grants R01 NS26115 and R21 MH77302, a Vanderbilt Kennedy Center Hobbs Discovery Grant (to D.M.M.), National Institutes of Health Grant F31 NS49743 (to J.D.W.), and National Institutes of Health Grant T32 GM08554.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910630107/-/DCSupplemental.
References
- 1.Petrozzi L, Ricci G, Giglioli NJ, Siciliano G, Mancuso M. Mitochondria and neurodegeneration. Biosci Rep. 2007;27:87–104. doi: 10.1007/s10540-007-9038-z. [DOI] [PubMed] [Google Scholar]
- 2.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 3.Orsucci D, Filosto M, Siciliano G, Mancuso M. Electron transfer mediators and other metabolites and cofactors in the treatment of mitochondrial dysfunction. Nutr Rev. 2009;67:427–438. doi: 10.1111/j.1753-4887.2009.00221.x. [DOI] [PubMed] [Google Scholar]
- 4.Brouillet E, Condé F, Beal MF, Hantraye P. Replicating Huntington's disease phenotype in experimental animals. Am J Physiol Prog Neurobiol. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 5.Bossy-Wetzel E, Petrilli A, Knott AB. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young KW, Piñon LGP, Bampton ETW, Nicotera P. Different pathways lead to mitochondrial fragmentation during apoptotic and excitotoxic cell death in primary neurons. J Biochem Mol Toxicol. 2010 doi: 10.1002/jbt.20343. 10.1002/jbt.20343. [DOI] [PubMed] [Google Scholar]
- 7.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of Huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formigli L, et al. Aponecrosis: Morphological and biochemical exploration of a syncretic process of cell death sharing apoptosis and necrosis. J Cell Physiol. 2000;182:41–49. doi: 10.1002/(SICI)1097-4652(200001)182:1<41::AID-JCP5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Wochna A, et al. A possible role of oxidative stress in the switch mechanism of the cell death mode from apoptosis to necrosis—studies on rho0 cells. Mitochondrion. 2007;7:119–124. doi: 10.1016/j.mito.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 12.Crane FL, Hatefi Y, Lester RL, Widmer C. Isolation of a quinone from beef heart mitochondria. Biochim Biophys Acta. 1957;25:220–221. doi: 10.1016/0006-3002(57)90457-2. [DOI] [PubMed] [Google Scholar]
- 13.Quinzii C, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollet J, et al. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest. 2007;117:765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier-Tourenne C, et al. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mollet J, et al. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am J Hum Genet. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiMauro S, Quinzii CM, Hirano M. Mutations in coenzyme Q10 biosynthetic genes. J Clin Invest. 2007;117:587–589. doi: 10.1172/JCI31423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Aguilera JC, Gaviklan A, Asencio C, Navas P. The role of ubiquinone in Caenorhabditis elegans. Ageing Res Rev. 2005;4:41–53. doi: 10.1016/j.arr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 21.Miller KG, et al. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavilán A, et al. C. elegans knockouts in ubiquinone biosynthesis genes result in different phenotypes during larval development. Biofactors. 2005;25:21–29. doi: 10.1002/biof.5520250104. [DOI] [PubMed] [Google Scholar]
- 23.Hihi AK, Gao Y, Hekimi S. Ubiquinone is necessary for Caenorhabditis elegans development at mitochondrial and non-mitochondrial sites. J Biol Chem. 2002;277:2202–2206. doi: 10.1074/jbc.M109034200. [DOI] [PubMed] [Google Scholar]
- 24.Asencio C, Rodríguez-Aguilera JC, Ruiz-Ferrer M, Vela J, Navas P. Silencing of ubiquinone biosynthesis genes extends life span in Caenorhabditis elegans. FASEB J. 2003;17:1135–1137. doi: 10.1096/fj.02-1022fje. [DOI] [PubMed] [Google Scholar]
- 25.Quinzii CM, DiMauro S, Hirano M. Human coenzyme Q10 deficiency. Neurochem Res. 2007;32:723–727. doi: 10.1007/s11064-006-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salviati L, et al. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: A CoQ10-responsive condition. Neurology. 2005;65:606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- 27.Ibrahim WH, Bhagavan HN, Chopra RK, Chow CK. Dietary coenzyme Q10 and vitamin E alter the status of these compounds in rat tissues and mitochondria. J Nutr. 2000;130:2343–2348. doi: 10.1093/jn/130.9.2343. [DOI] [PubMed] [Google Scholar]
- 28.Lalani SR, et al. Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Arch Neurol. 2005;62:317–320. doi: 10.1001/archneur.62.2.317. [DOI] [PubMed] [Google Scholar]
- 29.Campagnola PJ, et al. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys J. 2002;82:493–508. doi: 10.1016/S0006-3495(02)75414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- 31.Mattson MP. Calcium and neurodegeneration. Aging Cell. 2007;6:337–350. doi: 10.1111/j.1474-9726.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- 32.Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- 33.Xu K, Tavernarakis N, Driscoll M. Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca(2+) release from the endoplasmic reticulum. Neuron. 2001;31:957–971. doi: 10.1016/s0896-6273(01)00432-9. [DOI] [PubMed] [Google Scholar]
- 34.Driscoll M, Gerstbrein B. Dying for a cause: Invertebrate genetics takes on human neurodegeneration. Nat Rev Genet. 2003;4:181–194. doi: 10.1038/nrg1018. [DOI] [PubMed] [Google Scholar]
- 35.Demaurex N, Distelhorst C. Cell biology. Apoptosis—the calcium connection. Science. 2003;300:65–67. doi: 10.1126/science.1083628. [DOI] [PubMed] [Google Scholar]
- 36.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: A complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- 37.Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 39.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 40.Breckenridge DG, et al. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanazawa T, et al. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung S, Gumienny TL, Hengartner MO, Driscoll M. A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat Cell Biol. 2000;2:931–937. doi: 10.1038/35046585. [DOI] [PubMed] [Google Scholar]
- 43.Tao W, Walke DW, Morgan JI. Oligomerized Ced-4 kills budding yeast through a caspase-independent mechanism. Biochem Biophys Res Commun. 1999;260:799–805. doi: 10.1006/bbrc.1999.0982. [DOI] [PubMed] [Google Scholar]
- 44.Bloss TA, Witze ES, Rothman JH. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature. 2003;424:1066–1071. doi: 10.1038/nature01920. [DOI] [PubMed] [Google Scholar]
- 45.Shaham S. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J Biol Chem. 1998;273:35109–35117. doi: 10.1074/jbc.273.52.35109. [DOI] [PubMed] [Google Scholar]
- 46.Barsoum MJ, et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichishita R, et al. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J Biochem. 2008;143:449–454. doi: 10.1093/jb/mvm245. [DOI] [PubMed] [Google Scholar]
- 48.Pich S, et al. The Charcot-Marie-Tooth type 2A gene product, Mfn2 , up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet. 2005;14:1405–1415. doi: 10.1093/hmg/ddi149. [DOI] [PubMed] [Google Scholar]
- 49.Wallace KB, Starkov AA. Mitochondrial targets of drug toxicity. Annu Rev Pharmacol Toxicol. 2000;40:353–388. doi: 10.1146/annurev.pharmtox.40.1.353. [DOI] [PubMed] [Google Scholar]
- 50.Atamna H, Frey WH., 2nd Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer's disease. Mitochondrion. 2007;7:297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Montero R, et al. Clinical, biochemical and molecular aspects of cerebellar ataxia and Coenzyme Q10 deficiency. Cerebellum. 2007;6:118–122. doi: 10.1080/14734220601021700. [DOI] [PubMed] [Google Scholar]
- 52.Martin JB, Gusella JF. Huntington's disease. Pathogenesis and management. N Engl J Med. 1986;315:1267–1276. doi: 10.1056/NEJM198611133152006. [DOI] [PubMed] [Google Scholar]
- 53.Zoghbi HY. Spinocerebellar ataxia type 1. Clin Neurosci. 1995;3:5–11. [PubMed] [Google Scholar]
- 54.Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrante RJ, et al. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci. 2002;22:1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kraemer BC, Burgess JK, Chen JH, Thomas JH, Schellenberg GD. Molecular pathways that influence human tau-induced pathology in Caenorhabditis elegans. Hum Mol Genet. 2006;15:1483–1496. doi: 10.1093/hmg/ddl067. [DOI] [PubMed] [Google Scholar]
- 58.Dueñas AM, Goold R, Giunti P. Molecular pathogenesis of spinocerebellar ataxias. Brain. 2006;129:1357–1370. doi: 10.1093/brain/awl081. [DOI] [PubMed] [Google Scholar]
- 59.Kwong JQ, Beal MF, Manfredi G. The role of mitochondria in inherited neurodegenerative diseases. J Neurochem. 2006;97:1659–1675. doi: 10.1111/j.1471-4159.2006.03990.x. [DOI] [PubMed] [Google Scholar]
- 60.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 63.McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: Multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.