Abstract
A variety of tumor-suppressor mechanisms exist to promote genome integrity and organismal survival. One such mechanism is cellular senescence. In response to replicative aging, DNA damage, and oncogenic stimuli, the p53 and Rb pathways are activated to prevent the proliferation of damaged cells by inducing senescence or apoptosis. We have performed a loss-of-function genetic screen in primary human cells to identify components of the senescence machinery. Here we describe BRD7 and BAF180 as unique regulators of replicative senescence in human cells. Both regulate p53 transcriptional activity toward a subset of its target genes required for replicative and oncogenic stress senescence induction, and BRD7 physically interacts with p53. BRD7 is a deletion target in human cancer, suggesting that loss of BRD7 may provide an additional mechanism to antagonize p53 function in cancer cells.
Keywords: cancer, PBAF, SWI/SNF, p21
Replicative senescence is a permanent cell-cycle arrest induced by telomere attrition in aged cells. This program is also engaged in response to activated oncogenes, and DNA damage is thought to be the unifying theme among the stimuli that have been shown to induce a senescence phenotype (1, 2). Numerous studies have implicated p53 as a major component of the senescence machinery, which is activated and stabilized by multiple mechanisms in response to DNA damage. The importance of p53 during senescence has been attributed at least in part to its transcriptional control of the CDK inhibitor p21, which induces cell-cycle arrest (3–7). How p53 activates transcription of p21 and other targets required for senescence is not yet clear, and there are likely to exist many more components involved in these responses.
In this study we searched genetically for previously unexplored regulators of senescence induction and identified a bromodomain protein, BRD7, as a regulator of replicative senescence. Bromodomains often recruit proteins to chromatin through their ability to recognize acetylated histones. BRD7 was originally identified as a gene whose mRNA was down-regulated in nasopharyngeal carcinoma (8). It was also identified as a unique component of the SWI/SNF polybromo-associated BRG1-associated factor (PBAF) complex (SWI/SNF complex B). BRD7 is specific to this complex, and is not found in the related BAF complex (SWI/SNF complex A). Although BAF and PBAF complexes share many components, some components are nonoverlapping and thought to impart distinct biological functions to each complex. Furthermore, BRD7 was shown to specifically affect PBAF-mediated gene expression regulation (9). Several lines of evidence suggest that the PBAF complex contributes to proliferation control. A previous study showed that the specifying component exclusive to the PBAF complex, BAF180 (polybromo-1), physically binds the p21 promoter (10). This observation was also made regarding BRG1, which functions interchangeably with structurally related BRM1 as one of the two SWI/SNF ATPase subunits in BAF complexes, although it is the only ATPase subunit used by the PBAF complex (11, 12). Here, we show that BRD7 is a critical mediator of the ability of p53 to transduce the replicative and oncogene-induced senescence signal to p21 to initiate the senescence program of differentiation.
Results
BRD7 Regulates Cell Proliferation and Replicative Senescence.
In an ongoing whole-genome shRNA screen in primary BJ fibroblasts to identify genes that regulate replicative senescence by allowing cells to continue proliferation after the control cells had largely senesced, we recovered multiple shRNAs against BRD7. To recapitulate its role in senescence, we introduced three independent BRD7 shRNAs into BJ fibroblasts. BRD7 depletion resulted in a striking increase in proliferation rate in both young and midpassage BJ fibroblasts, suggesting that BRD7 may function in proliferation control before the onset of senescence (Fig. 1A, Upper). BRD7 depletion significantly delays replicative senescence, as these cells continued to actively proliferate while control firefly luciferase (FF) shRNA expressing cells ceased proliferation and stained positive for senescence-associated (SA) β-galactosidase (Fig. 1B and Fig. S1A). BRD7 depletion also extended replicative lifespan in BJs expressing the HPV E7 oncoprotein, which binds and inhibits Rb, suggesting that BRD7 controls proliferation independently of Rb (Fig. 1A, Lower). These phenotypes are observed with all three BRD7 shRNAs, and the strength of the proliferative phenotype correlates with the level of BRD7 depletion (Fig. 1C and Fig. S1B).
Fig. 1.
BRD7 depletion increases proliferation and delays senescence in primary fibroblasts. (A) Population-doubling (PD) curves in primary BJ fibroblasts (Upper) and BJs expressing the HPV E7 oncoprotein (Lower) and the indicated shRNAs. (B) Senescence-associated β-galactosidase staining quantification with error bars indicating SEM in primary human BJ fibroblasts infected with retroviruses expressing the indicated shRNAs. (C) BRD7 depletion measured by Western blot in primary human BJ fibroblasts infected with retroviruses expressing the indicated shRNAs. (D) Senescence-associated β-galactosidase staining quantification on day 16 posttransduction with RasV12D or vector alone; error bars indicate SEM. Population doublings in human BJ fibroblasts were measured on day 15 posttransduction with RasV12D or vector alone in primary BJ fibroblasts expressing the indicated shRNAs.
The observed delay in replicative senescence did not appear to be a result of induction of telomerase activity or hTERT expression as measured by telomeric repeat-amplification protocol assay and qRT-PCR, respectively (Fig. S2). Because of their increased proliferation rate, BRD7-depleted cells underwent more population doublings than control cells, suggesting that they may be defective in their response to the expected telomere attrition resulting from these further doublings.
Several studies have demonstrated that oncogenes, such as activated Ras, can induce senescence through the p53 and Rb pathways. BRD7 depletion resulted in significant resistance to RasV12D-induced senescence as indicated by cell proliferation and β-galactosidase staining (Fig. 1D and Fig. S3). This finding suggests that BRD7 is required for a function common to both replicative and oncogene-induced senescence.
BRD7 Is Required for p53 Transcriptional Activity.
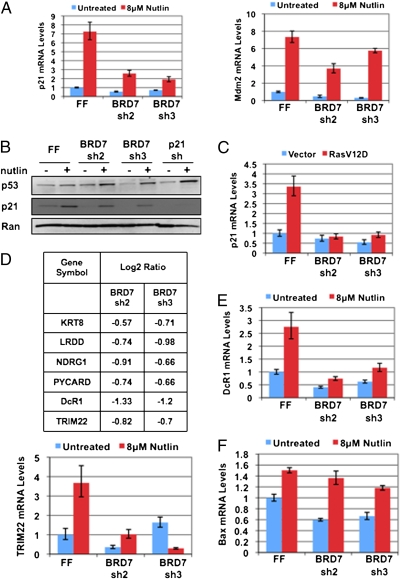
Because of the previous link between SWI/SNF complexes and p21 expression and the previous identification of BRD7 in PBAF complexes, we examined p21 mRNA levels by qRT-PCR in response to treatment with the Mdm2 inhibitor nutlin-3a, which stabilizes p53. BRD7 depletion reduces both the basal and induced expression of p21 in response to p53 activation (Fig. 2A, Left), although p53 was stabilized as expected (Fig. 2B), suggesting that BRD7 acts downstream of p53 stabilization. Mdm2 expression was also decreased in these experiments, suggesting that p21 is not the only p53 target gene controlled by BRD7 (Fig. 2A, Right). BRD7-depleted cells also showed decreased p21 and Mdm2 expression following transduction with RasV12D (Fig. 2C and Fig. S4), indicating that BRD7 is required for expression of multiple p53 target genes during both replicative and oncogene-induced senescence. In addition, expression of an shRNA#2-resistant BRD7 cDNA was able to restore p21 expression in response to nutlin-3a treatment, confirming that this phenotype is caused specifically by depletion of BRD7 (Fig. S5).
Fig. 2.
BRD7 regulates p53 activity toward a subset of its target genes. (A) Quantitative RT-PCR analysis of p21 and Mdm2 expression upon BRD7 depletion with and without 8 μM nutlin-3a treatment for 6 h. Error bars indicate SEM. The mRNA levels are normalized to β-actin and relative to an FF control shRNA. (B) Western blot analysis of p53 and p21 protein levels following treatment with 8 μM nutlin-3a for 6 h in cells expressing the indicated shRNAs. (C) Analysis of the effects of the indicated shRNAs on p21 expression by qRT-PCR in response to RasV12D transduction. (D) Gene-expression profiling in BJ fibroblasts in the presence and absence of 8 μM nutlin-3a treatment for 6 h and BRD7 depletion with two independent shRNAs. Additional p53 target genes that were down-regulated upon BRD7 depletion are listed as indicated in the table. (E and F) Quantitative RT-PCR analysis of the indicated p53 target genes expression upon BRD7 depletion with and without 8 μM nutlin-3a treatment for 6 h. Error bars indicate SEM. The mRNA levels are normalized to β-actin and are relative to an FF control shRNA.
Analysis of BRD7 and p53-Mediated Regulation of Gene Expression.
To broadly analyze the role of BRD7 in gene expression, we performed gene-expression profiling to compare gene expression between control cells and cells expressing one of two distinct BRD7 shRNAs (BRD7 sh2 and sh3), in the presence and absence of nutlin-3a treatment (Fig. 2D). We considered genes that met a log2 ratio criteria of ± 0.5 in both BRD7 shRNA samples when compared with control firefly luciferase (FF) shRNA expressing cells to be differentially regulated. In both conditions, BRD7 was the most significantly down-regulated gene. We compared BRD7 down-regulated genes in the nutlin-3a treated samples with a curated list of genes that have met several biological criteria to confirm that they are p53 target genes (13). Expression of several additional p53 target genes was down-regulated in BRD7-depleted cells in response to nutlin-3a treatment, such as DcR1, TRIM22, NDRG1, and PYCARD, suggesting that, like p21, BRD7 is required for their transactivation by p53 (Fig. 2D). We confirmed this by qRT-PCR for DcR1 and TRIM22 (Fig. 2E). However, several p53 target genes, such as Bax, did not appear to require BRD7 for their expression (Fig. 2F); thus, BRD7 is required only for a subset of p53 target genes.
BRD7 Interacts with p53 Independently of the BRD7 Bromodomain.
Transcription factors that cooperate on promoters often interact, and we were able to coimmunoprecipitate p53 and BRD7, suggesting that they exist in a biological complex (Fig. 3A). Previous work has shown that BRD7 can bind to acetylated histones in vitro, in particular histone H3 lysine 14, through its bromodomain (14). To examine whether the bromodomain is required for the interaction between BRD7 and p53, we deleted the BRD7 bromodomain (BRD7 ΔBromo) (Fig. 3B and Fig. S6) and found the interaction with p53 remained intact. This observation is consistent with the model that BRD7 could be modulating p53 transcriptional activity by simultaneously binding chromatin via its bromodomain while also interacting with p53.
Fig. 3.
BRD7 coimmunoprecipitates with p53, and BAF180 is also required for replicative senescence and p21 expression. (A) Coimmunoprecipitation of HA-tagged BRD7 and endogenous p53 from primary BJs expressing HA-tagged BRD7 using anti-p53 antibodies, with 15% input loaded. (B) Schematic showing structure of BRD7 and the bromodomain deletion mutant. Coimmunoprecipitation of HA-tagged BRD7 ΔB and p53 from primary BJs expressing HA-tagged BRD7 ΔB, with 15% input loaded. (C) Population-doubling curves and senescence-associated β-galactosidase staining quantification with error bars indicating SEM in primary human BJ fibroblasts with three independent shRNAs against BAF180. (D) Quantitative RT-PCR for p21 and Mdm2 expression in BAF180 depleted cells in response to 8 μM nutlin-3a treatment for 6 h, with error bars indicating SEM. The mRNA levels are normalized to β-actin and are relative to an FF control shRNA.
BAF180 Is Required for Replicative Senescence.
BAF180 was also recovered from our screen, suggesting that SWI/SNF regulation of the p21 promoter is relevant during senescence in primary cells. Indeed, BAF180 depletion using three independent shRNAs resulted in both increased proliferation and delayed senescence (Figs. 3C and Fig. S7). BAF180 depletion also resulted in decreased basal and induced p21 expression in response to nutlin-3a (Fig. 3D). This finding is consistent with the previously reported requirement for BAF180 in p53-dependent p21 expression in response to irradiation (10), and provides a mechanistic explanation for the increased proliferation and delayed senescence observed in BAF180-depleted cells. Decreased expression of Mdm2 and DcR1 in response to nutlin-3a was also observed, suggesting that, like BRD7, BAF180 is also required for expression of multiple p53 target genes (Figs. 3D and Fig. S8). The previous study, which linked BAF180 to p21 expression, also provided evidence for BAF180 deletion in breast cancer (10), which supports the idea that this complex may be targeted in human cancer to disable p53 activity and thus the senescence response.
BRD7 Overexpression Induces Premature Senescence.
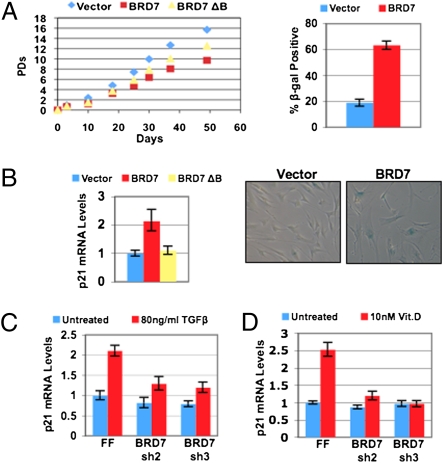
Overexpression of BRD7 slows proliferation and results in premature senescence in BJs fibroblasts (Fig. 4A). Expression of p21 was also elevated in BRD7-overexpressing cells (Fig. 4B), providing a mechanistic explanation for their slowed proliferation and premature senescence. However, overexpression of the BRD7 ΔBromo mutant did not slow proliferation to the same extent as BRD7 (Fig. 4A), and p21 mRNA levels were not significantly increased compared with control cells (Fig. 4B). Furthermore, nutlin-3a treatment resulted in reduced p21 expression in BRD7ΔBromo expressing cells, whereas wild-type BRD7-expressing cells displayed higher levels of p21 (Fig. S9). This result suggests that although interaction with p53 does not require the BRD7 bromodomain as shown above (Fig. 3B), the bromodomain may still have a functional role in mediating efficient p21 expression by p53. This finding is consistent with the idea that BRD7 may bridge p53 to chromatin via its bromodomain, possibly explaining our observation that overexpression of wild-type BRD7, but not BRD7 ΔBromo, is able to induce ectopic p21 expression and slow proliferation.
Fig. 4.
Forced expression of BRD7 induces premature senescence in primary fibroblasts, and BRD7 regulates p21 in both p53-dependent and -independent contexts. (A) Population-doubling curves in BJ fibroblasts expressing vector alone, BRD7 and BRD7 ΔBromo, and senescence-associated β-galactosidase staining of vector alone and BRD7 expressing BJ fibroblasts. (B) Quantitative RT-PCR analysis of p21 expression in BJ fibroblasts infected with the indicated retroviruses. Error bars indicate SEM. The mRNA levels are normalized to β-actin and are relative to vector alone containing control cells. (C) Quantitative RT-PCR for p21 expression in HCT116 p53−/− colon cancer cells, in response to 80 ng/mL TGF-β for 24 h. (D) Quantitative RT-PCR for p21 expression in human mammary epithelial cells in response to 10 nM 1α,25(OH)2D3 (calcitriol, the biologically active form of vitamin D) for 24 h. Error bars indicate SEM. The mRNA levels are normalized to β-actin and relative to an FF control shRNA.
BRD7 Is Frequently Deleted in Human Cancer.
A recent study of gene copy number in a large collection of human tumors and cancer cell lines from various tissues identified genes that are specifically deleted in cancer (15). We analyzed BRD7 in this database and found it to be frequently deleted in a tumor set consisting of 3,131 samples and cell lines from 54 cancer subtypes (Fig. S10). BRD7 falls within the peak of this deletion region with statistical significance, supporting the hypothesis that BRD7 is a tumor suppressor. Furthermore, BRD7 expression is frequently silenced by methylation of its promoter in nasopharyngeal carcinoma tumor samples (16). Interestingly, the PBAF-associated ATPase BRG1 is also mutated in cancer cell lines from several different tissues (17, 18). Furthermore, similarly to BRD7, BRG1 was also shown to be deleted in a recent copy-number analysis study (15), falling within identified peak regions with statistical significance in both a general lung cancer set of 774 samples and a nonsmall cell lung cancer set of 733 samples (Fig. S10). This finding is consistent with a previous report that ≈30% of human nonsmall cell lung cancer cell lines lack expression of BRG1 and also BRM1, the other SWI/SNF ATPase (19). Taken together, these data implicate multiple SWI/SNF components in tumor suppression.
BRD7 Regulates p21 Expression in p53-Dependent and Independent Transcription.
BAF180 regulates p21 in response to both p53 and Smad2/3/4, suggesting that PBAF may work with multiple transcription factors (10). The p21 gene is regulated by the vitamin D (VDR) and retinoic acid receptors in a p53-independent manner (20, 21). Furthermore, the PBAF complex has been shown to be required for nuclear hormone receptor transcription, such as for VDR (22). Therefore, we examined whether BRD7 depletion affects p21 induction in these contexts. BRD7 depletion in HCT116 cells significantly decreased p21 induction in response to TGF-β, which also occurred in p53 null-HCT116 cells, indicating p53 independence (Fig. 4C). BRD7 was also required for p21 induction by 1α,25(OH)2D3 (vitamin D) in human mammary epithelial cells (Fig. 4D). Taken together, our data indicate that BRD7 is required for p21 expression in both p53-dependent and p53-independent manners.
Discussion
BRD7 and SWI/SNF in p53-Dependent and Independent Transcription.
We have shown that BRD7 is required for both replicative and oncogene-induced senescence through the role of BRD7 in controlling p53-dependent transcription. BRD7 is required for p53 transcriptional activity toward multiple target genes, including p21, but has no negative effect on p53 abundance or stabilization in response to a variety of inducing agents, including the Mdm2 inhibitor nutlin-3a. This regulation is likely to be direct as BRD7 coimmunoprecipitates with p53. Enhanced BRD7 expression both reduces cell proliferation rates and accelerates replicative senescence. Thus, BRD7 levels may be rate-limiting for senescence induction. In addition, during the course of our study, Agami and colleagues (23) identified BRD7 as being important for oncogene-induced senescence, supporting our findings.
We have also shown that the PBAF component BAF180 is also required for replicative senescence, suggesting that the role of BRD7 in senescence control is mediated by its presence in the PBAF complex. Our data suggests that BRD7 could act by bridging p53 to a favorable chromatin environment for its transcriptional activity.
BRD7 is required for both p53-dependent and independent regulation of p21. Both vitamin D- and TGF-β-mediated induction of p21 depend upon BRD7 levels. Thus, although BRD7 deletion may serve as an additional mechanism to disrupt p53 function in cancer cells, there may be an advantage for disruption of BRD7, even in the context of p53 loss. Furthermore, given our data on BRD7 regulation of p53-independent p21 expression, it is likely that BRD7 may function in other differentiation programs that may be relevant to human disease.
BRD7 in Tumor Suppression.
Senescence is a bona fide tumor-suppression mechanism in vivo, and senescence also occurs as a response to chemotherapy. Our analysis of existing databases suggests that BRD7 is likely to be a deletion target in cancer, and it follows that cancer cells that inactivate BRD7 may be able to bypass these tumor-suppressive mechanisms, including sensitivity to chemotherapy. Our results and previous studies now suggest that several members of the PBAF complex, including BAF180 and BRD7, may be disrupted in human cancer, and our experiments strongly suggest that this is a result of compromised p53 function in both cases. Taken together, these results strongly suggest that loss of SWI/SNF function contributes to cancer progression, at least in part because of complete loss of PBAF-mediated regulation of p53. Our data suggest that loss of BAF180 or BRD7 would have similar consequences, as these two components are both specific to the PBAF complex and are required for PBAF-regulated gene expression (9, 22, 24).
Interestingly, a recent study found that BRD7 expression is negatively regulated by c-Myc via direct binding of c-Myc to the BRD7 promoter (25). This finding suggests that c-Myc-driven tumors may also have disrupted BRD7 function, thereby compromising part of p53 tumor-suppressor functions, which predicts that re-establishment of BRD7 expression levels in c-Myc-driven tumors may be capable of reversing the tumorigenic phenotype by restoration of p53 activity.
Materials and Methods
Cell Culture.
Human diploid BJ fibroblasts were obtained from ATCC and were maintained in 3% O2 conditions. Cells were grown in DMEM with 15% FBS, penicillin-strepomycin, 2 mM L-glutamine, and 0.1 mM nonessential amino acids (Invitrogen). Retroviral gene delivery techniques were done as previously described using 293T cells (26). Infection populations were selected with 1 μg/mL puromycin (Clontech) for at least 5 d. Nutlin-3a was obtained from Cayman Chemical. Human mammary epithelial cells were grown in MEGM media (Lonza). HCT116 cells were grown in McCoy's medium (Invitrogen) with 10% FBS. TGF-β peptide was obtained from R&D Systems. Calcitriol (1α,25(OH)2D3) was obtained from Sigma-Aldrich.
Retroviral Vectors.
The following vectors were used: MSCV-mir30 based shRNA constructs as previously described (27–29), MSCV-N-terminal HA-FLAG-puro, pHAGE-C-terminal-HA-FLAG-Neo, pWZL-hygro-HrasV12D, MSCV-neo-E7. The shRNA 22-mer targeting sequences are as follows: BRD7 sh1: AGATTAAATCAGAATCATCCTT, BRD7 sh2: TTTCATGGTACTAAAATCCATT, BRD7 sh3: TTATCTTCAAGCATATCTTTGT, p21 shRNA: TTGTTTAAATAATTCTAATGCC, BAF180 sh1: TTAGAACTCGCTTGTAGATGGC, BAF180 sh2: TAAATCAAGCCGACGGTAGCGA, and BAF180 sh3: TATAGAGTTCATGGCACACGGC. Retroviruses were packaged using 293T cells and Transit-293 (Mirus) transfection reagent according to the manufacturer's instructions.
Growth Curves and SA-β-galactosidase Assay.
Population doublings were monitored using a Coulter Counter, and cells were fed every 3 d when not split. The SA-β-galactosidase assay was performed using the Senescent Cell Staining Kit (Sigma) according to the manufacturer's instructions. Experiments were carried out in triplicate and at least 100 cells were scored in each field, with error bars indicating SEM.
Immunoblotting, Antibodies, and Immunoprecipitation.
Cell lysates were fractionated on 4 to 20% Tris-glycine SDS/PAGE gels (Invitrogen). Antibodies were as follows: human p53 (DO-1, Calbiochem), human p21 (OP64, Calbiochem), human Mdm2 (Santa Cruz), HA (HA.11, Covance), BRD7 (Cell Signaling), Ran (BD Biosciences), and BAF180 (A301-591A, Bethyl), HRP-conjugated human and mouse secondary antibodies (Jackson Laboratories), which were detected by chemilumiscence ECL reagent (Pierce). All immunoprecipitations were performed in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Nonidet P-40, with Complete-Mini protease inhibitor mixture tablets (Roche) and PhosSTOP phosphatase inhibitor mixture tablets (Roche). Antibodies were used as follows: HA (HA.11, Covance), p53 (DO-1, Calbiochem), normal rabbit IgG (Sigma), and AffiniPure Goat Anti-mouse IgG light chain specific secondary antibody (Jackson Laboratories). Two micrograms of each antibody was used for immunoprecipitations.
Quantitative RT-PCR and Gene-Expression Analysis.
Total RNA was isolated using the RNAeasy Mini Kit. (Qiagen), and cDNA was synthesized using SuperScript III (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR were performed in triplicate or quadruplicate using the Platinum Sybr Green Kit (Invitrogen) on an Applied Biosystems Fast 7500 machine, using β-actin as the endogenous normalization control, and data are shown relative to control FF shRNA containing cells. Primer sequences are available from the authors upon request. Gene expression analysis was performed using an Agilent 4 × 44 two-color gene expression array and Quick Amp Labeling protocol (Agilent). Probes that did not meet a processed signal value criteria of 100 or above in the control samples were filtered out. Genes considered to be down-regulated in BRD7-depleted cells had to meet a criteria of a log2 ratio of −0.5 or below in both BRD7 shRNA samples. This dataset is available from the National Center for Biotechnology Information Gene Expression Omnibus database with accession number GSE22607.
Telomerase Activity Assay.
Telomerase activity was measured using the TRAPeze-RT Kit (Chemicon) and Titanium Taq (Chemicon) according to the manufacturer's instructions on an Applied Biosystems Fast 7500 machine. Reactions were performed in triplicate containing 1,000 cell equivalents.
Supplementary Material
Acknowledgments
We thank Ji Luo for helpful discussions of the manuscript, members of the Elledge laboratory for advice and reagents, and Bert Vogelstein (Johns Hopkins Medical School, Baltimore), Scott Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), and Guang Hu (National Institute of Environmental Health Sciences, Research Triangle Park, NC) for reagents. This work was supported by grants from National Institutes of Health and Department of Defense (to S.J.E.) A.S. was supported by the Pathology Department at the Massachusetts General Hospital (T32CA09216) and by the Burroughs Wellcome Fund Career Award for Medical Scientists, and is an Irma T. Hirschl scholar. S.J.E. is an investigator with The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: A transcriptional microarray analysis has been deposited in the NCBI GEO database with accession number GSE22607.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009559107/-/DCSupplemental.
References
- 1.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 3.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211(1):90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 4.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 5.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 6.Tahara H, Sato E, Noda A, Ide T. Increase in expression level of p21sdi1/cip1/waf1 with increasing division age in both normal and SV40-transformed human fibroblasts. Oncogene. 1995;10:835–840. [PubMed] [Google Scholar]
- 7.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 8.Peng C, et al. BRD7 suppresses the growth of Nasopharyngeal Carcinoma cells (HNE1) through negatively regulating beta-catenin and ERK pathways. Mol Cell Biochem. 2007;303(1-2):141–149. doi: 10.1007/s11010-007-9466-x. [DOI] [PubMed] [Google Scholar]
- 9.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia W, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Zhang J, Chen X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem. 2007;282:37429–37435. doi: 10.1074/jbc.M706039200. [DOI] [PubMed] [Google Scholar]
- 12.Roberts CW, Orkin SH. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer. 2004;4(2):133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 13.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 14.Peng C, et al. The transcriptional regulation role of BRD7 by binding to acetylated histone through bromodomain. J Cell Biochem. 2006;97:882–892. doi: 10.1002/jcb.20645. [DOI] [PubMed] [Google Scholar]
- 15.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, et al. Promoter methylation inhibits BRD7 expression in human nasopharyngeal carcinoma cells. BMC Cancer. 2008;8:253. doi: 10.1186/1471-2407-8-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decristofaro MF, et al. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186(1):136–145. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Wong AK, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 19.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: Correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 20.Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10(2):142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Suh KS, Lo AM, De Luca LM. p21WAF1/CIP1 is a common transcriptional target of retinoid receptors: Pleiotropic regulatory mechanism through retinoic acid receptor (RAR)/retinoid X receptor (RXR) heterodimer and RXR/RXR homodimer. J Biol Chem. 2007;282:29987–29997. doi: 10.1074/jbc.M701700200. [DOI] [PubMed] [Google Scholar]
- 22.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 23.Drost J, et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 24.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, et al. Transcriptional regulation of BRD7 expression by Sp1 and c-Myc. BMC Mol Biol. 2008;9:111. doi: 10.1186/1471-2199-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 28.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 29.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.