Ephexin4 is a RhoG-specific guanine nucleotide exchange factor that interacts with the EphA2 receptor in breast cancer cells.
Abstract
EphA2, a member of the Eph receptor family, is frequently overexpressed in a variety of human cancers, including breast cancers, and promotes cancer cell motility and invasion independently of its ligand ephrin stimulation. In this study, we identify Ephexin4 as a guanine nucleotide exchange factor (GEF) for RhoG that interacts with EphA2 in breast cancer cells, and knockdown and rescue experiments show that Ephexin4 acts downstream of EphA2 to promote ligand-independent breast cancer cell migration and invasion toward epidermal growth factor through activation of RhoG. The activation of RhoG recruits its effector ELMO2 and a Rac GEF Dock4 to form a complex with EphA2 at the tips of cortactin-rich protrusions in migrating breast cancer cells. In addition, the Dock4-mediated Rac activation is required for breast cancer cell migration. Our findings reveal a novel link between EphA2 and Rac activation that contributes to the cell motility and invasiveness of breast cancer cells.
Introduction
EphA2, a member of Eph family receptor tyrosine kinases, is frequently overexpressed in a variety of human cancers, including breast cancers (Merlos-Suárez and Batlle, 2008; Pasquale, 2008). Overexpression of EphA2 is associated with an aggressive and metastatic cellular phenotype in breast cancers, and recent studies have revealed that EphA2 acts as a downstream effector of EGF receptors to promote cancer cell motility and invasion, independently of the ligand ephrin stimulation (Zelinski et al., 2001; Macrae et al., 2005; Larsen et al., 2007; Brantley-Sieders et al., 2008; Miao et al., 2009). Conversely, stimulation of EphA2 with its ligand ephrinA1 in cancer cells inhibits cell proliferation and migration (Miao et al., 2009). However, the mechanisms underlying the oncogenic effects of EphA2 remain poorly understood.
Rho family small GTPases play pivotal roles in the regulation of the actin cytoskeleton and cell migration and also contribute to many steps in cancer initiation and progression (Etienne-Manneville and Hall, 2002; Sahai and Marshall, 2002; Vega and Ridley, 2008). Among Rho GTPases, Rac is activated at the leading edge of motile cells and induces the formation of actin-rich lamellipodia protrusions, which serves as a major driving force of cell movement (Etienne-Manneville and Hall, 2002). Rac also plays a key role in the cancer cell movement and formation of protrusions in invading cancer cells (Kurisu et al., 2005; Sanz-Moreno et al., 2008; Yamazaki et al., 2009). The major downstream proteins for Rac that mediate actin polymerization in lamellipodia protrusions are the WAVE family proteins, the activators of the Arp2/3 complex (Miki et al., 1998; Kurisu et al., 2005; Sanz-Moreno et al., 2008). Activated Arp2/3 complex induces rapid polymerization of actin and the formation of the branched actin filaments present in lamellipodia (Pollard and Borisy, 2003).
Activation of Rho family GTPases requires GDP–GTP exchange catalyzed by various guanine nucleotide exchange factors (GEFs). The major class of GEFs is the Dbl family GEFs that contain the Dbl homology (DH)–pleckstrin homology (PH; DH-PH) tandem domain and mediate the GDP–GTP exchange through the DH domain. The second class of GEFs for Rho family GTPases is the Dock family GEFs that have no DH-PH tandem domain. Instead, they contain a new conserved domain that directly interacts with Rho GTPase and mediates its GDP–GTP exchange (Brugnera et al., 2002; Côté and Vuori, 2002; Meller et al., 2002). Presently, 11 mammalian Dock family members have been identified and are classified into four subfamilies, the Dock180 subfamily (Dock180, Dock2, and Dock5), Dock4 subfamily (Dock3/MOCA and Dock4), Dock9 subfamily (Dock9/Zizimin1, Dock10/Zizimin3, and Dock11/Zizimin2), and Dock7 subfamily (Dock6, Dock7, and Dock8; Côté and Vuori, 2002; Meller et al., 2005). They activate specific members of Rho GTPases; the Dock180 and Dock4 subfamilies specifically activate Rac, whereas the Zizimin subfamily activates Cdc42 (Kiyokawa et al., 1998; Nishihara et al., 1999; Meller et al., 2002; Namekata et al., 2004; Hiramoto et al., 2006). In contrast, Dock7 subfamily members activate both Rac and Cdc42 (Miyamoto et al., 2007; Yamauchi et al., 2008). Dock family members play key roles in a variety of important cellular functions, including cell migration, phagocytosis, and neuronal axon and dendrite morphogenesis (Meller et al., 2005; Côté and Vuori, 2007; Miyamoto and Yamauchi, 2010). In addition, several recent studies have identified their roles in cancer cell migration and invasion. Dock180 promotes glioma cell invasion, whereas Dock3 and Dock10 mediate different modes of cell movement and invasion in melanoma cells (Jarzynka et al., 2007; Gadea et al., 2008; Sanz-Moreno et al., 2008).
The small GTPase RhoG is a key upstream regulator of Rac in migrating cells (Katoh and Negishi, 2003; Hiramoto et al., 2006; Katoh et al., 2006; Elfenbein et al., 2009). RhoG activates Rac through its effector ELMO (Katoh and Negishi, 2003). ELMO forms a complex with Dock180 or Dock4, and they serve as a functional GEF for Rac in intact cells (Gumienny et al., 2001; Brugnera et al., 2002; Hiramoto et al., 2006). The interaction of RhoG with ELMO induces translocation of the ELMO–Dock180 or ELMO–Dock4 complex from the cytoplasm to the plasma membrane and activates Rac1 to promote lamellipodium formation and cell migration (Katoh and Negishi, 2003; Hiramoto et al., 2006; Katoh et al., 2006). However, the upstream regulators of RhoG activity in the regulation of cell migration still remain obscure.
Ephexin is a subfamily of Dbl family GEFs that interacts directly with EphA receptors (Shamah et al., 2001; Sahin et al., 2005). At least five members of the Ephexin subfamily (Ephexin1–5) have been reported, and Ephexin1 (ARHGEF27/NGEF), Ephexin2 (ARHGEF19/WGEF), Ephexin3 (ARHGEF5/TIM1), and Ephexin5 (ARHGEF15/Vsm–Rho GEF) activate RhoA (Shamah et al., 2001; Ogita et al., 2003; Wang et al., 2004; Xie et al., 2005). The function of Ephexin1 is well characterized, and it has been reported that Ephexin1 regulates axon guidance and spine morphogenesis through the interaction with EphA4 (Sahin et al., 2005; Fu et al., 2007). In contrast, the functions and biochemical properties of Ephexin4 (ARHGEF16/neuroblastoma) still remain unknown. In this study, we identified Ephexin4 as a GEF for RhoG. Ephexin4 interacts with EphA2 and mediates ligand-independent promotion of cell migration and invasion through the activation of RhoG in breast cancer cells. In addition, the ELMO2–Dock4 complex is involved in the downstream signaling pathway of RhoG. Our findings provide a novel signal transduction pathway that contributes to increased migration and invasion of breast cancers and thus may provide information useful for developing new therapeutic strategies that delay or prevent the process of breast cancer cell invasion.
Results
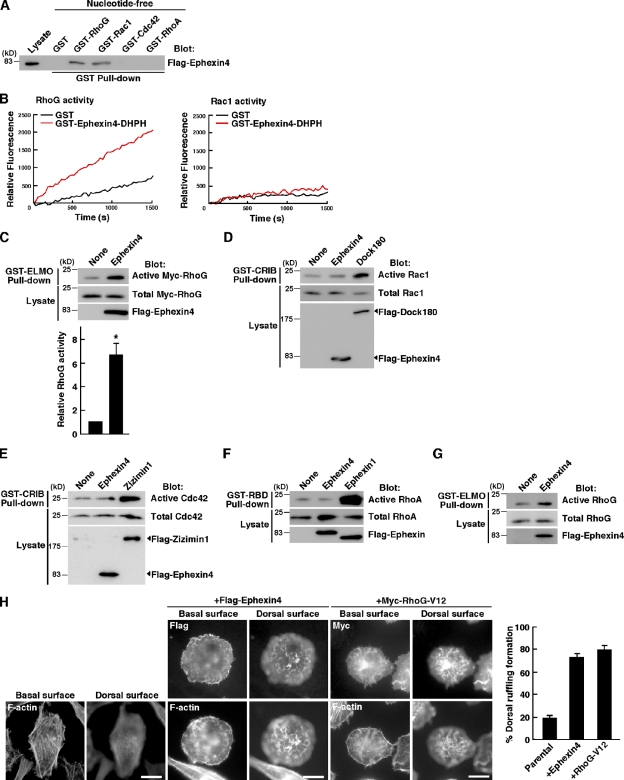
The DH domain of Ephexin4 shares high amino acid sequence homology with that of SGEF (60%), which is known as a RhoG GEF (Ellerbroek et al., 2004). Therefore, we first examined the interaction of Ephexin4 with RhoG and other well-characterized Rho family GTPases by pull-down assays with GST-fused Rho GTPases. Flag-tagged Ephexin4 expressed in HEK293T cells bound to the nucleotide-free forms of RhoG and Rac1 but not to those of RhoA and Cdc42 (Fig. 1 A). To identify RhoG, Rac1, or both as a substrate for Ephexin4, purified GST-fused DH-PH domain of Ephexin4 was incubated with purified RhoG or Rac1 in vitro, and nucleotide exchange activity was measured using an N-methylanthraniloyl (mant)-GTP fluorescence-based assay. Although the DH-PH domain of Ephexin4 did not exchange nucleotide on Rac1, it possessed GEF activity of RhoG in vitro (Fig. 1 B). We next examined whether Ephexin4 activates RhoG in cells by using the ability of purified GST-fused N-terminal RhoG-binding region of ELMO to specifically interact with GTP-bound active RhoG (Katoh and Negishi, 2003). Expression of Ephexin4 in HEK293T cells increased the amount of active exogenously expressed Myc-tagged RhoG about sixfold over the basal level (Fig. 1 C). However, we could observe no obvious increase in the activities of Rac1 and Cdc42 in HEK293T cells by expression of Ephexin4 in pull-down assays with GST-fused Cdc42/Rac1 interactive binding (CRIB) domain of Pak to precipitate GTP-bound active Rac1 or Cdc42 (Fig. 1, D and E; Dock180 and Zizimin1 were used as positive controls for activation of Rac1 and Cdc42, respectively). Other Ephexin subfamily members activate RhoA (Shamah et al., 2001; Ogita et al., 2003; Wang et al., 2004; Xie et al., 2005), but we could not detect the increase in RhoA activity in cells expressing Ephexin4, as measured by pull-down assays with GST-fused Rho-binding domain (RBD) of Rhotekin (Fig. 1 F; Ephexin1 was used as a positive control for RhoA activation). To confirm the activation of RhoG by Ephexin4, we used HeLa cells, which express a high amount of endogenous RhoG, and expression of Flag-Ephexin4 in HeLa cells enhanced endogenous RhoG activity (Fig. 1 G). In addition, HeLa cells expressing Flag-Ephexin4 or Myc-tagged constitutively active RhoG (Myc–RhoG-V12) showed extensive membrane ruffling at dorsal surfaces (∼80% of the transfected cells), which is a typical morphology of RhoG activation (Fig. 1 H; Blangy et al., 2000). However, many untransfected parental HeLa cells formed stress fibers but did not form dorsal ruffling. These results indicate that Ephexin4 activates RhoG in cells.
Figure 1.
Ephexin4 activates RhoG. (A) Flag-tagged Ephexin4 expressed in HEK293T cells was used in pull-down assays with nucleotide-free forms of GST-fused Rho family GTPases. Bound proteins were analyzed by immunoblotting with anti-Flag antibody. (B) GST-fused DH-PH domain of Ephexin4 purified from E. coli was incubated with purified RhoG or Rac1, and nucleotide exchange activity was measured in vitro using a mant-GTP fluorescence-based assay. The change in the rate of mant-GTP incorporation into GST-RhoG or -Rac1 was monitored. Nucleotide exchange on GST-Rac1 was stimulated by purified Dock4 that was used as a positive control (not depicted). (C) Cell lysates from HEK293T cells cotransfected with Flag-Ephexin4 and Myc-RhoG were incubated with GST-ELMO, and bound Myc-RhoG was detected with anti-Myc antibody. Relative Myc-RhoG activity was determined by the amount of GTP-bound Myc-RhoG bound to GST-ELMO normalized to the amount of Myc-RhoG in cell lysates analyzed by ImageJ software. Data are presented as the means ± SEM from three independent experiments (*, P < 0.05; t test). (D) The Rac1 activity in HEK293T cells transfected with Flag-Ephexin4 or Flag-Dock180 was analyzed by the GST-CRIB pull-down assay. (E) The Cdc42 activity in HEK293T cells transfected with Flag-Ephexin4 or Flag-Zizimin1 was analyzed by the GST-CRIB pull-down assay. (F) The RhoA activity in HEK293T cells transfected with Flag-Ephexin4 or Flag-Ephexin1 was analyzed by the GST–Rhotekin-RBD pull-down assay. (G) HeLa cells were transfected with Flag-Ephexin4, and endogenous RhoG activity was analyzed by the GST-ELMO pull-down assay. (H) Ephexin4 induces dorsal ruffling in HeLa cells. HeLa cells transfected with Flag-Ephexin4 or Myc–RhoG-V12 were subjected to immunofluorescent staining for Ephexin4 (anti-Flag), RhoG-V12 (anti-Myc), and F-actin (phalloidin). Images were captured at the basal and dorsal surfaces of the cells. Cells with dorsal ruffling were quantified, and results were expressed as a percentage of the number of the transfected cells. Data are presented as the means ± SEM from three independent experiments in which at least 100 cells were counted. Bars, 20 µm.
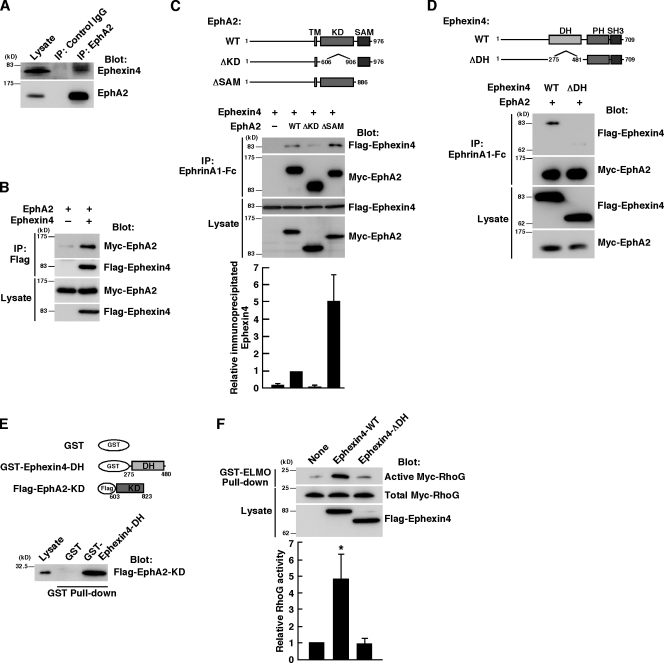
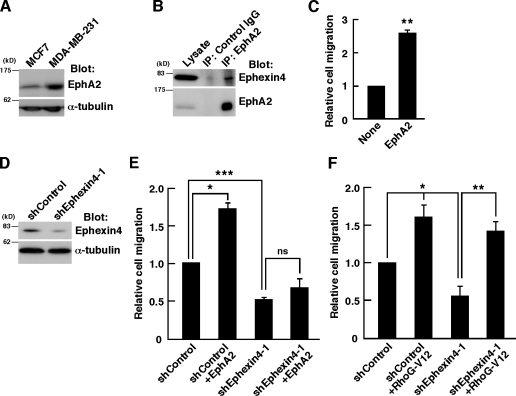
We raised an antibody against Ephexin4 and found that Ephexin4 was expressed in highly invasive MDA-MB-231 breast cancer cells (Fig. 2 A). It has been well noticed that EphA2 is highly expressed in invasive human breast cancer cell lines, including MDA-MB-231 cells (Zelinski et al., 2001; Macrae et al., 2005). To address the possibility that the interaction between endogenous Ephexin4 and EphA2 occurred in breast cancer cells, we performed immunoprecipitation from MDA-MB-231 cell lysates with anti-EphA2 antibody. Immunoblot analysis of the immunoprecipitates demonstrates the specific interaction between Ephexin4 and EphA2 (Fig. 2 A). We also observed the interaction between Flag-tagged Ephexin4 and Myc-tagged EphA2 expressed in HEK293T cells by immunoprecipitation with anti-Flag antibody (Fig. 2 B). To identify the region responsible for the interaction between EphA2 and Ephexin4, we constructed deletion mutants of EphA2 and Ephexin4. The cytoplasmic region of Eph receptors contains a protein tyrosine kinase domain and a sterile α motif (SAM) domain, and the interaction between Ephexin1 and EphA4 is mediated through the DH-PH motif of Ephexin1 and the kinase domain of EphA4 (Shamah et al., 2001). HEK293T cells were cotransfected with Flag-tagged Ephexin4 and Myc-tagged wild-type (WT) or deletion mutants of EphA2 (EphA2-ΔKD, lacking the kinase domain; EphA2-ΔSAM, lacking the SAM domain), and EphA2 was immunoprecipitated from the cell lysates with recombinant ephrinA1-Fc chimera. Ephexin4 was coimmunoprecipitated with WT EphA2 (EphA2-WT). However, deletion of the kinase domain reduced the interaction (Fig. 2 C). In contrast, the interaction between Ephexin4 and EphA2 was enhanced by deletion of the SAM domain, suggesting a possible inhibitory role of the SAM domain in their interaction. Deletion of the DH domain of Ephexin4 (Ephexin4-ΔDH) also reduced the interaction (Fig. 2 D). The interaction between the kinase domain of EphA2 and the DH domain of Ephexin4 was confirmed by a pull-down assay with Flag-tagged kinase domain of EphA2 and GST-fused DH domain of Ephexin4 (Fig. 2 E). In contrast, the interaction between Ephexin4 and EphA2 was independent of the ligand ephrinA1 stimulation (unpublished data), which is a result similar to that observed in the interaction between Ephexin1 and EphA4 (Sahin et al., 2005; Fu et al., 2007).
Figure 2.
Ephexin4 interacts with EphA2. (A) Cell lysates from MDA-MB-231 cells were immunoprecipitated with control or anti-EphA2 antibody, and bound proteins and total cell lysates were analyzed by immunoblotting with anti-EphA2 and anti-Ephexin4 antibodies. (B) HEK293T cells were transfected with Myc-EphA2 alone or together with Flag-Ephexin4, and the cell lysates were immunoprecipitated with anti-Flag antibody. Bound proteins and total cell lysates were analyzed by immunoblotting with anti-Flag and anti-EphA2 antibodies. (C and D) HEK293T cells were cotransfected with Flag-Ephexin4 (WT and ΔDH) and Myc-EphA2 (WT, ΔKD, and ΔSAM) as indicated. The cell lysates were immunoprecipitated with ephrinA1-Fc, and bound proteins and total cell lysates were analyzed by immunoblotting with anti-Flag and anti-Myc antibodies. Numbers indicate amino acid position within the sequence. Relative immunoprecipitated Ephexin4 was determined by the amount of immunoprecipitated Flag-Ephexin4 bound to Myc-EphA2 normalized to the amount of Flag-Ephexin4 in cell lysates analyzed by ImageJ software. Data are presented as the means ± SEM from four independent experiments. KD, kinase domain; SH3, Src-homology 3; TM, transmembrane. (E) Flag–EphA2-KD expressed in HEK293T cells was used in pull-down assays with GST-fused Ephexin4-DH. Bound proteins were analyzed by immunoblotting with anti-Flag antibody. (F) Cell lysates from HEK293T cells cotransfected with Myc-RhoG and Flag–Ephexin4-WT or Ephexin4-ΔDH were incubated with GST-ELMO, and bound Myc-RhoG was detected with anti-Myc antibody. Relative Myc-RhoG activity was determined by the amount of GTP-bound Myc-RhoG bound to GST-ELMO normalized to the amount of Myc-RhoG in cell lysates analyzed by ImageJ software. Data are presented as the means ± SEM from four independent experiments (*, P < 0.05; t test). IP, immunoprecipitation.
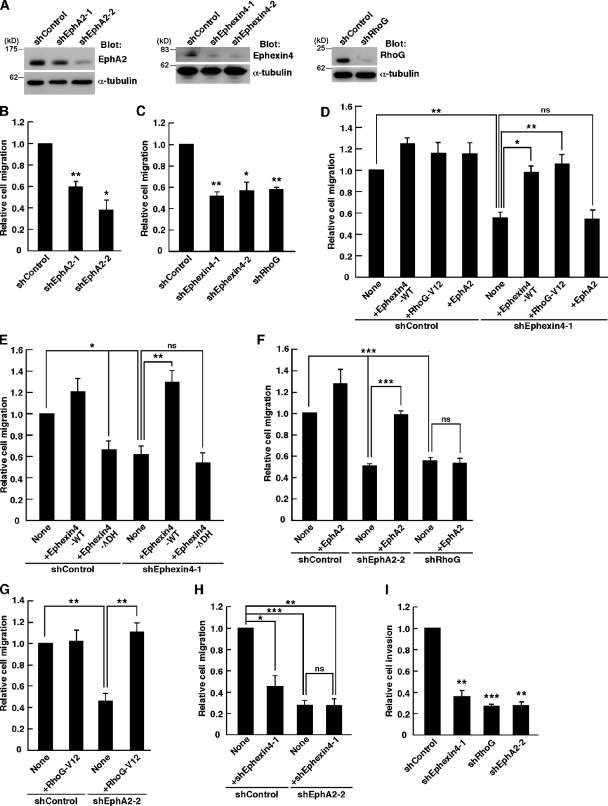
EphA2 is overexpressed in many cancer cell lines and promotes cell migration in a ligand-independent manner (Zelinski et al., 2001; Macrae et al., 2005; Larsen et al., 2007; Brantley-Sieders et al., 2008; Miao et al., 2009). To investigate the roles of Ephexin4 and RhoG in the promotion of cancer cell migration mediated by EphA2, we performed RNAi-mediated knockdown of Ephexin4, RhoG, and EphA2 in MDA-MB-231 cells using short hairpin RNA (shRNA) expression vectors, which effectively reduced the amounts of the endogenous proteins (Fig. 3 A). We evaluated the effect of knockdown of these proteins on the migration of MDA-MB-231 cells in Transwell migration assays by using EGF as a chemoattractant because MDA-MB-231 cells show enhanced cell migration and invasion in response to EGF (Hynes and Lane, 2005). Knockdown of EphA2 by shEphA2-1 or shEphA2-2, targeting different sequences of human EphA2 mRNA, significantly impaired the ability of MDA-MB-231 cells to migrate toward EGF-containing medium (Fig. 3 B), suggesting that EphA2 mediates a ligand-independent promotion of chemotactic migration in MDA-MB-231 cells in response to EGF stimulation. We next investigated the effect of Ephexin4 or RhoG knockdown on the migration of MDA-MB-231 cells toward EGF-containing medium and found that knockdown of Ephexin4 by shEphexin4-1 or shEphexin4-2, targeting different regions of human Ephexin4 mRNA, or knockdown of RhoG by RhoG-specific shRNA (shRhoG; Katoh et al., 2006) also significantly blocked the migration (Fig. 3 C). Expression of control Luciferase shRNA (shControl) had no effect on the migration of MDA-MB-231 cells compared with untransfected cells (unpublished data). Because the shEphexin4-1 targets Ephexin4 mRNA 3′ untranslated region, we could perform rescue experiments by cotransfection with the plasmid expressing Flag-tagged Ephexin4, which contains only the coding region of Ephexin4. Cotransfection with Flag-Ephexin4 (+Ephexin4-WT) or constitutively active RhoG (+RhoG-V12) completely rescued the defect in cell migration caused by shEphexin4-1 (Fig. 3 D). However, Flag–Ephexin4-ΔDH, which did not activate RhoG (Fig. 2 F), failed to rescue the defect by knockdown of Ephexin4 and also suppressed migration of the control cells (Fig. 3 E), suggesting that Ephexin4 promotes migration of MDA-MB-231 cells through activation of RhoG. In contrast, expression of Myc-tagged EphA2 (+EphA2), which could rescue the cell migration defect caused by shEphA2-2 (shEphA2-2 targets the 3′ untranslated region of EphA2 mRNA), had little effect on cell migration in Ephexin4 or RhoG knockdown cells (Fig. 3, D and F). In addition, the cell migration defect caused by shEphA2-2 was rescued by expression of RhoG-V12 (Fig. 3 G). Collectively, these results suggest that Ephexin4 and RhoG act downstream of EphA2 in the promotion of cell migration in MDA-MB-231 cells. Simultaneous knockdown of EphA2 and Ephexin4 by shEphA2-2 and shEphexin4-1 had no additive inhibitory effect on cell migration (Fig. 3 H). This result also supports the idea that EphA2 and Ephexin4 act in the same pathway. Because MDA-MB-231 cells are well known as highly invasive cells, we used Matrigel invasion assays to examine whether EphA2, Ephexin4, and RhoG also contribute to the invasion of MDA-MB-231 cells and found that knockdown of EphA2, Ephexin4, and RhoG markedly suppressed the EGF-induced Matrigel invasion in MDA-MB-231 cells (Fig. 3 I).
Figure 3.
Ephexin4 and RhoG mediate promotion of cell migration and invasion by EphA2 in MDA-MB-231 cells. (A) Cell lysates from MDA-MB-231 cells transfected with the indicated shRNAs were analyzed by immunoblotting with antibodies against Ephexin4, EphA2, RhoG, and α-tubulin. (B and C) Transwell migration assays using MDA-MB-231 transfected with YFP together with the indicated shRNAs in the presence of 10 ng/ml EGF for 4–6 h. Data are presented as the means ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; t test). (D–H) Transwell migration assays using MDA-MB-231 cells transfected with YFP and the indicated plasmids in the presence of EGF for 6 h. Data are presented as the means ± SEM from three or four independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant; t test). (I) Matrigel invasion assays using MDA-MB-231 cells cotransfected with YFP and the indicated shRNAs in the presence of EGF for 24 h. Data are presented as the means ± SEM from three independent experiments (**, P < 0.01; ***, P < 0.001; t test).
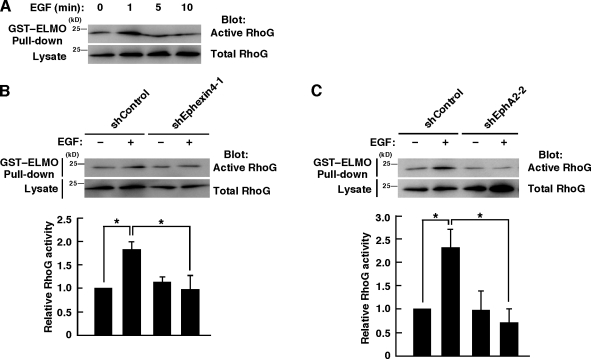
We next examined whether EphA2 and Ephexin4 regulate RhoG activity in MDA-MB-231 cells after stimulation with EGF by the pull-down assay with GST-fused ELMO. Stimulation with EGF induced a rapid increase in the amount of GTP-bound RhoG within 1 min, and then the level decreased within 5 min (Fig. 4 A). Knockdown of Ephexin4 or EphA2 in MDA-MB-231 cells significantly suppressed the activation of RhoG induced by EGF stimulation (Fig. 4, B and C). These results suggest that EphA2 and Ephexin4 mediate RhoG activation in response to EGF stimulation in MDA-MB-231 cells.
Figure 4.
EphA2 and Ephexin4 mediate EGF-induced RhoG activation in MDA-MB-231 cells. (A) MDA-MB-231 cells were stimulated with EGF for the indicated times, and RhoG activity was analyzed by the GST-ELMO pull-down assay. (B and C) MDA-MB-231 cells transfected with the indicated plasmids were stimulated with or without EGF for 1 min, and RhoG activity was analyzed by the GST-ELMO pull-down assay. Data are presented as the means ± SEM from four or five independent experiments (*, P < 0.05; t test).
To verify the involvement of Ephexin4 in the EphA2-mediated promotion of cell migration, we used the noninvasive breast cancer cell line MCF7, which expresses a moderate level of endogenous EphA2 compared with that in MDA-MB-231 (Fig. 5 A). In addition, Ephexin4 is expressed in MCF7 cells, and the interaction with EphA2 was also detected by immunoprecipitation (Fig. 5 B). Ectopic expression of Myc-tagged EphA2 in MCF7 cells promoted cell migration in the absence of ephrin stimulation (Fig. 5 C). However, knockdown of Ephexin4 by shEphexin4-1 reduced migration of MCF7 cells and completely suppressed the EphA2-induced promotion of cell migration (Fig. 5, D and E). Similar results were obtained in cervical carcinoma HeLa cells, which also endogenously express EphA2 and Ephexin4 (Fig. S1). In contrast, migration of MCF7 cells was also promoted by expression of RhoG-V12, and it was not affected by knockdown of Ephexin4 (Fig. 5 F). These results support the conclusion that Ephexin4 is required for the ligand-independent promotion of cell migration by EphA2 through RhoG activation.
Figure 5.
Ectopic expression of EphA2 in MCF7 cells promotes cell migration through Ephexin4. (A) Expression of EphA2 in MCF7 and MDA-MB-231 breast cancer cell lines was analyzed by immunoblotting with anti-EphA2 and anti–α-tubulin antibodies. (B) Cell lysates from MCF7 cells were immunoprecipitated with control or anti-EphA2 antibody, and bound proteins and total cell lysates were analyzed by immunoblotting with anti-EphA2 and anti-Ephexin4 antibodies. IP, immunoprecipitation. (C) Transwell migration assays using MCF7 cells transfected with GFP alone or together with Myc-EphA2 for 6 h. Data are presented as the means ± SEM from four independent experiments (**, P < 0.01; t test). (D) Cell lysates from MCF7 cells transfected with control or Ephexin4-1 shRNA were analyzed by immunoblotting with anti-Ephexin4 and anti–α-tubulin antibodies. (E and F) Transwell migration assays using MCF7 cells transfected with YFP and the indicated plasmids for 24 h. Data are presented as the means ± SEM from three or four independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant; t test).
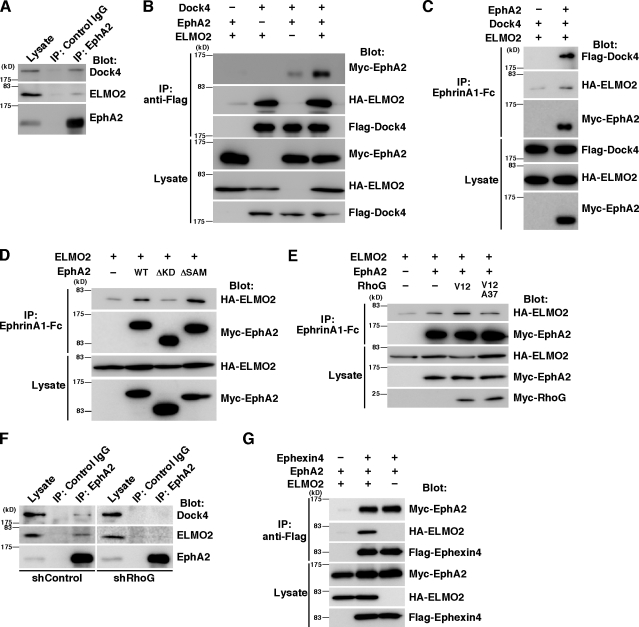
RhoG activates Rac1 and promotes cell migration through its effector ELMO and the ELMO-binding protein Dock4 (Hiramoto et al., 2006). In a coimmunoprecipitation experiment with anti-EphA2 antibody, we found that EphA2 formed a complex with ELMO2 and Dock4 in MDA-MB-231 cells (Fig. 6 A). To confirm the interaction of EphA2 with ELMO2 and Dock4, Myc-EphA2 was coexpressed with HA-ELMO2, Flag-Dock4, or both in HEK293T cells, and the cell lysates were immunoprecipitated with anti-Flag antibody. The interaction between Myc-EphA2 and Flag-Dock4 without HA-ELMO2 expression was very weak, but it was clearly detected in the presence of HA-ELMO2 (Fig. 6 B). The interaction between EphA2 and ELMO2–Dock4 was also observed by immunoprecipitation with ephrinA1-Fc in HEK293T cells cotransfected with HA-ELMO2, Flag-Dock4, and Myc-EphA2 (Fig. 6 C). Collectively, our results suggest that EphA2 forms a ternary complex with ELMO2 and Dock4. The weak interaction between EphA2 and Dock4 without ELMO2 expression may be caused by an endogenous ELMO2-mediated interaction. However, our results cannot rule out the possibility that EphA2 can bind to Dock4 without ELMO2. Immunoprecipitation analysis with deletion mutants of EphA2 showed that the kinase domain of EphA2 was important for the interaction with ELMO2 (Fig. 6 D). Interestingly, the interaction between EphA2 and ELMO2 was enhanced by coexpression with Myc–RhoG-V12 but not by coexpression with Myc–RhoG-V12A37, a mutant of RhoG-V12 which fails to interact with ELMO2 (Fig. 6 E; Katoh and Negishi, 2003). These results suggest that activated RhoG binds to ELMO2 and recruits ELMO2 and Dock4 to form a complex with EphA2. An immunoprecipitation analysis with anti-EphA2 antibody shows that interaction of endogenous EphA2 with ELMO2–Dock4 was reduced by knockdown of RhoG in MDA-MB-231 cells (Fig. 6 F). This result also supports the conclusion that RhoG regulates the interaction of ELMO2 with EphA2. In contrast, HA-ELMO2 and Myc-EphA2 were coimmunoprecipitated with Flag-Ephexin4 in HEK293T cells transfected with Flag-Ephexin4, Myc-EphA2, and HA-ELMO2 (Fig. 6 G), suggesting that Ephexin4 was also present in the same complex. In our immunoprecipitation experiments using Fc-fused ephrinA1, the bands of HA-ELMO2 were weakly detected in the absence of overexpressed EphA2 (Fig. 6, C–E, first lanes). This reason is unknown, but HA-ELMO2 was also pulled down by control Fc alone (Fig. S2).
Figure 6.
ELMO2 and Dock4 form a complex with EphA2. (A) Cell lysates from MDA-MB-231 cells were immunoprecipitated with control or anti-EphA2 antibody, and bound proteins and total cell lysates were analyzed by immunoblotting with antibodies against EphA2, ELMO2, and Dock4. (B) Cell lysates from HEK293T cells transfected with Flag-tagged Dock4, HA-tagged ELMO2, and Myc-tagged EphA2 were immunoprecipitated with anti-Flag antibody, and bound proteins and total cell lysates were analyzed by immunoblotting with antibodies against Flag, HA, and Myc. (C–E) Cell lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with ephrinA1-Fc, and bound proteins and total cell lysates were analyzed by immunoblotting. (F) Cell lysates from MDA-MB-231 cells transfected with control or RhoG shRNA were immunoprecipitated with control or anti-EphA2 antibody, and bound proteins and total cell lysates were analyzed by immunoblotting with antibodies against EphA2, ELMO2, and Dock4. (G) Cell lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with anti-Flag antibody, and bound proteins and total cell lysates were analyzed by immunoblotting. IP, immunoprecipitation.
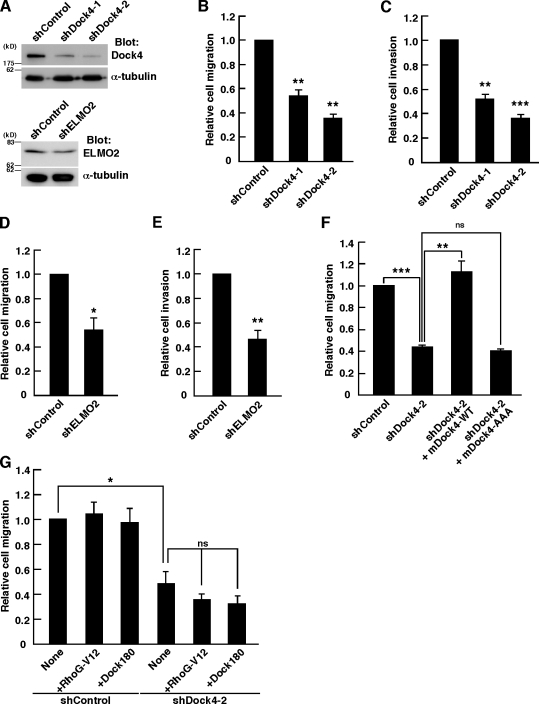
To investigate the role of Dock4 and ELMO2 in migration and invasion of MDA-MB-231 cells, we constructed two shRNAs targeting different sequences of human Dock4 (shDock4-1 and -2) and an shRNA targeting human ELMO2 (shELMO2), which effectively reduced the endogenous protein levels in MDA-MB-231 cells (Fig. 7 A). Expression of shDock4-1 or shDock4-2 significantly suppressed the migration and invasion of MDA-MB-231 cells toward EGF-containing medium (Fig. 7, B and C). Knockdown of ELMO2 also suppressed the EGF-induced migration and invasion in MDA-MB-231 cells (Fig. 7, D and E). Thus, Dock4 and ELMO2 are involved in the EGF-induced migration and invasion of MDA-MB-231 cells. We next performed rescue experiments by cotransfection with mouse Dock4 WT (mDock4-WT), which was resistant to the human Dock4 shRNA shDock4-2. Expression of mDock4-WT completely rescued the impaired cell migration caused by shDock4-2. However, cotransfection with a mutant of mouse Dock4 (mDock4-AAA), which contains mutations in the Rac GEF domain and has no ability to activate Rac (Hiramoto et al., 2006), had no effect (Fig. 7 F). These results suggest that Dock4 mediates the migration and invasion of MDA-MB-231 cells through the activation of Rac. However, expression of RhoG-V12, which could rescue the cell migration defect caused by knockdown of EphA2 or Ephexin4 (Fig. 3, D and G), did not rescue in Dock4 knockdown cells (Fig. 7 G), supporting a model in which RhoG acts upstream of Dock4. Although Dock180 is another member of Dock family GEFs that activates Rac, expression of Flag-Dock180 had little effect on migration of Dock4 knockdown cells (Fig. 7 G), suggesting that this function is specific to Dock4.
Figure 7.
Dock4-mediated activation of Rac is required for the cell migration and invasion in MDA-MB-231 cells. (A) Cell lysates from MDA-MB-231 cells transfected with control, Dock4, or ELMO2 shRNA were analyzed by immunoblotting with anti-Dock4, anti-ELMO2, and anti–α-tubulin antibodies. (B and D) Transwell migration assays using MDA-MB-231 cells transfected with YFP alone or together with the indicated shRNAs in the presence of EGF for 4 h. Data are presented as the means ± SEM from three or four independent experiments (*, P < 0.05; **, P < 0.01; t test). (C and E) Matrigel invasion assays using MDA-MB-231 cells transfected with YFP alone or together with the indicated shRNAs in the presence of EGF for 24 h. Data are presented as the means ± SEM from three independent experiments (**, P < 0.01; ***, P < 0.001; t test). (F and G) Transwell migration assays using MDA-MB-231 cells transfected with YFP and the indicated plasmids in the presence of EGF for 6 h. Data are presented as the means ± SEM from three or four independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant; t test).
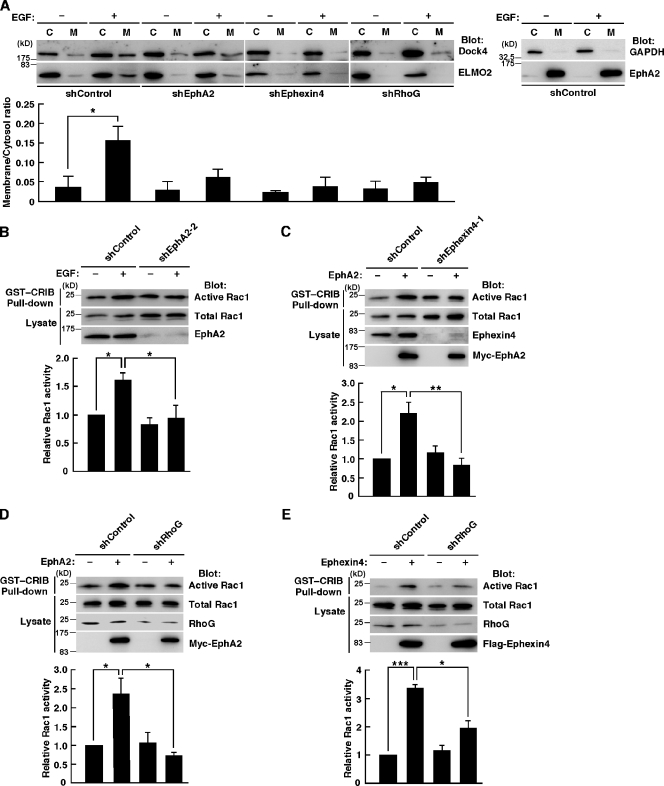
Dock family members localize mainly in the cytoplasm, and the translocation to the plasma membrane is essential for their signaling and functions (Côté et al., 2005). We previously showed that interaction of active RhoG with ELMO2 recruits the ELMO2–Dock4 complex to the plasma membrane (Hiramoto et al., 2006). Therefore, to examine whether ELMO2 and Dock4 function downstream of the EphA2–Ephexin4–RhoG signaling pathway, we prepared membrane and cytosol fractions from cellular homogenates of MDA-MB-231 cells expressing shRNA for the control Luciferase, EphA2, RhoG, or Ephexin4 in the absence and presence of chemotactic stimulus EGF and analyzed the distributions of ELMO2 and Dock4 by immunoblotting with anti-Dock4 and anti-ELMO2 antibodies. In the control cells, EGF stimulation enhanced the levels of ELMO2 and Dock4 in the membrane fractions. However, knockdown of EphA2, RhoG, or Ephexin4 suppressed the EGF-induced membrane recruitment of ELMO2 and Dock4 (Fig. 8 A). The efficiency of cell fractionation was shown by immunoblotting with anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; cytosol protein) and anti-EphA2 (membrane protein) antibodies, and EGF stimulation had no effect on their distributions (Fig. 8 A). Collectively, our results suggest that EphA2- and Ephexin4-mediated RhoG activation recruits the ELMO2–Dock4 complex to the plasma membrane in response to EGF stimulation in MDA-MB-231 cells. There is Dock4 but not ELMO2 in the membrane fraction under RhoG knockdown conditions. It is possible that Dock4 is involved in other cellular functions independent of RhoG activation, for example the regulation of cell–cell adhesion and receptor endocytosis (Yajnik et al., 2003; Upadhyay et al., 2008; Kawada et al., 2009). Next, to examine whether Rac was indeed activated upon EGF stimulation and that knockdown of EphA2 prevented it, we measured Rac activity in MDA-MB-231 cells expressing the control Luciferase or EphA2 shRNA in the absence and presence of EGF by pull-down assays with GST-fused CRIB domain of Pak. In the control cells, stimulation with EGF induced a rapid increase in the amount of active Rac1. However, knockdown of EphA2 significantly suppressed the Rac1 activation by EGF (Fig. 8 B). Thus, EphA2 mediates Rac activation in response to EGF stimulation.
Figure 8.
EphA2 and Ephexin4 mediate the membrane recruitment of the ELMO2–Dock4 complex and activation of Rac1. (A) The control (shControl), EphA2 (shEphA2), Ephexin4 (shEphexin4), or RhoG (shRhoG) knockdown MDA-MB-231 cells were stimulated with or without EGF for 1 min, and the cellular homogenates separated into the cytosol (C) and the membrane (M) fractions were analyzed by immunoblotting with anti-ELMO2 and anti-Dock4 antibodies. The membrane/cytosol ratio of Dock4 was analyzed by ImageJ software. Data are presented as the means ± SEM from three independent experiments (*, P < 0.05; t test). Immunoblotting with anti-GAPDH (cytosol protein) and anti-EphA2 (membrane protein) antibodies shows efficiency of cell fractionation (right). (B) MDA-MB-231 cells transfected with the indicated plasmids were stimulated with or without EGF for 1 min, and Rac1 activity was analyzed by the GST-CRIB pull-down assay. Data are presented as the means ± SEM from five independent experiments (*, P < 0.05; t test). (C–E) HeLa cells were transfected with the indicated plasmids, and Rac1 activity was analyzed by the GST-CRIB pull-down assay. Data are presented as the means ± SEM from five (C), four (D), or three (E) independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; t test).
To further confirm that Rac activation occurs downstream of EphA2, HeLa cells were transfected with Myc-tagged EphA2, and the Rac activity was measured. Expression of EphA2 significantly increased Rac1 activity, which was suppressed by knockdown of Ephexin4 or RhoG (Fig. 8, C and D). These results suggest that EphA2 activates Rac1 through Ephexin4 and RhoG.
In Fig. 1 D, expression of Flag-tagged Ephexin4 induced no obvious increase in the Rac1 activity in HEK293T cells. A possible reason for this might be because of the low level of RhoG expression in HEK293T cells (unpublished data). Therefore, we examined whether Rac1 was activated after overexpression of Ephexin4 in HeLa cells, which express a high amount of endogenous RhoG. In HeLa cells, expression of Flag-Ephexin4 significantly increased Rac1 activity, which was suppressed by knockdown of RhoG (Fig. 8 E). These results support a model in which Rac1 is activated downstream of Ephexin4 and RhoG.
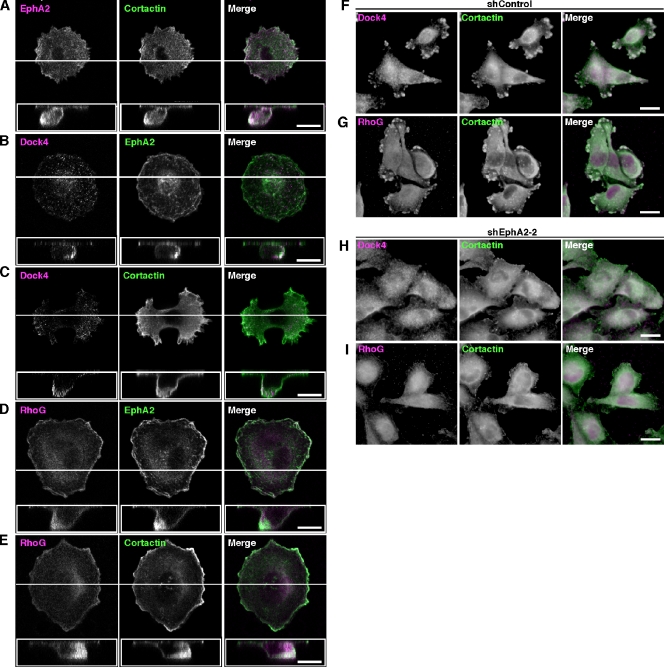
To determine whether EphA2 cooperates with Dock4 in migrating cells, we investigated the subcellular localization of EphA2 and Dock4 in MDA-MB-231 cells migrating toward EGF-containing medium through the Transwell filters. Cortactin is well known as a critical component of actin-rich protrusions in migrating and invading cancer cells and has a crucial role in promoting cell motility and invasion (Weaver, 2008). EphA2 was colocalized with cortactin at the tip of protrusion in migrating MDA-MB-231 cells through the pore of the Transwell filter (Fig. 9 A). Immunofluorescence staining with anti-Dock4 antibody revealed that Dock4 was colocalized with EphA2 and cortactin at the tip of cell protrusion into the pore (Fig. 9, B and C). We could not obtain clear images of the localization of Ephexin4, probably because our Ephexin4 antibody was not suitable for immunofluorescence staining of endogenous Ephexin4 (unpublished data). Instead, RhoG was also colocalized with EphA2 and cortactin (Fig. 9, D and E). Because knockdown of EphA2 suppresses the formation of protrusions and migration of the cells, we examined the effect of EphA2 knockdown on colocalization of Dock4 and RhoG with cortactin in MDA-MB-231 cells on glass coverslips. In the control cells expressing Luciferase shRNA, Dock4 and RhoG were colocalized with cortactin at the cell periphery (Fig. 9, F and G). In contrast, diffuse Dock4 and RhoG staining was observed in EphA2 knockdown cells, and they were not colocalized with cortactin (Fig. 9, H and I). The specificity of Dock4 or RhoG staining with anti-Dock4 or anti-RhoG antibody was verified with Dock4 or RhoG knockdown MDA-MB-231 cells (Fig. S3). Thus, these findings suggest that EphA2 cooperates with RhoG and Dock4 at the tips of cortactin-rich protrusions to promote cell migration.
Figure 9.
EphA2, Dock4, and RhoG are colocalized with cortactin at the tip of protrusion in migrating MDA-MB-231 cells. (A–E) MDA-MB-231 cells were seeded on the Transwell filters in the presence of 10 ng/ml EGF under the filter and incubated for 2 h. Then cells were subjected to immunofluorescent staining. Images of the z-plane are shown in white boxes, and selected positions of z-plane images are indicated with white lines. About 80 cells were examined under each condition. (F–I) Control (F and G) or EphA2 knockdown (H and I) MDA-MB-231 cells were seeded on coverslips and subjected to immunofluorescent staining. Bars: (A–E) 10 µm; (F–I) 20 µm.
Discussion
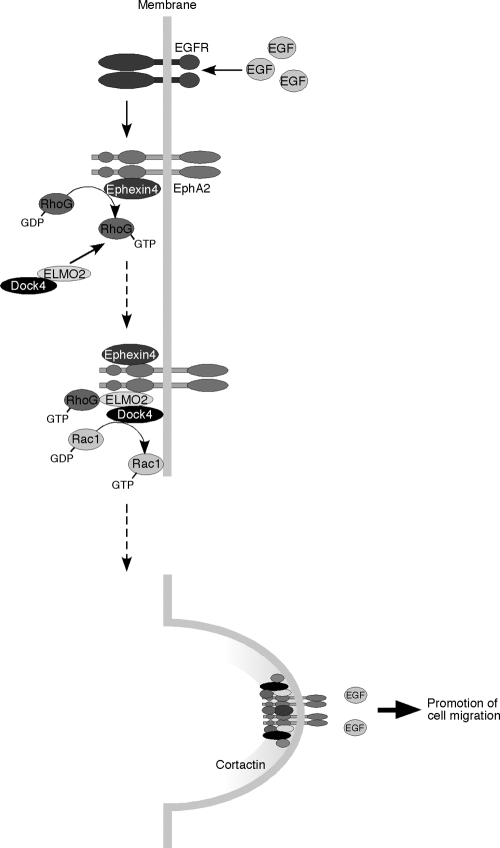
EphA2 is a member of Eph family receptor tyrosine kinases that is strongly expressed in highly invasive breast cancers (Merlos-Suárez and Batlle, 2008; Pasquale, 2008). EphA2 acts as a downstream effector of EGF receptors to promote cancer cell motility and invasion, independently of the ligand ephrin stimulation (Zelinski et al., 2001; Macrae et al., 2005; Larsen et al., 2007; Brantley-Sieders et al., 2008; Miao et al., 2009). In this study, we show that Ephexin4 is a novel binding partner for EphA2 that mediates ligand-independent promotion of cell migration and invasion in breast cancer cells overexpressing EphA2. EphA2 and Ephexin4 mediate the activation of RhoG in response to EGF stimulation, and activated RhoG binds to ELMO2 and recruits ELMO2 and its binding partner Dock4 from the cytoplasm to the plasma membrane to promote the formation of a complex with EphA2 and induce Dock4-mediated activation of Rac. We also found that EphA2 and Dock4 are colocalized with cortactin at the tips of protrusions in migrating breast cancer cells. Cortactin is overexpressed in cancer cells and promotes cancer cell motility and invasion by the formation of actin-rich protrusions associated with degradation of the extracellular matrix (Rothschild et al., 2006; Weaver, 2008; DesMarais et al., 2009). In contrast, Rac plays a key role in the cancer cell movement and the formation of protrusions in invading cancer cells (Kurisu et al., 2005; Sanz-Moreno et al., 2008; Vega and Ridley, 2008; Yamazaki et al., 2009), and the intracellular localization of cortactin is regulated by the activation of Rac1 (Weed et al., 1998). Collectively, these findings support a model in which recruitment of the ELMO2–Dock4 complex to EphA2 receptor through Ephexin4-mediated RhoG activation triggers a local activation of Rac by Dock4, which can cause the formation of cortactin-rich protrusions, leading to the promotion of cell polarization and migration (Fig. 10). Our findings also suggest that EphA2 functions as a scaffold downstream of EGF receptor to promote the formation of a signaling complex that enhances the cell motility and invasiveness of breast cancer cells.
Figure 10.
The schematic model of EphA2-mediated promotion of cell migration in response to EGF stimulation in breast cancer cells. EphA2 binds to Ephexin4 and induces the activation of RhoG in response to EGF stimulation. Activated RhoG recruits ELMO2 and Dock4 to form a complex with EphA2, leading to Rac activation and promotion of cell migration.
Ephexin1 interacts with EphA4 and regulates EphA4-mediated axon guidance and dendritic spine morphogenesis (Shamah et al., 2001; Sahin et al., 2005; Fu et al., 2007). Ephexin1 interacts with the kinase domain of EphA4 through the DH-PH motif of Ephexin1 (Shamah et al., 2001). The interaction between Ephexin4 and EphA2 also requires the DH domain of Ephexin4 and the kinase domain of EphA2, suggesting that Ephexin1 and Ephexin4 interact with EphA receptors through a similar manner, although it is unknown whether there are binding specificities between Ephexin subfamily GEFs and EphA receptors. Our results also demonstrate that the interaction between Ephexin4 and EphA2 is enhanced by the deletion of the SAM domain of EphA2, suggesting that the SAM domain negatively regulates their interaction. In a recent study, several growth factors, including EGF, stimulate phosphorylation of EphA2 at a residue of serine 897 through Akt, and this phosphorylation is required for the EphA2-mediated ligand-independent promotion of cell migration and invasion (Miao et al., 2009). Because serine 897 of EphA2 is located in a linker region between the kinase and SAM domains, phosphorylation of EphA2 at serine 897 by EGF-induced Akt activation may trigger a conformational change within the cytoplasmic region of EphA2 to promote the interaction with Ephexin4 and the Ephexin4-dependent RhoG activation. However, the tyrosine phosphorylation of Ephexin1 or Ephexin5/Vsm–Rho GEF by EphA4 enhances the GEF activity toward RhoA in response to ephrinA1 stimulation (Ogita et al., 2003; Sahin et al., 2005). Thus, although Ephexin family members bind to EphA receptors through a similar region, they appear to differ in the regulation of the GEF activity. In contrast to the ligand-independent signaling by EphA2, stimulation of EphA2 with its ligand ephrinA1 in cancer cells induces EphA2 receptor internalization and degradation, which is also mediated through the activation of Rac (Walker-Daniels et al., 2002; Zhuang et al., 2007). The Vav family of Rac-specific GEFs plays an important role in the regulation of ephrin-Eph endocytosis, and ephrinA1 stimulation causes tyrosine phosphorylation of EphA2 in the juxtamembrane domain and recruits Vav family GEFs to the phosphorylated EphA2 receptor (Cowan et al., 2005; Hunter et al., 2006). Thus, it is possible that EphA2 couples different GEFs in ligand-dependent and -independent signaling pathways.
Dock4 belongs to the Dock family of GEFs for Rho family small GTPases and positively regulates cell migration by specifically activating Rac (Côté and Vuori, 2002; Hiramoto et al., 2006; Upadhyay et al., 2008; Kawada et al., 2009). We show that Dock4 is expressed in breast cancer cells and is required for EGF-stimulated cell migration and invasion. Dock family members, including Dock4, localize mainly in the cytoplasm, and the translocation to the plasma membrane is critical for the activation of Rho family GTPases in cells and their cellular functions (Côté et al., 2005; Hiramoto et al., 2006; Meller et al., 2008; Kuramoto et al., 2009). We previously reported that RhoG regulates Dock4 through the interaction with ELMO2 and recruitment of the ELMO2–Dock4 complex from the cytoplasm to the plasma membrane (Hiramoto et al., 2006). In this study, knockdown of EphA2 or Ephexin4 in MDA-MB-231 breast cancer cells suppresses the activation of RhoG and RhoG-dependent translocation of ELMO2 and Dock4 to the plasma membrane induced by EGF stimulation. Thus, EphA2 and Ephexin4 are key upstream regulators for the RhoG–ELMO2–Dock4 signaling pathway to control cell migration and invasion in MDA-MB-231 breast cancer cells. Phylogenetic analysis indicates that Dock3 and Dock4 belong to the same subfamily within the Dock family members (Côté and Vuori, 2002). Dock3 is expressed in melanoma cells and also positively regulates cell migration and invasion through activation of Rac (Sanz-Moreno et al., 2008). The Dock3-mediated regulation of cell movement in melanoma cells requires NEDD9, a member of the p130Cas, and a Rac downstream effector WAVE2 (Sanz-Moreno et al., 2008). Thus, it will be interesting in future studies to investigate whether NEDD9 and WAVE family proteins are involved in the Dock4-mediated signaling in breast cancer cells.
Our results show that activation of RhoG contributes to increased migration and invasion of breast cancer cells. In addition to its role in cell migration, RhoG plays an important role in cell proliferation and survival (Roux et al., 1997; Murga et al., 2002; Yamaki et al., 2007; Fujimoto et al., 2009). Therefore, the EphA2-mediated RhoG activation may also contribute to other malignant cellular behaviors in cancer cells overexpressing EphA2.
Materials and methods
Plasmids and antibodies
The expression plasmid encoding Flag-Dock180 (pCXN2) was a gift from M. Matsuda (Kyoto University, Sakyo-ku, Kyoto, Japan). pCAG vector encoding EYFP was a gift from J. Miyazaki (Osaka University, Suita, Osaka, Japan) and T. Saito (Chiba University, Inage-ku, Chiba, Japan). Human Ephexin4 and EphA2 were obtained from HeLa cells, and mouse Ephexin1 was obtained from mouse brain by RT-PCR, and they were sequenced completely. WT Ephexin4 was subcloned into pCXN2-Flag or pCAG-EYFP-CAG vector (Saito and Nakatsuji, 2001), and Ephexin4-ΔDH (aa 1–275 and 481–709) and Ephexin1 were subcloned into pCXN2-Flag vector. The DH domain (aa 275–480) and DH-PH domain (aa 276–633) of Ephexin4 were subcloned into pGEX-4T-2 (GE Healthcare). WT EphA2, EphA2-ΔKD (aa 1–606 and 906–976), and EphA2-ΔSAM (aa 1–886) were subcloned into pcDNA3.1 (+) vector containing Ig κ leader sequences–Myc (Iwasato et al., 2007; Takeuchi et al., 2009). The kinase domain of EphA2 (aa 603–823) was subcloned into pCXN2-Flag vector. Plasmids encoding Myc-tagged human RhoG-WT, RhoG-V12 (G12V), RhoG-V12A37 (G12V and F37A), Flag-tagged mouse Dock4-WT, Dock4-AAA (M1475A, S1476A, and P1477A), and mouse Zizimin1 were generated as described previously (Katoh and Negishi, 2003; Hiramoto et al., 2006; Katoh et al., 2006; Kuramoto et al., 2009). Plasmids encoding GST-fused human RhoG, Rac1, Cdc42, and RhoA were generated as described previously (Katoh et al., 2002; Katoh and Negishi, 2003). Ephexin4 shRNAs were designed to target 19 or 21 nt of the human Ephexin4 transcript (shEphexin4-1, 5′-GGAGGCACCAATAGCGATTAT-3′; and shEphexin4-2, 5′-GCGGAGAGCTGTTCTTAGT-3′). Dock4, ELMO2, and EphA2 shRNAs were designed to target 19 nt of the human Dock4 transcript (shDock4-1, 5′-GCATATACCCTCCTCTTAT-3′; and shDock4-2, 5′-CCGCAAGGTCTCTCAGTTA-3′), the human ELMO2 transcript (shELMO2, 5′-TTCATCGCACCTAATAAAT-3′), and the human EphA2 transcript (shEphA2-1, 5′-GCAAGGTGCACGAATTCCA-3′; and shEphA2-2, 5′-CAGCCTTCGGACAGACATA-3′), respectively. All of the targets of each transcript were designed with no significant homology to any mammalian gene sequence and were expressed by using an shRNA expression vector pSilencer-hygro (Applied Biosystems) or pCAG-EYFP-hU6 as described previously (Katoh et al., 2006; Iwasato et al., 2007; Fujimoto et al., 2009). RhoG shRNA or control shRNA was designed to target 19 nt of the human RhoG or Luciferase, respectively, as described previously (Katoh et al., 2006; Yamaki et al., 2007).
A rabbit pAb for Ephexin4 was raised against bacterially expressed GST-fused peptide, corresponding to residues 690–708 of human Ephexin4, and the specific antibody was purified with the peptide-conjugated affinity column. The antibody against Ephexin4 was used to detect endogenous Ephexin4 by immunoblotting at a concentration of 1 µg/ml. The antibody for Dock4 was used as described previously (Ueda et al., 2008). The other antibodies were purchased commercially: mouse mAbs against Myc (9E10), RhoA (26C4), and Dock4 (R6Y), rabbit pAbs against EphA2 (C-20), cortactin (H-191) and HA (Y-11), and a rat mAb against RhoG (1F3 B3 E5; Santa Cruz Biotechnology, Inc.); mouse mAbs against Flag (M2) and α-tubulin (Sigma-Aldrich); a mouse mAb against Rac1 (BD); mouse mAbs against EphA2 (D7) and cortactin (4F11; Millipore); a rat mAb against HA (3F10; Roche); a rabbit pAb against Myc (MBL); a goat pAb against ELMO2 (Abcam); control mouse IgG (Jackson ImmunoResearch Laboratories, Inc.); secondary antibodies conjugated to horseradish peroxidase (Dako); and secondary antibodies conjugated to Alexa Fluor 488 or 594 (Invitrogen). F-actin was visualized with Alexa Fluor 488–conjugated phalloidin (Invitrogen).
Cell culture and transfection
MDA-MB-231 cells were purchased from American Type Culture Collection. MCF7 cells were gift from S. Yonehara (Kyoto University). HeLa, HEK293T, and MCF7 cells were grown in DME supplemented with 4 mM glutamine, 100 U/ml penicillin, 0.2 mg/ml streptomycin, and 10% FBS, and MDA-MB-231 cells were grown in DME with high glucose (Invitrogen) containing 10% FBS and 5% NuSerum (BD). Cells were cultured under humidified air containing 5% CO2 at 37°C. Cells were transfected with the indicated plasmids using Lipofectamine Plus (Invitrogen; for HEK293T cells) or Lipofectamine 2000 (for HeLa, MCF7, and MDA-MB-231 cells), according to the manufacturer’s instructions. In some experiments, MDA-MB-231 and HeLa cells transfected with the shRNA expression vector were used after selection with 200 µg/ml and 300 µg/ml hygromycin-B, respectively.
Pull-down assay, immunoprecipitation, and immunoblotting
In vitro binding assays were performed using lysates of HEK293T cells transfected with Flag-tagged Ephexin4 and nucleotide-free forms of recombinant GST-fused Rho GTPases prepared from Escherichia coli. To prepare nucleotide-free GTPases, purified proteins were incubated in buffer C (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.1% Triton X-100, 5% glycerol, 1 mM PMSF, and 1 mM DTT) containing 5 mM EDTA for 20 min at 25°C and kept on ice for 10 min. HEK293T cells transfected with Flag-tagged Ephexin4 were lysed with buffer C, and the cell lysates were then centrifuged at 16,000 g for 10 min at 4°C. The supernatants were incubated with 25 µg GST or GST-fused GTPases and glutathione-Sepharose beads for 1 h at 4°C. After the beads were washed with ice-cold buffer C, the bound proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting.
For immunoprecipitation, HEK293T cells cotransfected with the indicated plasmids were lysed for 10 min with ice-cold cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1% Triton X-100, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin). After centrifugation for 10 min at 16,000 g, the supernatants were incubated with anti-Flag (M2) antibody or recombinant mouse ephrinA1-Fc (R&D Systems) for 2 h and then with protein G–Sepharose (GE Healthcare) for 1 h. Then the beads were washed with the cell lysis buffer. For detecting endogenous binding proteins, MDA-MB-231 or MCF7 cells were lysed for 10 min with ice-cold cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1% Triton X-100, 10 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin). After centrifugation for 10 min at 16,000 g, the supernatants were incubated with anti-EphA2 (D7) antibody for 1.5 h, followed by incubation with protein G–Sepharose for 1 h. Then the beads were washed with the cell lysis buffer.
For immunoblot analysis, proteins were separated by SDS-PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 3% low fat milk in Tris-buffered saline and then incubated with primary antibodies. The primary antibodies were detected with horseradish peroxidase–conjugated secondary antibodies and an ECL detection kit (GE Healthcare).
Measurement of Rho GTPase activity in cells
Measurement of RhoG, Rac1, Cdc42, or RhoA activity in cells was performed according to the modified methods of Benard et al. (1999; for RhoG, Rac1, and Cdc42) and Ren et al. (1999; for RhoA). The CRIB domain of Pak (aa 70–150), the N-terminal RhoG-binding domain of ELMO2 (ELMO-NT; aa 1–362), and the RhoA-binding domain of Rhotekin (aa 2–89) were expressed in E. coli as fusion proteins with GST, purified on glutathione-Sepharose beads, and isolated from the beads with 16 mM reduced glutathione. The purified proteins were dialyzed with 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, and 0.1 mM DTT and stored at −80°C. Protein concentration was determined by comparing with BSA standards after SDS-PAGE and by staining with Coomassie brilliant blue. To determine RhoG, Rac1, or Cdc42 activity in HEK293T or HeLa cells, transfected cells were serum-starved and then lysed with the ice-cold cell lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 10% glycerol, 1 mM DTT, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin) containing 5 µg GST-CRIB or 20 µg GST–ELMO-NT. To measure RhoG or Rac1 activity in MDA-MB-231 cells, cells were stimulated with 10 ng/ml EGF for the indicated times before cell lysis. Cell lysates were then centrifuged for 5 min at 10,000 g at 4°C, and the supernatant was incubated with glutathione-Sepharose beads for 30 min at 4°C. The beads were washed with lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting. For measurement of RhoA activity in HEK293T cells, transfected cells were serum-starved and lysed with ice-cold cell lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 30 mM MgCl2, 1% Triton X-100, 10% glycerol, 1 mM DTT, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin) containing 20 µg of GST-fused RBD of mouse Rhotekin. The cell lysates were centrifuged for 5 min at 16,000 g at 4°C, and the supernatants were incubated with glutathione-Sepharose beads for 50 min at 4°C. The beads were washed with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 30 mM MgCl2, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin), and bound proteins were analyzed by SDS-PAGE and immunoblotting.
In vitro guanine nucleotide exchange assay
Fluorescence spectroscopic analysis of mant-GTP incorporation into GST-RhoG or -Rac1 was performed using a microplate reader (GENios; Tecan) at 20°C. 2 µM GST-RhoG or -Rac1 was prepared and allowed to equilibrate in exchange buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 50 mg/ml BSA, and 0.8 µM mant-GTP). After equilibration, normalized amounts (400 nM) of purified GST or GST-fused DH-PH domain of Ephexin4 were added to the assay mixtures, and the change of mant-GTP fluorescence (excitation = 360 nm; emission = 440 nm) was monitored. Experiments were performed in triplicate for each reaction.
Transwell cell migration assay
MDA-MB-231 or MCF7 cells were transfected with GFP or YFP alone or together with the indicated plasmids and incubated for 24–72 h. The cells were detached with PBS containing EDTA and then resuspended in serum-free DME. The cells were replated at a density of 1–3 × 104 cells onto the upper chamber of a Transwell filter (Costar; 8-µm pore size). The cultured medium (for MCF7 cells) or the medium supplemented with 10 ng/ml EGF (Sigma-Aldrich; for MDA-MB-231 cells) was added to the lower chamber. At 4–6 (for MDA-MB-231 cells) or 6–24 h (for MCF7 cells) after plating, cells were fixed with 4% PFA in PBS. Nonmigrated cells on the upper side of the filter were removed with a cotton swab. In parallel, cells were separately plated to culture wells without the Transwell filters for estimating the total number of attached cells. Relative cell migration was determined by the number of the GFP- or YFP-positive migrated cells normalized to the total number of the GFP- or YFP-positive cells adhering to the plate. For each experiment, the number of cells in 20 random fields on the underside of the filter was counted, and three or four independent filters were analyzed.
Matrigel cell invasion assay
MDA-MB-231 cells were transfected with YFP alone or together with the indicated plasmids and incubated for 24–72 h. The cells were detached with PBS containing EDTA and then resuspended in serum-free DME. The cells were then replated at a density of 3 × 104 cells onto the upper chamber of a Matrigel invasion filter (BD; 8-µm pore size). The medium supplemented with 10 ng/ml EGF was added to the lower chamber. At 24 h after plating, cells were fixed with 4% PFA in PBS. Nonmigrated cells on the upper side of the filter were removed with a cotton swab. In parallel, cells were also separately plated to culture wells without the Matrigel filters for estimating the total number of attached cells. Relative cell invasion was determined by the number of the YFP-positive invaded cells normalized to the total number of the YFP-positive cells adhering to the plate. For each experiment, the number of cells in 20 random fields on the underside of the filter was counted, and three independent filters were analyzed.
Immunofluorescence microscopy
Cells on coverslips were fixed with 4% paraformaldehyde in PBS for 15 min and washed with PBS five times. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% FBS in PBS for 30 min to block nonspecific antibody binding. Then cells were incubated with primary antibodies in PBS or Can Get Signal Immunostain Immunoreaction Enhancer SolutionA (TOYOBO) for 24 h. After wash with PBS at once, cells were incubated with secondary antibodies conjugated with Alexa Fluor 488 or 594 in PBS for 1 h, washed with PBS for 30 min, and mounted in ProLong Gold antifade reagent (Invitrogen). For F-actin staining in HeLa cells, cells were incubated with Alexa Flour 594–conjugated phalloidin in PBS for 1 h, washed with PBS for 30 min, and mounted in 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS. Images were captured at RT using IM50 software (Leica) and a microscope (Eclipse E800; Nikon) with a 40× NA 0.75 objective (Nikon) and a digital camera (DC350F; Leica).
For immunofluorescence staining of migrating cells, MDA-MB-231 cells were replated at a density of 3 × 103 cells onto the upper chamber of the Transwell filter. The medium supplemented with 10 ng/ml EGF was added to the lower chamber. At 2 h after plating, cells were fixed with 4% PFA in PBS for 20 min and washed with PBS. The membrane of Transwell filter was cut off with a scalpel and kept floated in the following methods. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% FBS in PBS for 30 min to block nonspecific antibody binding. Then cells were incubated with primary antibodies in PBS or Can Get Signal Immunostain Immunoreaction Enhancer SolutionA for 24 h. After wash with PBS at once, cells were incubated with secondary antibodies conjugated with Alexa Fluor 488 or 594 in PBS for 1 h, washed with PBS for 30 min, and mounted in ProLong Gold antifade reagent. To obtain a z-plane image, optical sections of images were captured through the cell in 0.50-µm steps at RT using a C1 laser-scanning confocal imaging system (EZ-C1 version 3.20 software; Nikon) and a microscope (Eclipse TE2000-U; Nikon) with a 60× NA 1.40 oil objective (Nikon) and a digital camera (DXM1200C; Nikon). All images were prepared with Photoshop 7.0 (Adobe).
Separation of membrane and cytosol fractions
MDA-MB-231 cells were stimulated with 10 ng/ml EGF for 1 min and then suspended in ice-cold buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, and 1 mM PMSF). After rapid freezing in liquid nitrogen and thawing in a water bath, cells were centrifuged at 16,000 g for 10 min at 4°C. The supernatant was removed and used as a cytosol fraction. After washing the pellet with buffer A, it was lysed with buffer A containing 1% Triton X-100 and centrifuged at 10,000 g for 10 min at 4°C. The supernatant was removed and used as a membrane fraction. Both cytosol and membrane fractions were separated by SDS-PAGE and analyzed by immunoblotting.
Data analysis
Densitometry analysis was performed with ImageJ free image analysis software (National Institutes of Health), and relative RhoG or Rac1 activity was determined by the amount of GTP-bound RhoG or Rac1 bound to GST-ELMO or -CRIB normalized to the amount of total RhoG or Rac1 in cell lysates, respectively. Statistical significance was established by t test using SPSS software (version 16.0; SPSS, Inc.).
Online supplemental material
Fig. S1 shows that Ephexin4 mediates promotion of cell migration by EphA2 in HeLa cells in Transwell migration assays. Fig. S2 shows immunoprecipitation with control Fc or ephrinA1-Fc in HEK293T cell lysates transfected with HA-ELMO2 alone or together with Myc-EphA2. Fig. S3 shows the specificity of Dock4 or RhoG staining with anti-Dock4 or anti-RhoG antibody using Dock4 or RhoG knockdown MDA-MB-231 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005141/DC1.
Acknowledgments
We thank Dr. M. Matsuda for the Dock180 expression plasmid, Drs. J. Miyazaki and T. Saito for the EYFP expression plasmid, Dr. S. Yonehara for MCF7 cells, and Drs. E. Kiyokawa, H. Sabe, and A. Hashimoto for technical help.
This work was supported in part by Grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Abbreviations used in this paper:
- CRIB
- Cdc42/Rac1 interactive binding
- DH
- Dbl homology
- GEF
- guanine nucleotide exchange factor
- PH
- pleckstrin homology
- RBD
- Rho-binding domain
- SAM
- sterile α motif
- shRNA
- short hairpin RNA
- WT
- wild type
References
- Benard V., Bohl B.P., Bokoch G.M. 1999. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274:13198–13204 10.1074/jbc.274.19.13198 [DOI] [PubMed] [Google Scholar]
- Blangy A., Vignal E., Schmidt S., Debant A., Gauthier-Rouvière C., Fort P. 2000. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J. Cell Sci. 113:729–739 [DOI] [PubMed] [Google Scholar]
- Brantley-Sieders D.M., Zhuang G., Hicks D., Fang W.B., Hwang Y., Cates J.M.M., Coffman K., Jackson D., Bruckheimer E., Muraoka-Cook R.S., Chen J. 2008. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. J. Clin. Invest. 118:64–78 10.1172/JCI33154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.C., Macara I.G., Madhani H., Fink G.R., Ravichandran K.S. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574–582 [DOI] [PubMed] [Google Scholar]
- Côté J.F., Vuori K. 2002. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115:4901–4913 10.1242/jcs.00219 [DOI] [PubMed] [Google Scholar]
- Côté J.F., Vuori K. 2007. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17:383–393 10.1016/j.tcb.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J.F., Motoyama A.B., Bush J.A., Vuori K. 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7:797–807 10.1038/ncb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C.W., Shao Y.R., Sahin M., Shamah S.M., Lin M.Z., Greer P.L., Gao S., Griffith E.C., Brugge J.S., Greenberg M.E. 2005. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron. 46:205–217 10.1016/j.neuron.2005.03.019 [DOI] [PubMed] [Google Scholar]
- DesMarais V., Yamaguchi H., Oser M., Soon L., Mouneimne G., Sarmiento C., Eddy R., Condeelis J. 2009. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskeleton. 66:303–316 10.1002/cm.20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfenbein A., Rhodes J.M., Meller J., Schwartz M.A., Matsuda M., Simons M. 2009. Suppression of RhoG activity is mediated by a syndecan 4–synectin–RhoGDI1 complex and is reversed by PKCα in a Rac1 activation pathway. J. Cell Biol. 186:75–83 10.1083/jcb.200810179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek S.M., Wennerberg K., Arthur W.T., Dunty J.M., Bowman D.R., DeMali K.A., Der C., Burridge K. 2004. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol. Biol. Cell. 15:3309–3319 10.1091/mbc.E04-02-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2002. Rho GTPases in cell biology. Nature. 420:629–635 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Fu W.Y., Chen Y., Sahin M., Zhao X.S., Shi L., Bikoff J.B., Lai K.O., Yung W.H., Fu A.K.Y., Greenberg M.E., Ip N.Y. 2007. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 10:67–76 10.1038/nn1811 [DOI] [PubMed] [Google Scholar]
- Fujimoto S., Negishi M., Katoh H. 2009. RhoG promotes neural progenitor cell proliferation in mouse cerebral cortex. Mol. Biol. Cell. 20:4941–4950 10.1091/mbc.E09-03-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea G., Sanz-Moreno V., Self A., Godi A., Marshall C.J. 2008. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr. Biol. 18:1456–1465 10.1016/j.cub.2008.08.053 [DOI] [PubMed] [Google Scholar]
- Gumienny T.L., Brugnera E., Tosello-Trampont A.C., Kinchen J.M., Haney L.B., Nishiwaki K., Walk S.F., Nemergut M.E., Macara I.G., Francis R., et al. 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 107:27–41 10.1016/S0092-8674(01)00520-7 [DOI] [PubMed] [Google Scholar]
- Hiramoto K., Negishi M., Katoh H. 2006. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 312:4205–4216 10.1016/j.yexcr.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Hunter S.G., Zhuang G., Brantley-Sieders D., Swat W., Cowan C.W., Chen J. 2006. Essential role of Vav family guanine nucleotide exchange factors in EphA receptor-mediated angiogenesis. Mol. Cell. Biol. 26:4830–4842 10.1128/MCB.02215-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N.E., Lane H.A. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 5:341–354 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- Iwasato T., Katoh H., Nishimaru H., Ishikawa Y., Inoue H., Saito Y.M., Ando R., Iwama M., Takahashi R., Negishi M., Itohara S. 2007. Rac-GAP alpha-chimerin regulates motor-circuit formation as a key mediator of EphrinB3/EphA4 forward signaling. Cell. 130:742–753 10.1016/j.cell.2007.07.022 [DOI] [PubMed] [Google Scholar]
- Jarzynka M.J., Hu B., Hui K.M., Bar-Joseph I., Gu W., Hirose T., Haney L.B., Ravichandran K.S., Nishikawa R., Cheng S.Y. 2007. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 67:7203–7211 10.1158/0008-5472.CAN-07-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., Negishi M. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 424:461–464 10.1038/nature01817 [DOI] [PubMed] [Google Scholar]
- Katoh H., Harada A., Mori K., Negishi M. 2002. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol. Cell. Biol. 22:2952–2964 10.1128/MCB.22.9.2952-2964.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., Hiramoto K., Negishi M. 2006. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 119:56–65 10.1242/jcs.02720 [DOI] [PubMed] [Google Scholar]
- Kawada K., Upadhyay G., Ferandon S., Janarthanan S., Hall M., Vilardaga J.P., Yajnik V. 2009. Cell migration is regulated by platelet-derived growth factor receptor endocytosis. Mol. Cell. Biol. 29:4508–4518 10.1128/MCB.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa E., Hashimoto Y., Kobayashi S., Sugimura H., Kurata T., Matsuda M. 1998. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 12:3331–3336 10.1101/gad.12.21.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto K., Negishi M., Katoh H. 2009. Regulation of dendrite growth by the Cdc42 activator Zizimin1/Dock9 in hippocampal neurons. J. Neurosci. Res. 87:1794–1805 10.1002/jnr.21997 [DOI] [PubMed] [Google Scholar]
- Kurisu S., Suetsugu S., Yamazaki D., Yamaguchi H., Takenawa T. 2005. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene. 24:1309–1319 10.1038/sj.onc.1208177 [DOI] [PubMed] [Google Scholar]
- Larsen A.B., Pedersen M.W., Stockhausen M.T., Grandal M.V., van Deurs B., Poulsen H.S. 2007. Activation of the EGFR gene target EphA2 inhibits epidermal growth factor-induced cancer cell motility. Mol. Cancer Res. 5:283–293 10.1158/1541-7786.MCR-06-0321 [DOI] [PubMed] [Google Scholar]
- Macrae M., Neve R.M., Rodriguez-Viciana P., Haqq C., Yeh J., Chen C., Gray J.W., McCormick F. 2005. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 8:111–118 10.1016/j.ccr.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Meller N., Irani-Tehrani M., Kiosses W.B., Del Pozo M.A., Schwartz M.A. 2002. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol. 4:639–647 10.1038/ncb835 [DOI] [PubMed] [Google Scholar]
- Meller N., Merlot S., Guda C. 2005. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 118:4937–4946 10.1242/jcs.02671 [DOI] [PubMed] [Google Scholar]
- Meller N., Westbrook M.J., Shannon J.D., Guda C., Schwartz M.A. 2008. Function of the N-terminus of zizimin1: autoinhibition and membrane targeting. Biochem. J. 409:525–533 10.1042/BJ20071263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suárez A., Batlle E. 2008. Eph-ephrin signalling in adult tissues and cancer. Curr. Opin. Cell Biol. 20:194–200 10.1016/j.ceb.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Miao H., Li D.Q., Mukherjee A., Guo H., Petty A., Cutter J., Basilion J.P., Sedor J., Wu J., Danielpour D., et al. 2009. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 16:9–20 10.1016/j.ccr.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T. 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17:6932–6941 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Yamauchi J. 2010. Cellular signaling of Dock family proteins in neural function. Cell. Signal. 22:175–182 10.1016/j.cellsig.2009.09.036 [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Yamauchi J., Sanbe A., Tanoue A. 2007. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 313:791–804 10.1016/j.yexcr.2006.11.017 [DOI] [PubMed] [Google Scholar]
- Murga C., Zohar M., Teramoto H., Gutkind J.S. 2002. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB. Oncogene. 21:207–216 10.1038/sj.onc.1205036 [DOI] [PubMed] [Google Scholar]
- Namekata K., Enokido Y., Iwasawa K., Kimura H. 2004. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 279:14331–14337 10.1074/jbc.M311275200 [DOI] [PubMed] [Google Scholar]
- Nishihara H., Kobayashi S., Hashimoto Y., Ohba F., Mochizuki N., Kurata T., Nagashima K., Matsuda M. 1999. Non-adherent cell-specific expression of DOCK2, a member of the human CDM-family proteins. Biochim. Biophys. Acta. 1452:179–187 10.1016/S0167-4889(99)00133-0 [DOI] [PubMed] [Google Scholar]
- Ogita H., Kunimoto S., Kamioka Y., Sawa H., Masuda M., Mochizuki N. 2003. EphA4-mediated Rho activation via Vsm-RhoGEF expressed specifically in vascular smooth muscle cells. Circ. Res. 93:23–31 10.1161/01.RES.0000079310.81429.C8 [DOI] [PubMed] [Google Scholar]
- Pasquale E.B. 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 133:38–52 10.1016/j.cell.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Ren X.D., Kiosses W.B., Schwartz M.A. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578–585 10.1093/emboj/18.3.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild B.L., Shim A.H., Ammer A.G., Kelley L.C., Irby K.B., Head J.A., Chen L., Varella-Garcia M., Sacks P.G., Frederick B., et al. 2006. Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 66:8017–8025 10.1158/0008-5472.CAN-05-4490 [DOI] [PubMed] [Google Scholar]
- Roux P., Gauthier-Rouvière C., Doucet-Brutin S., Fort P. 1997. The small GTPases Cdc42Hs, Rac1 and RhoG delineate Raf-independent pathways that cooperate to transform NIH3T3 cells. Curr. Biol. 7:629–637 10.1016/S0960-9822(06)00289-2 [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2002. RHO-GTPases and cancer. Nat. Rev. Cancer. 2:133–142 10.1038/nrc725 [DOI] [PubMed] [Google Scholar]
- Sahin M., Greer P.L., Lin M.Z., Poucher H., Eberhart J., Schmidt S., Wright T.M., Shamah S.M., O’Connell S., Cowan C.W., et al. 2005. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 46:191–204 10.1016/j.neuron.2005.01.030 [DOI] [PubMed] [Google Scholar]
- Saito T., Nakatsuji N. 2001. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240:237–246 10.1006/dbio.2001.0439 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 135:510–523 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- Shamah S.M., Lin M.Z., Goldberg J.L., Estrach S., Sahin M., Hu L., Bazalakova M., Neve R.L., Corfas G., Debant A., Greenberg M.E. 2001. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 105:233–244 10.1016/S0092-8674(01)00314-2 [DOI] [PubMed] [Google Scholar]
- Takeuchi S., Yamaki N., Iwasato T., Negishi M., Katoh H. 2009. β2-chimaerin binds to EphA receptors and regulates cell migration. FEBS Lett. 583:1237–1242 10.1016/j.febslet.2009.03.032 [DOI] [PubMed] [Google Scholar]
- Ueda S., Fujimoto S., Hiramoto K., Negishi M., Katoh H. 2008. Dock4 regulates dendritic development in hippocampal neurons. J. Neurosci. Res. 86:3052–3061 10.1002/jnr.21763 [DOI] [PubMed] [Google Scholar]
- Upadhyay G., Goessling W., North T.E., Xavier R., Zon L.I., Yajnik V. 2008. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene. 27:5845–5855 10.1038/onc.2008.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F.M., Ridley A.J. 2008. Rho GTPases in cancer cell biology. FEBS Lett. 582:2093–2101 10.1016/j.febslet.2008.04.039 [DOI] [PubMed] [Google Scholar]
- Walker-Daniels J., Riese D.J., II, Kinch M.S. 2002. c-Cbl-dependent EphA2 protein degradation is induced by ligand binding. Mol. Cancer Res. 1:79–87 [PubMed] [Google Scholar]
- Wang Y., Suzuki H., Yokoo T., Tada-Iida K., Kihara R., Miura M., Watanabe K., Sone H., Shimano H., Toyoshima H., Yamada N. 2004. WGEF is a novel RhoGEF expressed in intestine, liver, heart, and kidney. Biochem. Biophys. Res. Commun. 324:1053–1058 10.1016/j.bbrc.2004.09.153 [DOI] [PubMed] [Google Scholar]
- Weaver A.M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157–166 10.1016/j.canlet.2008.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed S.A., Du Y., Parsons J.T. 1998. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J. Cell Sci. 111:2433–2443 [DOI] [PubMed] [Google Scholar]
- Xie X., Chang S.W., Tatsumoto T., Chan A.M., Miki T. 2005. TIM, a Dbl-related protein, regulates cell shape and cytoskeletal organization in a Rho-dependent manner. Cell. Signal. 17:461–471 10.1016/j.cellsig.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Yajnik V., Paulding C., Sordella R., McClatchey A.I., Saito M., Wahrer D.C., Reynolds P., Bell D.W., Lake R., van den Heuvel S., et al. 2003. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 112:673–684 10.1016/S0092-8674(03)00155-7 [DOI] [PubMed] [Google Scholar]
- Yamaki N., Negishi M., Katoh H. 2007. RhoG regulates anoikis through a phosphatidylinositol 3-kinase-dependent mechanism. Exp. Cell Res. 313:2821–2832 10.1016/j.yexcr.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Yamauchi J., Miyamoto Y., Chan J.R., Tanoue A. 2008. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell Biol. 181:351–365 10.1083/jcb.200709033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D., Kurisu S., Takenawa T. 2009. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene. 28:1570–1583 10.1038/onc.2009.2 [DOI] [PubMed] [Google Scholar]
- Zelinski D.P., Zantek N.D., Stewart J.C., Irizarry A.R., Kinch M.S. 2001. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 61:2301–2306 [PubMed] [Google Scholar]
- Zhuang G., Hunter S., Hwang Y., Chen J. 2007. Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-Kinase-dependent Rac1 activation. J. Biol. Chem. 282:2683–2694 10.1074/jbc.M608509200 [DOI] [PubMed] [Google Scholar]