Abstract
Translational regulation plays a critical role in the control of cell growth and proliferation. A key player in translational control is eIF4E, the mRNA 5′ cap-binding protein. Aberrant expression of eIF4E promotes tumorigenesis and has been implicated in cancer development and progression. The activity of eIF4E is dysregulated in cancer. Regulation of eIF4E is partly achieved through phosphorylation. However, the physiological significance of eIF4E phosphorylation in mammals is not clear. Here, we show that knock-in mice expressing a nonphosphorylatable form of eIF4E are resistant to tumorigenesis in a prostate cancer model. By using a genome-wide analysis of translated mRNAs, we show that the phosphorylation of eIF4E is required for translational up-regulation of several proteins implicated in tumorigenesis. Accordingly, increased phospho-eIF4E levels correlate with disease progression in patients with prostate cancer. Our findings establish eIF4E phosphorylation as a critical event in tumorigenesis. These findings raise the possibility that chemical compounds that prevent the phosphorylation of eIF4E could act as anticancer drugs.
Keywords: PTEN, translational control
Aberrations in the control of mRNA translation initiation have been documented in many tumor types (1–4). Translation initiation is controlled in part by eIF4E, the mRNA 5′ cap-binding protein. eIF4E is a proto-oncogene, inasmuch as its overexpression in immortalized rodent fibroblasts or human epithelial cells causes transformation (5, 6), and in mouse models its overexpression engenders tumor formation (7, 8). eIF4E is phosphorylated by the MNK1/2 serine/threonine kinases, which are activated in response to mitogenic and stress signaling downstream of ERK1/2 and p38 MAP kinase, respectively (9, 10). eIF4E phosphorylation at serine 209 by MNK1/2 promotes its transformation activity (11, 12). To study the role of eIF4E phosphorylation in tumorigenesis in the whole organism, we generated a knock-in (KI) mouse in which eIF4E serine 209 was mutated to alanine. Here, we show that mouse embryonic fibroblasts (MEFs) isolated from eIF4ES209A/S209A embryos display a marked resistance to oncogene-induced transformation. Furthermore, the mutant mice are viable, but are resistant to development of Pten loss–induced prostate cancer, and this resistance is associated with a decrease in MMP3, CCL2, VEGFC, and BIRC2 proteins. Moreover, eIF4E is highly phosphorylated in hormone-refractory prostate cancer, which correlates with poor clinical outcome. These results demonstrate the importance of eIF4E phosphorylation in tumorigenesis and validate the eIF4E phosphorylation pathway as a potential therapeutic target for cancer.
Results
Ser209 Is the Only Phosphorylation Site in eIF4E.
To address the role of eIF4E phosphorylation in tumorigenesis, a knock-in (KI) mouse in which serine 209 was replaced by an alanine residue was generated. The strategy and targeting vector construction for the generation, selection, and genotyping of the S209A mice is shown in Fig. S1. The eIF4ES209A/S209A mice (referred to as KI mice hereafter) showed no obvious phenotype. To determine whether S209 is the only phosphorylation site on eIF4E, orthophosphate labeling of MEFs isolated from WT and KI littermate embryos was performed. Phosphorous 32–radiolabeled eIF4E was detected by immunoprecipitation in only WT MEFs (Fig. 1A). As expected, TPA stimulation, which activates MNK (13), induced a twofold increase in eIF4E phosphorylation (Fig. 1A). Thus, mutating S209 abrogates eIF4E phosphorylation.
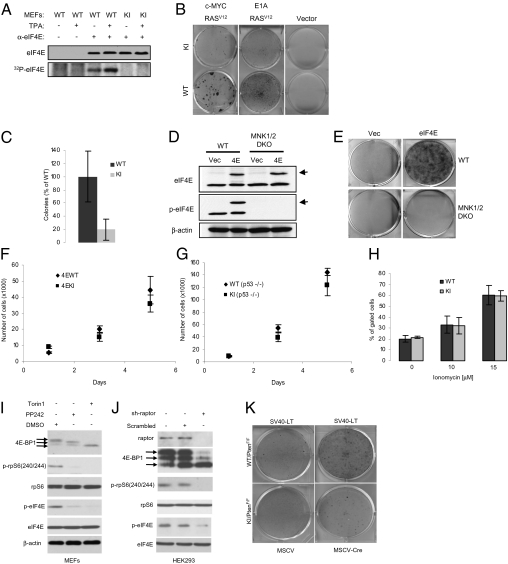
Fig. 1.
KI MEFs are resistant to malignant transformation. (A) Untreated or TPA-treated (100 ng/mL) MEFs were labeled with 32P-orthophosphate for 2 h as described in Materials and Methods. Cells were lysed and the supernatant was incubated with control (preimmune serum) or eIF4E antibody. Immunoprecipitated proteins were resolved by SDS/PAGE followed by autoradiography and Western blotting. (B) MEFs infected with the indicated retroviruses were grown on a monolayer and focus formation was determined after 10 d in culture by methylene blue staining. Similar results were obtained with eight independent pairs of MEFs. (C) WT and KI MEFs infected with c-MYC and RASV12 expression vectors were grown in soft agar and the total number of colonies consisting of more than eight cells were counted (six wells for each pair of MEFs). WT MEFs formed significantly more colonies than KI MEFs (two-tailed Student t test, P = 0.037). (D) Cell lysates from WT and MNK1/2 DKO MEFs expressing HA-eIF4E were resolved by SDS/PAGE followed by Western blotting. Arrows indicate the slower migrating band corresponding to the HA-tagged eIF4E. (E) MEFs infected with the indicated retroviruses were grown on a monolayer and focus formation was determined after 10 d in culture by methylene blue staining. (F and G) Cells (1 × 105) were seeded on d 0 and counted on d 1, 3, and 5. Data points represent mean ± SD of three independent experiments. (H) Cells (1 × 106) were seeded and subjected to treatment for 24 h with the indicated concentrations of ionomycin. Floating and attached cells were collected, and the double-positive Annexin V/PI subpopulation of cells was gated. No significant differences were observed between WT and KI MEFs (two-tailed Student t test: not treated, P = 0.511; 10 μM, P = 0.329; 15 μM, P = 0.952). Similar results were obtained in three independent experiments. (I) MEFs were serum starved for 6 h in the presence of vehicle (DMSO), PP242 (2.5 μM), or Torin1 (250 nM), serum-stimulated for 30 min, and the phosphorylation and levels of indicated proteins were determined by Western blotting. (J) Raptor was silenced in HEK293 cells by shRNA (sh-raptor). As a control, cells were infected with a lentivirus encoding a scrambled shRNA. Expression and the phosphorylation levels of the indicated proteins were determined by Western blotting. (K) WT/PTENF/F and KI/PTENF/F MEFs infected with the indicated retroviruses were grown as a monolayer and focus formation was determined after 10 d in culture by methylene blue staining. Similar results were obtained from two independent experiments each done in triplicate.
eIF4E-KI MEFs Are Resistant to RAS-Induced Transformation.
RAS is an upstream activator of MNK1 and MNK2 through ERK-1 and -2 (9, 10). Therefore, it was pertinent to determine whether transformation by RAS is facilitated by eIF4E phosphorylation. To this end, the two-oncogene transformation assay was performed with retroviruses expressing RASV12 (RAS containing the activating mutation G12V) together with c-MYC or E1A. Experiments were carried out in primary MEFs between passages three and five. Strikingly, KI MEFs infected with a combination of retroviruses expressing RASV12 and c-MYC formed approximately fivefold fewer foci than WT MEFs (Fig. 1B). A similar difference in transformation efficiency was observed when MEFs were infected with retroviruses expressing the adenovirus E1A together with RASV12 (Fig. 1B). WT and KI MEFs infected with an empty retrovirus failed to form foci (Fig. 1B). Anchorage-independent growth of WT and KI MEFs transduced with activated RASV12 and c-MYC was determined by colony formation in soft agar. The assay was performed with three different pairs of MEFs, each isolated from embryos of a different pregnant mouse. KI MEFs formed four- to 10-fold fewer colonies than WT MEFs (Fig. 1C). These results demonstrate that eIF4E phosphorylation on serine 209 is required for efficient transformation by RAS. To further demonstrate the importance of eIF4E phosphorylation for cellular transformation, we performed transformation assays using immortalized WT or MNK1/2 DKO MEFs and HA-tagged eIF4E as the transforming oncogene. HA-tagged eIF4E was expressed at similar levels in WT and MNK1/2 DKO MEFs, and as expected no phosphorylation of endogenous or HA-tagged eIF4E was detected by Western blotting in MNK1/2 DKO MEFs (Fig. 1D). WT MEFs overexpressing HA-eIF4E formed foci after 10 d in culture. Strikingly, MNK1/2 DKO MEFs were completely resistant to transformation by HA-eIF4E (Fig. 1E). These findings clearly demonstrate that eIF4E transforming activity is absolutely dependent on phosphorylation by the MNK1/2 kinases.
Abrogating eIF4E Phosphorylation Does Not Impair Cell Proliferation.
The resistance of KI MEFs to transformation could be explained by an inherently reduced proliferative rate of KI MEFs compared with WT MEFs. To investigate this possibility, growth curves using primary MEFs at passage three were performed. No difference in growth between WT and KI MEFs over a 5-d period was detected (Fig. 1F). Primary MEFs are slow-growing cells and consequently a small effect on growth could have been missed. We therefore crossed the WT and the KI mice with p53-null mice to generate immortalized WT and KI MEFs, which grow faster than primary MEFs. Similar growth curves were obtained for both WT and KI p53-null MEFs (Fig. 1G). Another possible explanation for the reduced transformation efficiency of KI MEFs is that they are more susceptible to apoptosis. To address this possibility, WT and KI primary MEFs were treated with the apoptosis inducing ionophore ionomycin. Apoptotic and necrotic cells were visualized by dual annexin V–propidium iodide staining. No difference in the percentage of apoptotic and necrotic cells was observed between WT and KI MEFs after ionomycin treatment (Fig. 1H). In addition, there was no difference in cell cycle progression or apoptosis between WT and KI cells transformed by c-MYC and RASV12 (Fig. S2 A–C). These results indicate that the resistance of KI MEFs to Ras-induced transformation cannot be explained by decreased proliferation or increased cell death.
eIF4E Phosphorylation Is Regulated by Mammalian Target of Rapamycin Complex 1 and Contributes to Transformation via the PI3K Pathway.
Another pathway that controls eIF4E activity is the PI3K pathway via the phosphorylation of the suppressor 4E-BPs by mammalian target of rapamycin complex 1 (mTORC1) (1, 14). The loss of the tumor suppressor PTEN leads to AKT, and subsequent mTORC1 activation (15). To determine whether eIF4E phosphorylation is impacted by mTORC1, we treated immortalized MEFs with two active-site inhibitors of mTORC1, Torin1 (16) and PP242 (17). Inhibition of the mTORC1 kinase was confirmed by a decrease in the phosphorylation of the small ribosomal subunit protein S6 (p-rpS6) and 4E-BP1 (Fig. 1I). Interestingly, eIF4E phosphorylation was drastically diminished following mTORC1 inhibition (Fig. 1I). To confirm that the decrease in eIF4E phosphorylation was not a result of nonspecific effects of the two inhibitors, we used shRNA-mediated knockdown of RAPTOR as an alternative approach for inhibiting mTORC1 activity in 293 cells. Consistent with the results obtained with Torin1 and PP242, lowering the amount of RAPTOR caused a marked decrease in eIF4E phosphorylation (Fig. 1J). These results can be readily explained by the fact that dephosphorylation of 4E-BPs increases their binding to eIF4E, thus preventing eIF4E binding to eIF4G, on which MNK docks (18). We investigated further the role of eIF4E phosphorylation control downstream of mTORC1 by generating WT and KI MEFs in which both alleles of PTEN were deleted via CRE-mediated recombination (15). MEFs were immortalized with SV40 largeT and subsequently infected with a CRE-expressing retrovirus to delete Pten. Loss of PTEN in combination with SV40 large T expression caused malignant transformation as determined by focus formation in WT MEFs, whereas KI MEFs were resistant to transformation, as only a few foci were detected (Fig. 1K). These results demonstrate that eIF4E phosphorylation is involved in PTEN loss–driven cellular transformation.
eIF4E-KI Mice Are Resistant to PTEN Loss-Induced Prostate Cancer.
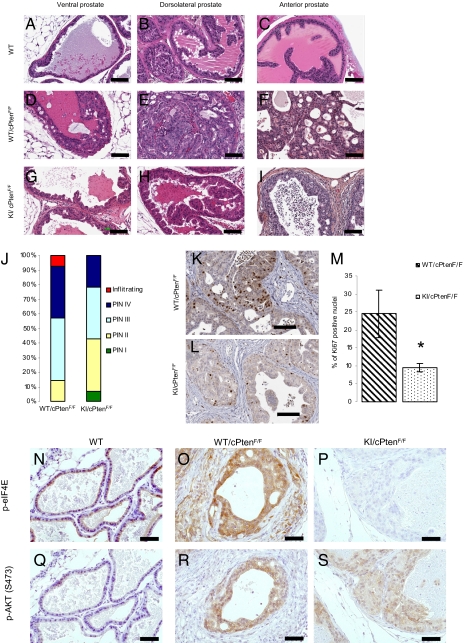
Because Pten deletion is expected to result in increased eIF4E phosphorylation, we investigated the importance of eIF4E phosphorylation in tumor formation in vivo in a mouse model of prostate cancer in which the tumor suppressor Pten is deleted in the prostate epithelium (PtenFlox/Flox; PB-Cre4) (15). These mice develop invasive carcinoma by 5 to 8 mo of age with complete penetrance (15). KI mice were crossed with Pten-conditional KO mice to generate eIF4E WT/PtenFlox/Flox/PB-Cre4 and eIF4E KI/PtenFlox/Flox/PB-Cre4, hereafter referred to as WT/cPtenF/F and KI/cPtenF/F, respectively. Seven mice of each genotype were killed between the ages of 5 and 7 mo (Table S1). Cancerous lesions, mainly prostatic intraepithelial neoplasia (PIN), were graded from I to IV (Fig. S4 shows representative sections) (19, 20). Analysis of H&E-stained sections of all prostate lobes in 5- to 7-mo-old mice shows that KI/cPtenF/F mice had a lower incidence of high grade PIN (represented by PIN IV lesions) than WT/cPtenF/F mice (Fig. 2 A–I). There was a significant difference in the grade of lesions observed between WT/cPtenF/F and KI/cPtenF/F mice (P = 0.038, Mann–Whitney U test). The distribution of the lesions in WT/cPtenF/F and KI/cPtenF/F is shown in Fig. 2J. It is noteworthy that no infiltrating adenocarcinoma was observed in KI/cPtenF/F. It is unlikely that the differences seen in the severity of the lesions between WT/cPtenF/F and KI/cPtenF/F are caused by a small delay in tumor appearance, as the median age at sacrifice time was 25 wk for the KI/cPtenF/F compared with 20 wk for the WT/cPtenF/F mice (Table S1). The differences in the severity of the lesions between the WT and KI groups were also determined by the number of proliferative cells as detected by Ki67 staining in the dorsolateral prostate (DLP) of mice from each genotype (Fig. 2 K–L). DLP was chosen because it is histologically and functionally the most closely related to the peripheral zone of the human prostate (21), which is the region where most prostate cancers occur in men (22, 23). There was a significant decrease (P = 0.013) in the number of Ki67-positive nuclei between WT/cPtenF/F (24.5 ± 6.6%) and KI/cPtenF/F (9.5 ± 1.1%) mice (Fig. 2M). In addition, eIF4E was highly phosphorylated on Ser209 in PIN IV lesions from WT/cPtenF/F mice (Fig. 2 N–P). AKTSer473 phosphorylation was increased to the same extent in WT/cPtenF/F and KI/cPtenF/F mice (Fig. 2 Q–S), suggesting that differences in lesion grade are not caused by a weaker activation of the PI3K pathway in the KI mice. Taken together, these results demonstrate that eIF4E phosphorylation plays an important role in Pten loss–induced tumorigenesis.
Fig. 2.
KI mice are resistant to Pten loss–induced prostate cancer. (A–I) Representative H&E staining of sections from the anterior (AP), ventral (VP), and DLP lobes of the prostate of WT, WT/cPtenF/F, and KI/cPtenF/F from 5- to 6-mo-old mice (no histological difference was observed between WT and KI mice without Cre; Fig. S3). (Scale bar: 100 μm.) (J) Histograms represent percentage of predominant lesions in a cohort of seven WT/cPtenF/F and seven KI/cPtenF/F. (K and L) Representative images of Ki67 staining of DLP sections from WT/cPtenF/F and KI/cPtenF/F mice (5–6 mo old). (Scale bar: 100 μm.) (M) Slides were scanned with the Aperio Scanscope and nuclear staining was quantified with the Image Scope software nuclear algorithm. At least 4,000 nuclei per mouse were analyzed (n = 4 mice per group). Student t test (two-tailed) was performed. (N–S) Ventral prostate sections from WT, WT/cPtenF/F, and KI/cPtenF/F mice were used for IHC with the indicated antibodies. (Scale bar: 50 μm.)
eIF4E Phosphorylation Increases the Translation Efficiency of a Subset of mRNAs Encoding Protumorigenic Factors.
To study the molecular basis that underlies the eIF4E-KI resistance to tumorigenesis at the translational level, polysome profiling was performed on WT and KI immortalized MEFs. The changes in distribution of mRNAs along a sucrose density gradient between WT and KI MEFs were studied using DNA oligonucleotide microarrays. mRNAs in KI MEFs that shifted to lighter fractions relative to WT MEFs in the density gradient, after correcting for total cytoplasmic mRNA levels, are expected to be translated less efficiently in KI MEFs. A list of 35 mRNAs that exhibited the most pronounced shifts is shown in Table S2. Among these mRNAs are the chemokines Ccl2 and Ccl7 that are implicated in tumor progression. Targeting CCL2 with a neutralizing antibody caused prostate tumor regression (24–26). The list also includes mRNAs encoding the matrix metalloproteinases (MMPs) MMP3 and MMP9, which are overexpressed in PC3 prostate cancer cells (27), and promote invasion and metastasis by rearrangement of the ECM (28, 29). Other mRNAs encode the inhibitor of apoptosis, baculoviral IAP repeat–containing protein 2 (BIRC2), and the growth factor VEGFC. To determine whether the differences in polysome sedimentation between WT and KI MEFs are reflected in changes in the abundance of protein, we examined VEGFC, BIRC2, MMP3, and nuclear factor of κ-light polypeptide gene enhancer in B-cells inhibitor-α (NFKBIA; also known as IκBα) proteins by Western blotting (Fig. 3A). A 1.7- to 4.8-fold reduction in the amount of these proteins was observed in KI MEFs compared with WT MEFs, whereas the amount of AKT, eIF4E, and β-actin remained unchanged. In addition, inhibiting Mnk1/2 kinases in two prostate cancer cell lines with the inhibitor CGP57380 (30) led to a decrease in the amount of VEGFC, BIRC2, and NFKBIA proteins (Fig. S5A). We also examined the amount of MMP3 in DLP extracts isolated from KI/cPtenF/F and WT/cPtenF/F mice. A fourfold decrease in MMP3 protein in the DLP of KI/cPtenF/F compared with WT/cPtenF/F was detected, whereas the protein was barely detected in DLP from WT mice (Fig. 3B). Considering that the KI mice develop fewer PIN IV lesions, characterized by a loss of the fibromuscular layer around the prostatic ducts (20), as seen by staining for smooth muscle actin (SMA; Fig. 3 C–E), MMP3 staining by immunohistochemistry (IHC) was performed. Consistent with the Western blotting data, MMP3 staining intensity was weaker in the prostate of KI/cPtenF/F compared with WT/cPtenF/F mice (Fig. 3 F–H). In addition, CCL2 staining in the prostate of KI/cPtenF/F was weaker compared with WT/cPtenF/F mice (Fig. 3 I–K). These results demonstrate that several mRNAs important for tumor progression are differentially translated in WT versus KI MEFs. Impairment in the remodeling of the extracellular matrix or differences in paracrine signaling could be possible explanations for the resistance of KI mice to Pten loss–induced tumorigenesis.
Fig. 3.
Differential expression of selected proteins in prostate from WT versus KI mice. (A) Serum starved WT and KI MEFs were serum-stimulated for 2 h and cell lysates were resolved by SDS/PAGE followed by Western blotting with the indicated antibodies. (B) DLP extracts were resolved by SDS/PAGE and Western blotting with the indicated antibodies was performed. (C–K) Sections of the prostate from WT, WT/cPtenF/F, and KI/cPtenF/F mice were used for IHC with the indicated antibodies. VP sections were used for detection of SMA and MMP3. CCL2 was detected in the anterior prostate. (Scale bar: 50 μm.)
eIF4E Phosphorylation Is Associated with High Gleason Score and Strongly Correlates with MMP3 Expression in a Cohort of Patients with Prostate Cancer.
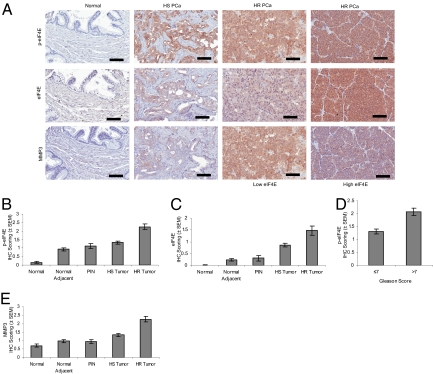
Next, we investigated the correlation between eIF4E phosphorylation in human prostate cancer and disease progression. A tissue microarray (TMA) constructed with human prostate cancer (PCa) samples including patients presenting primary and hormone-refractory PCa (HR PCa) was used (31, 32). Both eIF4E and phospho-eIF4E staining intensity increased gradually from normal to PIN, to hormone-sensitive (HS), and further to HR PCa tumors (Fig. 4A shows representative staining). Increased phospho-eIF4E and total eIF4E staining was significantly associated with HR tumors (Fig. 4 B and C). Statistical analysis of the association between phospho-eIF4E staining and Gleason grading demonstrated that tumors presenting with a Gleason score greater than 7 displayed a statistically significant increase in staining compared with tumors with a Gleason score of 7 or lower (Fig. 4D). Thus, there is a strong association between the degree of eIF4E phosphorylation and progression of prostate cancer to its deadliest stage. To determine whether MMP3 could also serve as a marker of prostate cancer progression, we stained the same TMA for the presence of MMP3 (Fig. 4A shows representative staining). MMP3 expression was significantly higher in hormone-sensitive and HR tumors compared with PIN and normal tissue (Fig. 4E). Furthermore, analysis using Spearman rank correlation demonstrated that MMP3 is significantly associated with phospho-eIF4E (ρ = 0.396) and eIF4E (ρ = 0.524) in tumor tissues (Table S3). In addition, we performed staining for p-ERK and p-AKT to examine the association between upstream signaling activity and eIF4E phosphorylation. As expected, eIF4E phosphorylation correlated with p-ERK (ρ = 0.266) and p-AKT (ρ = 0.321). Strikingly, MMP3 was not significantly associated with p-ERK (ρ = 0.006) and p-AKT (ρ = 0.110), suggesting a stronger association between p-eIF4E and MMP3 than with upstream MAPK and PI3K signaling.
Fig. 4.
p-eIF4E correlates with progression to HR PCa. eIF4E, phospho-eIF4E, and MMP3 were detected by IHC in human prostate cancer TMAs. The intensity of staining was scored from 0 to 4 for each core and data were analyzed as described in Materials and Methods. (A) Representative images of staining intensity obtained for eIF4E and p-eIF4E. (Scale bar: 100 μm.) (B and C) eIF4E and phospho-eIF4E immunoreactivity shows a gradual increase in intensity from the normal, normal adjacent, PIN, HS tumor, and HR tumor tissues. Significant statistical differences were found between the different histopathological groups (P < 0.001, Kruskal-Wallis test). (D) The mean intensity difference of phospho-eIF4E in cases with Gleason score greater than 7 compared with Gleason scores of 7 or lower is statistically significant (P < 0.05, Mann–Whitney U test). (E) MMP3 immunoreactivity is gradually increased in intensity from PIN to HS tumor and HR tumor tissues. Significant statistical differences exist between the different histopathological groups (P < 0.001, Kruskal-Wallis test).
Discussion
We show that eIF4E phosphorylation promotes prostate tumor development and progression in mice. mRNAs that are less well translated in the absence of eIF4E phosphorylation include those that encode for proteins involved in the remodeling of the ECM, inhibition of apoptosis, and cellular growth and proliferation. The decrease in MMP3 and the chemokine CCL2 is consistent with the reduction in invasiveness observed in the KI mice prostate tumors. eIF4E phosphorylation is strongly associated with high Gleason score (>7) and HR prostate cancer in a patient cohort, which predicts poor survival (33, 34). It is well established that eIF4E is overexpressed in many human tumors, including prostate cancer (35), and this was also shown here by using TMAs of patients with prostate cancer. However, an increase in the amount of eIF4E is not sufficient for transformation, as eIF4E must be phosphorylated by MNKs. The Oncomine database (36) documents that MNK2 is overexpressed 1.5- to 4.4-fold in HR and metastatic prostate tumors (37–39). Moreover, MNK activity promotes proliferation in prostate cancer cells (40). This is expected to contribute to the increase in eIF4E phosphorylation seen in tumors with high Gleason scores, considering that MNK2 constitutively phosphorylates eIF4E (41).
Previous work has demonstrated that eIF4E phosphorylation is not required for translation under standard growth conditions (41). However, many studies documented a positive correlation between increased eIF4E phosphorylation and cell proliferation and enhanced translation (42, 43). Here, we identified mRNAs that are preferentially responsive to eIF4E phosphorylation for maximal translation. Future work will be needed to identify the mechanisms that render the translation of these mRNAs more sensitive to eIF4E phosphorylation. Several studies have demonstrated that p-eIF4E binds with lower affinity to the cap structure (44–46). Although this finding is counterintuitive, it was proposed that the decreased affinity could stimulate translation by releasing eIF4E from the cap, akin to the mechanism of promoter clearing during transcription initiation (47).
In a preclinical mouse model of prostate cancer, inhibition of mTOR and ERK signaling pathways using a combination therapy of rapamycin and PD0325901 impaired tumor growth (48). We demonstrated that the phosphorylation of eIF4E is regulated by these two signaling pathways; therefore, inhibition of eIF4E phosphorylation as a treatment for cancer is a very intriguing idea, considering that it does not have a conspicuous effect on normal proliferation.
Materials and Methods
IHC.
IHC was performed using the Vectastain Elite ABC kit (Vector Laboratories) according to the manufacturer's instructions. Antibodies were used at the following dilutions: p-eIF4E 1:500 (Novus Biologicals), MMP3 1:250 (Abcam), CCL2 1:100 (Novus Biologicals), eIF4E 1:400 (CST), p-AKT S473 1:50 (CST), and Ki67 1:2,000 (Novocastra). SMA detection was performed with the Mouse-On-Mouse kit (Vector Laboratories) according to the manufacturer's instructions. SMA antibody (Dako) was used at 1:500.
Western Blotting.
Primary antibodies were used at the following dilutions: pan-AKT 1:1,000 (CST), p-AKT S473 1:1,000 (CST), eIF4E 1:1,000 (CST), NFKBIA 1:1,000 (CST), BIRC2 1:1,000 (Novus Biologicals), p-eIF4E 1:5,000 (Novus Biologicals), MMP3 1:500 (Abcam), β-actin 1:10,000 (Sigma), VEGFC 1:250 (Abcam), RAPTOR 1:1,000 (CST), 4E-BP1 1:1,000 (CST), rpS6 1:5,000 (Santa Cruz Biotechnology), and p-rpS6[240/244] 1:1,000 (CST).
Transformation Assays.
c-Myc-, E1A-, RasV12-, SV40 largeT-, CRE-, and HA-eIF4E–expressing retroviruses were generated by transfecting packaging Phoenix cells with the following plasmids: pBabe-c-Myc, pBabe-HA-eIF4E, pLpc-E1A/RasV12, pWzl-RasV12, pMSCV-CRE, and pSV40T (Myc-, E1A-, and RasV12-expressing plasmids were gifts of Scott Lowe, Cold Spring Harbor Laboratory, NY; pSV40T was a gift of Julian Downward, London Research Institute, UK). MEFs were infected with retroviruses three times over a 48-h period and pools of infected cells were selected by two rounds of selection with puromycin for pBabe, hygromycin for pWzl, or G418 for pMSCV.
Supplementary Material
Acknowledgments
We thank Rachelle Lee Dillon for assistance with microscope image analysis, Mustapha Riad for help with histopathological analysis, Rikiro Fukunaga for providing the MNK1/2 DKO mice, and Mark Livingstone and Ivan Topisirovic for comments on the manuscript. This work was funded by National Cancer Institute of Canada (Canada Cancer Society) Grant 016208 (to N.S.) and National Institutes of Health/National Cancer Institute Grant CA84292 (to P.P.P.). L.F. is a Research Fellow of the Terry Fox Foundation through Award 19676 from the National Cancer Institute of Canada. O.L. is supported by a fellowship from the Knut and Alice Wallenberg foundation. F.S. holds the Université de Montréal Chair in Prostate Cancer. I.H.K. holds an award from the Fonds de la recherche en santé du Québec.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE17451).
*This Direct Submission article had a prearranged editor.
See Commentary on page 13975.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005320107/-/DCSupplemental.
References
- 1.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 2.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 3.Clemens MJ. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene. 2004;23:3180–3188. doi: 10.1038/sj.onc.1207544. [DOI] [PubMed] [Google Scholar]
- 4.Schneider RJ, Sonenberg N. Translational control in cancer development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 401–432. [Google Scholar]
- 5.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 6.Avdulov S, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 8.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 9.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64:8639–8642. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 12.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waskiewicz AJ, et al. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- 15.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyronnet S, et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couto SS, et al. Simultaneous haploinsufficiency of Pten and Trp53 tumor suppressor genes accelerates tumorigenesis in a mouse model of prostate cancer. Differentiation. 2009;77:103–111. doi: 10.1016/j.diff.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JH, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price D. Comparative aspects of development and structure in the prostate. Natl Cancer Inst Monogr. 1963;12:1–27. [PubMed] [Google Scholar]
- 22.Haffner J, et al. Peripheral zone prostate cancers: Location and intraprostatic patterns of spread at histopathology. Prostate. 2009;69:276–282. doi: 10.1002/pros.20881. [DOI] [PubMed] [Google Scholar]
- 23.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol. 1988;12:897–906. doi: 10.1097/00000478-198812000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Li X, et al. A destructive cascade mediated by CCL2 facilitates prostate cancer growth in bone. Cancer Res. 2009;69:1685–1692. doi: 10.1158/0008-5472.CAN-08-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loberg RD, et al. Targeting CCL2 with systemic delivery of neutralizing antibodies induces prostate cancer tumor regression in vivo. Cancer Res. 2007;67:9417–9424. doi: 10.1158/0008-5472.CAN-07-1286. [DOI] [PubMed] [Google Scholar]
- 26.Van Damme J, Proost P, Lenaerts JP, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S, Singh UP, Grizzle WE, Lillard JW., Jr CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest. 2004;84:1666–1676. doi: 10.1038/labinvest.3700181. [DOI] [PubMed] [Google Scholar]
- 28.Wilson TJ, Singh RK. Proteases as modulators of tumor-stromal interaction: primary tumors to bone metastases. Biochim Biophys Acta. 2008;1785:85–95. doi: 10.1016/j.bbcan.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowlett RM, et al. MNK kinases regulate multiple TLR pathways and innate proinflammatory cytokines in macrophages. Am J Physiol Gastrointest Liver Physiol. 2008;294:G452–G459. doi: 10.1152/ajpgi.00077.2007. [DOI] [PubMed] [Google Scholar]
- 31.Le Page C, Koumakpayi IH, Alam-Fahmy M, Mes-Masson AM, Saad F. Expression and localisation of Akt-1, Akt-2 and Akt-3 correlate with clinical outcome of prostate cancer patients. Br J Cancer. 2006;94:1906–1912. doi: 10.1038/sj.bjc.6603184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diallo JS, et al. NOXA and PUMA expression add to clinical markers in predicting biochemical recurrence of prostate cancer patients in a survival tree model. Clin Cancer Res. 2007;13:7044–7052. doi: 10.1158/1078-0432.CCR-07-1224. [DOI] [PubMed] [Google Scholar]
- 33.Lau WK, Bergstralh EJ, Blute ML, Slezak JM, Zincke H. Radical prostatectomy for pathological Gleason 8 or greater prostate cancer: Influence of concomitant pathological variables. J Urol. 2002;167:117–122. [PubMed] [Google Scholar]
- 34.Oefelein MG, Agarwal PK, Resnick MI. Survival of patients with hormone refractory prostate cancer in the prostate specific antigen era. J Urol. 2004;171:1525–1528. doi: 10.1097/01.ju.0000118294.88852.cd. [DOI] [PubMed] [Google Scholar]
- 35.Graff JR, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69:3866–3873. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 39.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Bianchini A, et al. Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis. 2008;29:2279–2288. doi: 10.1093/carcin/bgn221. [DOI] [PubMed] [Google Scholar]
- 41.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flynn A, Proud CG. Insulin and phorbol ester stimulate initiation factor eIF-4E phosphorylation by distinct pathways in Chinese hamster ovary cells overexpressing the insulin receptor. Eur J Biochem. 1996;236:40–47. doi: 10.1111/j.1432-1033.1996.00040.x. [DOI] [PubMed] [Google Scholar]
- 43.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 44.Scheper GC, et al. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem. 2002;277:3303–3309. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- 45.Slepenkov SV, Darzynkiewicz E, Rhoads RE. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: Evidence for a one-step binding mechanism. J Biol Chem. 2006;281:14927–14938. doi: 10.1074/jbc.M601653200. [DOI] [PubMed] [Google Scholar]
- 46.Zuberek J, et al. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003;9:52–61. doi: 10.1261/rna.2133403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–5359. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinkade CW, et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008;118:3051–3064. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frederickson RM, Montine KS, Sonenberg N. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol Cell Biol. 1991;11:2896–2900. doi: 10.1128/mcb.11.5.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller E. Apoptosis measurement by annexin v staining. Methods Mol Med. 2004;88:191–202. doi: 10.1385/1-59259-406-9:191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.