Abstract
Despite expanding data sets and advances in phylogenomic methods, deep-level metazoan relationships remain highly controversial. Recent phylogenomic analyses depart from classical concepts in recovering ctenophores as the earliest branching metazoan taxon and propose a sister-group relationship between sponges and cnidarians (e.g., Dunn CW, Hejnol A, Matus DQ, et al. (18 co-authors). 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749). Here, we argue that these results are artifacts stemming from insufficient taxon sampling and long-branch attraction (LBA). By increasing taxon sampling from previously unsampled nonbilaterians and using an identical gene set to that reported by Dunn et al., we recover monophyletic Porifera as the sister group to all other Metazoa. This suggests that the basal position of the fast-evolving Ctenophora proposed by Dunn et al. was due to LBA and that broad taxon sampling is of fundamental importance to metazoan phylogenomic analyses. Additionally, saturation in the Dunn et al. character set is comparatively high, possibly contributing to the poor support for some nonbilaterian nodes.
Keywords: multigene analysis, EST, Metazoa, Ctenophora, Porifera, long-branch attraction, saturation
Resolving the relationships of deep branching metazoan lineages is critical if we are to understand early animal evolution. Unraveling these relationships through the analysis of large-scale molecular data sets has recently given birth to the field of phylogenomics (e.g., Philippe et al. 2005). Despite significant advances in this field, recent studies have generated contradictory results regarding relationships within and between early diverging metazoan lineages: cnidarians, ctenophores (comb jellies), sponges, placozoans (anatomically the simplest extant metazoans), and bilaterians. Placozoans have historically been regarded by some as relicts of the metazoan ancestor (see summary by Schierwater 2005), and some recent analyses place Placozoa at the base of a group of nonbilaterian animals (Dellaporta et al. 2006; Schierwater et al. 2009). However, recent whole-genome (Srivastava et al. 2008) and phylogenomic (Philippe et al. 2009) analyses including Trichoplax recovered sponges as the sister group to all other metazoans in accordance with morphological analyses (Ax 1996). Such contradictory hypotheses regarding nonbilaterian metazoan relationships prevent a consensus view of metazoan evolution, a goal that is of fundamental importance if we hope to fully understand the early evolution of animals (for an overview see Erpenbeck and Wörheide 2007).
A recent phylogenomic analysis adds further controversy to this debate (Dunn et al. 2008) (c.f., Hejnol et al. 2009). Their outcome is highly unusual as sponges form a clade with the Cnidaria, while the ctenophores (despite being morphologically derived) are proposed to be the earliest branching metazoan taxon. As suggested by Philippe et al. (2009), we hypothesized that a long-branch attraction (LBA) artifact was responsible for these controversial findings due to insufficient ingroup sampling and an inappropriate choice of outgroup taxa. Furthermore, the Placozoa are conspicuously absent from the Dunn et al. (2008) data set, and sponges are represented by only one Demospongiae and one Homoscleromorpha with no representatives of the remaining two extant sponge classes: Calcarea (Calcispongiae or calcareous sponges) and Hexactinellida (glass sponges). Sparse taxon sampling is a common pitfall of phylogenetic analyses (Lecointre et al. 1993) and is largely responsible for the lack of a robustly supported nonbilaterian metazoan phylogeny (Erpenbeck and Wörheide 2007). With a largely different gene set (only 45 genes in common with the 150 gene set of Dunn et al. 2008) and an increased sampling of nonbilaterian species, Philippe et al. (2009) obtained monophyletic sponges as the first-diverging metazoan lineage and a sister-group relationship between the Cnidaria and the Ctenophora.
To test whether insufficient sampling of nonbilaterian taxa and inappropriate outgroup choice adversely influenced the analyses performed by Dunn et al. (2008), we reanalyzed their 64-taxon matrix cleared of instable taxa (leaf stability <90%) and with the following major modifications (cf. Baurain et al. 2007):
1) Ingroup taxon sampling was increased by the addition of nonbilaterian expressed sequence tag and genomic sequences. These included: 12 additional sponge taxa representing all four major sponge lineages; 1 additional ctenophore; 5 additional cnidarians (see supplementary table S1, Supplementary Material online), and Trichoplax adhaerens (Placozoa).
2) We removed outgroup taxa with long branches. Long branches in the outgroup can strongly influence the topology of early branching ingroups (Philippe and Laurent 1998; Rota-Stabelli and Telford 2008). The long branches of the fungal outgroup are not visible in the cladogram of the PhyloBayes analysis (CAT + Γ4) of Dunn et al. (see their fig. 2) but are evident in their supplementary figure S1 (Supplementary Material online). Consequently, we analyzed our data set with two sets of outgroups. First, using only choanoflagellates, the most likely sister group to all Metazoa (Carr et al. 2008), consisting of Monosiga ovata (shortest branch of outgroup taxa of Dunn et al.), Monosiga brevicollis (complete genome data), and Proterospongia sp.. Second, with more distant outgroups, such as those used by Dunn et al. (2008) (see supplementary fig. S1 and supplementary data, Supplementary Material online, for a detailed taxon list and methods used).
Furthermore, we eliminated errors (e.g., frameshifts) and refined the alignment of Dunn et al. (2008), for example, by reducing missing data and removing 2,150 ambiguously aligned positions (see supplementary data, Supplementary Material online, for detailed procedures). Our extended data set with the choanoflagellate-only outgroup consists of 80 taxa and 19,002 characters. Using this data set we performed Bayesian phylogenetic analyses under the CAT + Γ4 model (Lartillot and Philippe 2004) and subsequent nonparametric bootstrapping (cf. Philippe et al. 2009).
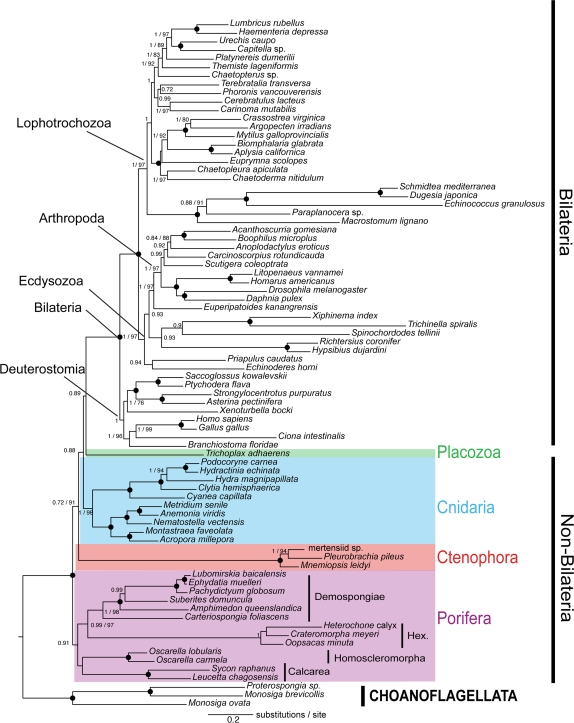
Contrary to Dunn et al. (2008), and also Hejnol et al. (2009), we recover sponges as the sister group to all other metazoan taxa (fig. 1). This is in congruence with earlier morphological (Ax 1996) and phylogenomic analyses (Philippe et al. 2009). In accordance with the latter study, we also recover sponges as a monophyletic group. The Homoscleromorpha, a taxon previously assigned to the Demospongiae (see Hooper and Van Soest 2002), are found to be the sister group to Calcarea as suggested by van Soest (1984) and Grothe (1989) based on morphology and subsequently by Dohrmann et al. (2008) based on ribosomal RNA (rRNA) data. Similarly, Hexactinellida and the remaining Demospongiae sensu stricto form a monophyletic group (Silicea sensu stricto).
FIG. 1.
Phylogenetic tree based on refinements to the Dunn et al. (2008) 64-taxon set reconstructed with PhyloBayes (Lartillot et al. 2009) under the CAT + Γ4 model. Choanoflagellates were set as outgroup and an additional 18 nonbilaterian taxa included. Posterior probabilities >0.7 are indicated followed by bootstrap support values >70. A large black dot indicates maximum support in posterior probabilities and Bayesian bootstraps (=1/100).
The basal position of ctenophores proposed by Dunn et al. (2008) was probably caused by the attraction of ctenophores to distant outgroup species, particularly fungi. In comparison, our reanalysis of the updated Dunn et al. (2008) data set with increased ingroup taxon sampling and a refined alignment indicates that LBA is reduced, independent of whether we use the choanoflagellate-only outgroup or more distant outgroups (see fig. 1; supplementary fig. S1, Supplementary Material online). This indicates that in-goup taxon sampling and probably to a lesser extent data refinement are the most important parameters affecting nonbilaterian relationships.
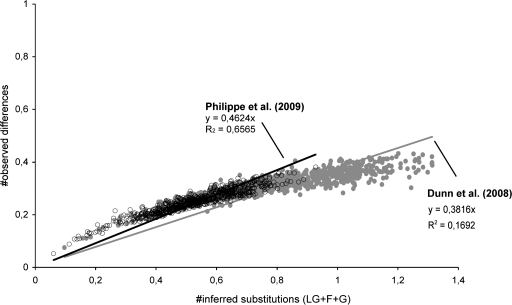
Results of our analyses indicate that sponges are the sister group to the remaining Metazoa, and Placozoa are sister group to the Bilateria. We also recover both monophyletic Ctenophores and Cnidaria, but they are paraphyletic with respect to Placozoa + Bilateria (fig. 1). This is in contrast to the findings of Philippe et al. (2009) that supported the “Coelenterata hypothesis” (cf. Haeckel 1866), that is, a monophyletic Cnidaria + Ctenophora clade and a sister-group relationship between Coelenterata and Bilateria. However, support values for the position of Ctenophora, Cnidaria, and Placozoa in our analysis are either not significant (posterior probabilities <0.9) or low (bootstrap support <70%). We suspected that character set of Dunn et al. contains a substantial amount of nonphylogenetic signal due to multiple substitutions. To test this, we conducted a saturation analysis of inferred substitutions against observed amino acid differences (fig. 2). This revealed a higher saturation in the original Dunn et al. (2008) character set (slope = 0.38×) compared with the character set of Philippe et al. (2009) (slope = 0.46×) (fig. 2). From this we conclude that despite increasing the number of nonbilaterian taxa by a factor of 3 (from 9 to 27), multiple substitutions have partly masked phylogenetic signal contributing to the incongruent results reported here with those of Philippe et al. (2009). However, with the expanded and refined data set reported here, none of these incongruencies are statistically significant, indicating that nonphylogenetic signal has been reduced with respect to the original character set of Dunn et al. (2008). Furthermore, Dunn et al. (2008) recovered high support for the sister-group relationship of ctenophores to the remaining Metazoa—based on our analyses here, this hypothesis should be rejected (with a bootstrap value of 91%).
FIG. 2.
Saturation plot of character sets. See Supplementary Material for method details. Gray line and filled dots: Dunn et al. (2008). Black line and open dots: Philippe et al. (2009).
The inclusion of additional taxa has little influence on the relationships within and between bilaterian crown groups. Three of the four differences between the findings of Dunn et al. (2008) and our results affect the relationships of a single sequence within their well-defined clades (Euprymna within Mollusca, Paraplanocera within Platyhelminthes, and Anoplodactylus among the chelicerate arthropods). None of these splits were strongly supported in the original Dunn et al. (2008) analysis. Additionally, we do not recover Panarthropoda due to a difference in the position of Tardigrada. Panarthropoda was also weakly supported in the Dunn et al. (2008) analysis (posterior probability values under WAG and CAT models were 0 and 0.86, respectively, and RAxML bootstrap support under the WAG model with 64 and 77 taxa was 4% and 2%, respectively).
Our results highlight the sensitivity of phylogenomic studies to ingroup taxon sampling and demonstrate the need for great care in the analysis and interpretation of large data sets. Character-rich analyses are thought to outperform character-poor analyses and have been suggested to be of greater importance than increased taxon sampling with regard to recovering robust metazoan phylogenies (Rokas and Carroll 2005). However, our analyses demonstrate the strong influence of taxon sampling, even though nonbilaterian taxa still remain underrepresented (Cnidaria: no Octocorallia, Ceriantharia, Cubozoa, or Staurozoa; Ctenophora: no Platyctenida, Beroida, or Cestida; just one placozoan strain etc.). The phylogenomic approach promises to reveal a well-resolved consensus metazoan tree, but it should not be assumed that a large data set will automatically produce a strong or correct phylogenetic signal (Jeffroy et al. 2006). A wide range of factors, such as saturation, LBA, the best fitting evolutionary model, and appropriate outgroup choice (Philippe et al. 2005), need to be carefully addressed before a fully resolved and robust animal tree of life will be realized.
Supplementary Material
Supplementary figure S1, supplementary table S1, and supplementary data are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This study was supported by the German Science Foundation (DFG) through the Priority program SPP 1174 “Deep Metazoan Phylogeny” (Projects Wo896/6-1, 2; Wi 2116/2-1, 2). M. Kube and his team at the Max-Planck Institute for Molecular Genetics (Berlin, Germany) are acknowledged for library construction and expressed sequence tag sequencing, as well as I. Ebersberger and his team at the Center for Integrative Bioinformatics (Vienna, Austria) for their bioinformatic processing. We thank C. Eckert for some tissue samples. H.P. gratefully acknowledges financial support by Natural Sciences and Engineering Research Council of Canada, the Canadian Research Chair Program and the Université de Montréal, and the Réseau Québecois de Calcul de Haute Performance for computational resources. M.M. acknowledges the French Ministry of Research (“ACI jeunes chercheurs” and ANR NT_NV_52 Genocnidaire), the Consortium National de Recherche en Génomique, the Genoscope, and the Groupement d'Intérêt Scientifique Institut de la Génomique Marine for financial support. D.J.J. is supported by DFG funding to the Courant Research Centre for Geobiology, Göttingen.
References
- Ax P. Das System der Metazoa. ein Lehrbuch der Phylogenetischen Systematik. Stuttgart (Germany): Gustav Fischer Verlag; 1996. [Google Scholar]
- Baurain D, Brinkmann H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol Biol Evol. 2007;24:6–9. doi: 10.1093/molbev/msl137. [DOI] [PubMed] [Google Scholar]
- Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Xu A, Sagasser S, Jakob W, Moreno MA, Buss LW, Schierwater B. Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci U S A. 2006;103:8751–8756. doi: 10.1073/pnas.0602076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann M, Janussen D, Reitner J, Collins A, Wörheide G. Phylogeny and evolution of glass sponges (Porifera: Hexactinellida) Syst Biol. 2008;57:388–405. doi: 10.1080/10635150802161088. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, et al. (18 co-authors) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Erpenbeck D, Wörheide G. On the molecular phylogeny of sponges (Porifera) In: Zhang Z-Q, Shear WA, editors. Linnaeus Tercentenary: Progress in Invertebrate Taxonomy. Zootaxa 1668. Auckland (New Zealand): Magnolia Press; 2007. pp. 107–126. [Google Scholar]
- Grothe F. On the phylogeny of homoscleromorphs. Berl Geowiss Abh A. 1989;106:155–164. [Google Scholar]
- Haeckel EH. Generelle Morphologie der Organismen. Berlin (Germany): G. Reimer; 1866. [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, et al. (17 co-authors) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc Lond B Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JNA, Van Soest RWM. Systema Porifera. Guide to the Supraspecific Classification of Sponges and Spongiomorphs (Porifera) New York: Plenum; 2002. [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lecointre G, Philippe H, Van Le HL, Le Guyader H. Species sampling has a major impact on phylogenetic inference. Mol Phylogenet Evol. 1993;2:205–224. doi: 10.1006/mpev.1993.1021. [DOI] [PubMed] [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Ann Rev Ecol Syst. 2005;36:541–562. [Google Scholar]
- Philippe H, Derelle R, Lopez P, et al. (20 co-authors) Phylogenomics restores traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr Opin Genet Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Telford MJ. A multi criterion approach for the selection of optimal outgroups in phylogeny: recovering some support for Mandibulata over Myriochelata using mitogenomics. Mol Phylogenet Evol. 2008;48:103–111. doi: 10.1016/j.ympev.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Schierwater B. My favorite animal, Trichoplax adhaerens. BioEssays. 2005;27:1294–1302. doi: 10.1002/bies.20320. [DOI] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus H, Hadrys H, Dellaporta S, Kolokotronis S, Desalle R, Penny D. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, et al. (21 co-authors) The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- van Soest RWM. Deficient Merlia normani from the Curaçao reefs, with a discussion on the phylogenetic interpretation of sclerosponges. Bijdr Dierkd. 1984;54:211–219. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.