SHIP2 is recruited early to clathrin-coated pits by the scaffold protein intersectin and dissociates before fission.
Abstract
Phosphatidylinositol (PI) 4,5-bisphosphate (PI(4,5)P2) and its phosphorylated product PI 3,4,5-triphosphate (PI(3,4,5)P3) are two major phosphoinositides concentrated at the plasma membrane. Their levels, which are tightly controlled by kinases, phospholipases, and phosphatases, regulate a variety of cellular functions, including clathrin-mediated endocytosis and receptor signaling. In this study, we show that the inositol 5-phosphatase SHIP2, a negative regulator of PI(3,4,5)P3-dependent signaling, also negatively regulates PI(4,5)P2 levels and is concentrated at endocytic clathrin-coated pits (CCPs) via interactions with the scaffold protein intersectin. SHIP2 is recruited early at the pits and dissociates before fission. Both knockdown of SHIP2 expression and acute production of PI(3,4,5)P3 shorten CCP lifetime by enhancing the rate of pit maturation, which is consistent with a positive role of both SHIP2 substrates, PI(4,5)P2 and PI(3,4,5)P3, on coat assembly. Because SHIP2 is a negative regulator of insulin signaling, our findings suggest the importance of the phosphoinositide metabolism at CCPs in the regulation of insulin signal output.
Introduction
Phosphatidylinositol (PI) 4,5-bisphosphate (PI(4,5)P2) and PI 3,4,5-triphosphate (PI(3,4,5)P3) are two phosphoinositides concentrated at the plasma membrane that play major regulatory roles in a variety of cellular functions. Their levels are tightly controlled by kinases, phosphatases, and phospholipases (Di Paolo and De Camilli, 2006; Vicinanza et al., 2008). Some phosphatases not only control cell surface–associated levels of these phosphoinositides but also couple endocytosis to their dephosphorylation, thus ensuring their preferential or selective retention at the plasma membrane. More specifically, synaptojanin 1 and 2, as well as OCRL (oculocerebrorenal syndrome of Lowe) and INPP5B, all contain an inositol 5-phosphatase domain, bind endocytic proteins, and are found at early stages of the endocytic pathway (McPherson et al., 1996; Shin et al., 2005; Hyvola et al., 2006; Perera et al., 2006; Erdmann et al., 2007). Additionally, synaptojanin 1 and OCRL, which contain binding sites for clathrin’s heavy chain and its adaptor AP-2, are recruited to endocytic clathrin-coated pits (CCPs; Perera et al., 2006; Erdmann et al., 2007; Choudhury et al., 2009; Mao et al., 2009). Studies of these enzymes, as well as evidence for the critical role of PI(4,5)P2 in the recruitment to the plasma membrane of endocytic clathrin adaptors and their accessory factors, led to the now well-established concept that PI(4,5)P2 plays an important role in CCP dynamics (Cremona et al., 1999; Haucke, 2005; Di Paolo and De Camilli, 2006; Zoncu et al., 2007) in addition to its classical signaling roles.
Endocytic clathrin adaptors also bind PI(3,4,5)P3 (Hao et al., 1997; Rapoport et al., 1997; Gaidarov and Keen, 1999; Itoh et al., 2001), an important mediator of the actions of insulin and other growth factors. Furthermore, inositol 5-phosphatases known to be located at endocytic CCPs, namely synaptojanin and OCRL (Perera et al., 2006; Erdmann et al., 2007; Mao et al., 2009), can act on PI(3,4,5)P3 in addition to PI(4,5)P2 (Woscholski et al., 1997; Zhang et al., 1998; Ooms et al., 2009). Thus, PI(3,4,5)P3 may contribute to clathrin coat dynamics besides having a role in signaling. Based on these considerations, we have investigated whether SHIP2, a broadly expressed inositol 5-phosphatase whose preferred substrate is PI(3,4,5)P3 but can also act on PI(4,5)P2 (Hejna et al., 1995; Taylor et al., 2000), has a role at endocytic CCPs.
Results and discussion
SHIP2 is localized at endocytic CCPs
Total internal reflection fluorescence microscopy (TIRFM) of COS-7 cells cotransfected with GFP-SHIP2 and with an mRFP fusion of the clathrin light chain (clathrin-mRFP) revealed that in >95% of cells expressing GFP-SHIP2 at low levels, this protein appeared in small diffraction-limited spots that overlapped with clathrin puncta (Figs. 1 A and 2 A; Gaidarov et al., 1999). Accordingly, SHIP2 also colocalized with mRFP-tagged epsin, an endocytic clathrin adaptor (Fig. 1 B; Chen et al., 1998). More than 80% of endocytic CCPs were positive for SHIP2. Only in a minority of cells was SHIP2 localized instead to focal adhesions, which is consistent with the known interaction of SHIP2 with focal adhesion proteins such as p130CAS and filamin (Dyson et al., 2001; Prasad et al., 2001). Both CCP and focal adhesion localizations of GFP-SHIP2 were observed in cells expressing higher levels of the protein. The presence of GFP-SHIP2 at CCPs was observed in all cells examined (C2C12, PtK2, mouse fibroblasts, and primary astrocytes; Fig. S1 A), whereas the highly homologous 5-phosphatase SHIP1 did not localize to CCPs (not depicted).
Figure 1.
Localization of SHIP2 at CCPs. (A and B) TIRFM images of COS-7 cells expressing GFP-SHIP2 and either clathrin-mRFP (A) or mRFP-epsin (B). (A) Insets show the boxed areas at high magnification. White arrows point to SHIP2-positive CCPs, whereas red arrowheads point to SHIP2-negative CCPs. (C and D) Snapshots of clathrin-mRFP and GFP-SHIP2 fluorescence at a single CCP (C) and mean time course of relative fluorescence intensity at CCPs (D). (D) Error bars show mean ± SD. (E) Snapshots of GFP-SHIP2 and cortactin-DsRed fluorescence at a CCP. Bars: (A and B) 5 µm; (C and E) 1 µm.
Figure 2.
The recruitment of SHIP2 to CCPs is mediated by intersectin. (A–F) COS-7 cells transiently expressing GFP fusions of SHIP2 constructs as indicated together with either clathrin-mRFP or mCherry-actin. 5-Ptase, 5-phosphatase. (G) SDS-PAGE gel (Coomassie blue staining) of the material affinity purified from a rat brain extract by pull-downs with GST or GST–SHIP2-1022–1078 as indicated (ITSN-L and -S, long and short isoforms of intersectin). (H) Anti–intersectin 1 Western blotting of material pulled down from rat brain lysate with GST, GST–SHIP2-1022–1078 (WT) or GST–SHIP2-1022–1078 with an F to A mutation at position 1054 (FA). The GST proteins used for the pull-down are shown below the Western blot (Coomassie blue staining). The dashed line indicates separate membranes. (I) Western blotting with anti–intersectin 1 or anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies of COS-7 cells transfected with control siRNA (ctrl) or intersectin 1–specific siRNA (KD). (J) TIRFM image of COS-7 cell transfected with control siRNA (ctrl) or intersectin 1–specific siRNA (KD) transiently expressing GFP-SHIP2 and clathrin-mRFP. Bars, 5 µm.
The dynamics of SHIP2 at CCPs were analyzed by TIRFM in COS-7 cells expressing low levels of GFP-SHIP2 and clathrin-mRFP. Clathrin spots, shown previously to represent individual CCPs (Gaidarov et al., 1999; Zoncu et al., 2007), formed continuously at the plasma membrane and abruptly disappeared as the result of fission and internalization. Their cumulative surface lifetimes were 1–2 min (Fig. 1, C and D), as we and others previously reported (Gaidarov et al., 1999; Perera et al., 2006). GFP-SHIP2 signal increased in parallel with clathrin-mRFP fluorescence but plateaued earlier than clathrin and disappeared slightly before clathrin (Fig. 1, C and D; and Video 1). Comparison of the dynamics of GFP-SHIP2 relative to cortactin-DsRed, which visits late-stage CCPs together with dynamin just before fission (Merrifield et al., 2005), revealed that cortactin appeared just as SHIP2 disappeared (Fig. 1 E), confirming that SHIP2 leaves CCPs before fission. Collectively, these results demonstrate that SHIP2 is a component of early-stage CCPs.
The localization of SHIP2 at CCPs is mediated by its proline-rich region
The catalytic domain of SHIP2 (see domain structure in Fig. 2 A) is flanked at the C-terminal side by a proline-rich region followed by a SAM (sterile α motif) domain (Ooms et al., 2009). Deletion of the SAM domain did not affect CCP localization (Fig. S1 C). However, a SHIP2 construct lacking the proline-rich region no longer localized to CCPs (Fig. 2 B). This construct localized instead to focal adhesions, as verified by colocalization with the tip of actin stress fibers (Fig. 2 C), and Mena, a focal adhesion protein (Fig. S2 D; Krause et al., 2003). Thus, SHIP2 is recruited to CCPs through its proline-rich region, and when the targeting signal present in this domain is missing, another targeting signal that directs the protein to focal adhesion predominates. The small fraction of cells that normally exhibit preferential focal adhesion localization of full-length SHIP2 may reflect a different functional state of these cells and suggests that the balance between these two types of SHIP2 localizations may be regulated. Analysis of additional SHIP2 truncation mutants mapped the CCP localization signal to aa 1022–1078 (Fig. 2 D and Fig. S2 E). This region does not include known clathrin or clathrin adaptor binding motifs.
Binding of a polyproline motif of SHIP2 to intersectin is responsible for CCP localization
To identify protein interactors of the aa 1022–1078 region of SHIP2 that may be responsible for CCP localization, pull-down experiments from a rat brain cytosolic extract were performed. SDS-PAGE analysis of the material specifically affinity purified by GST–SHIP2-1022–1078 revealed ∼190- and ∼150-kD bands, which mass spectrometry identified as the long (L) and short (S) forms of intersectin 1 (Fig. 2 G). This identification, which is consistent with a recent yeast two hybrid–based study of a direct interaction of SHIP2 with intersectin 1 (Xie et al., 2008), was confirmed by Western blotting of the affinity-purified material (Fig. 2 H).
Because intersectin 1 is a scaffold adaptor protein concentrated at CCPs (McPherson, 2002), its role in subcellular targeting of SHIP2 was investigated by TIRFM. In COS-7 cells, both GFP–intersectin 1–L and –S colocalized with clathrin-mRFP as expected (Fig. S2, A and B). SHIP2 also colocalized at these puncta (Fig. S2 C). However, when levels of intersectin 1 were reduced by ∼90% via siRNA-mediated knockdown (KD; Fig. 2 I), SHIP2 localization at CCPs was no longer observed (Fig. 2 J), although the clathrin signal itself remained robust. Under these conditions, SHIP2 had a mainly diffuse localization with some accumulation at focal adhesions. Because localization of intersectin 1 at CCPs was not perturbed in SHIP2 KD cells (Fig. S2 D), these results indicate that intersectin 1 regulates the CCP localization of SHIP2. This possibility is consistent with the recent report that intersectin, as shown in this study for SHIP2 (Fig. 1), is an early component of CCPs that dissociates from them before fission (Henne et al., 2010).
Binding of SHIP2 to intersectin 1 was shown to involve its SH3 domains (Xie et al., 2008). Upon inspection of the 1022–1078 aa sequence of SHIP2 for proline-based putative SH3-binding motifs using the Eukaryotic Linear Motif server (http://elm.eu.org/) and SH3-Hunter websites (http://cbm.bio.uniroma2.it/SH3-Hunter/), several candidate SH3-binding sites were identified, including the motif PPPDF (aa 1050–1054), which is similar to the previously recognized SH3 binding motif PPPDY (Mongioví et al., 1999). Pull-downs with GST–SHIP2-1022–1078 from brain extracts revealed that intersectin binding was disrupted by an F to A mutation at position 1054 (Fig. 2 H). Moreover, full-length GFP–SHIP2-F1054A mutant did not localize at CCPs (Fig. 2 E) and produced the focal adhesion pattern as well as a diffuse localization (Fig. 2 F). We conclude that the sequence of SHIP2 centered around the PPPDF sequence plays a key role in the localization of SHIP2 to CCPs.
SHIP2 is a negative regulator of CCP growth
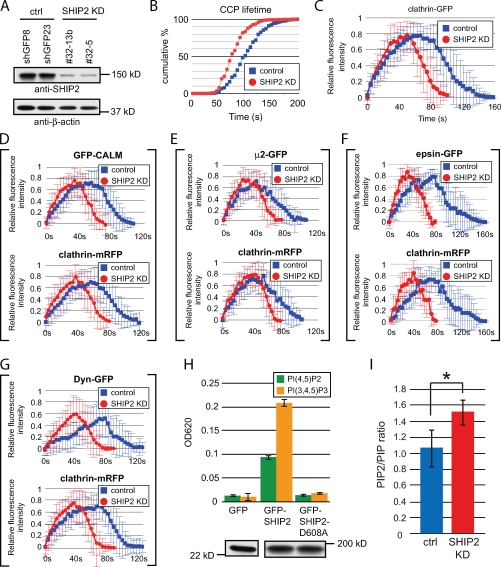
We next explored a potential role of SHIP2 on CCP dynamics. COS-7 cells were stably transfected with control short hairpin RNA (shRNA) or shRNA directed against SHIP2 to knock down this phosphatase (Fig. 3 A). TIRFM of these cells after transient expression of GFP–clathrin light chain revealed that the mean CCP lifetime in SHIP2 KD cells (77 s; n = 1,648; Fig. 3 B, red; and Video 2) was 25% shorter than in control cells (102 s; n = 697; Fig. 3 B, blue; and Video 3). This change was accounted for, at least in part, by a faster rate of increase in the GFP-clathrin signal (Fig. 3 C), suggesting that SHIP2 negatively regulates the speed of maturation of CCPs.
Figure 3.
Loss of SHIP2 affects PI(4,5)P2 levels and accelerates endocytic CCP dynamics. (A) Anti-SHIP2 and anti–β-actin Western blots of COS-7 cells stably expressing control shRNA (ctrl) or SHIP2-specific shRNA (SHIP2 KD). (B) Cumulative histograms of CCP lifetime (GFP–clathrin light chain signal) in control cells (n = 697) and SHIP2 KD cells (n = 1,648). (C) Time course of relative fluorescence intensity of GFP-clathrin in control cells and SHIP2 KD cells. (D–G) Time course of relative fluorescence intensity of mRFP and GFP fluorescence in cells double transfected with clathrin-mRFP and with GFP fusions of clathrin adaptors and dynamin as indicated. (H, top) PI(4,5)P2 and PI(3,4,5)P3 phosphatase activity (Malachite green assay) in anti-GFP immunoprecipitates from COS-7 cells expressing GFP, GFP-SHIP2, or GFP–SHIP2-D608A (catalytically dead). (bottom) Anti-GFP Western blot of the material used in the phosphatase assay. (I) PIP2/PIP ratio in control and SHIP2 KD cells as determined by HPLC (*, P < 0.05; t test). (C–I) Error bars show mean ± SD.
Similar results were obtained by monitoring the effect of SHIP2 KD on the recruitment to the pits of three clathrin adaptors. The adaptor AP-2, as demonstrated by imaging its GFP-tagged μ2 subunit, and the monomeric adaptors epsin1 (epsin1-GFP) and CALM (clathrin assembly lymphoid myeloid leukemia; GFP-CALM) accumulated more rapidly and peaked earlier at CCPs of SHIP2 KD cells than in control cells (Fig. 3, D–F), with a time course that precisely paralleled that of clathrin (Fig. S3). An earlier recruitment to the CCPs was also observed for dynamin (dynamin-GFP; Fig. 3 G), a critical factor for endocytic CCP fission, which is consistent with the overall shorter lifetime of the CCPs.
SHIP2 and phosphoinositide levels
PI(4,5)P2, which binds all endocytic clathrin adaptors as well as dynamin, is a master regulator of endocytic clathrin coat assembly (Gaidarov and Keen, 1999; Ford et al., 2001; Collins et al., 2002; Haucke, 2005; Zoncu et al., 2007; Thieman et al., 2009). Although the preferred substrate for the inositol 5-phosphatase activity of SHIP2 was reported to be PI(3,4,5)P3 (Damen et al., 1996; Pesesse et al., 1998), PI(4,5)P2 is also a substrate (Taylor et al., 2000), as we have confirmed by an in vitro phosphatase assay (Fig. 3 H). A physiological and major role of SHIP2 in the control of PI(4,5)P2 levels was supported by the finding that the PI bisphosphate (PIP2)/PI phosphate (PIP) ratio in SHIP2 KD cells was ∼40% higher than in control cells (Fig. 3 I). Thus, a potential explanation for the faster growth of endocytic CCPs in SHIP2 KD cells is that higher levels of PI(4,5)P2 promote a faster recruitment of the endocytic clathrin adaptors and their accessory proteins (Fig. 3, C–G).
Clathrin adaptors can also bind PI(3,4,5)P3 (Gaidarov et al., 1996; Hao et al., 1997; Rapoport et al., 1997; Gaidarov and Keen, 1999; Itoh et al., 2001; Collins et al., 2002). However, PI(3,4,5)P3 levels were at the limit of detectability in both control and SHIP2 KD cells kept in normal serum-containing medium, unless cells were subjected to acute serum stimulation after serum deprivation. Under such conditions, a PI(3,4,5)P3 peak was readily detected, and its levels were ∼20% higher in SHIP2 KD cells (Fig. 4 A). Thus, global changes in PI(3,4,5)P3 levels are unlikely to account for changes in the dynamics of CCPs under our standard culture conditions. It remains possible that local pools of PI(3,4,5)P3 generated by PI 3-kinases localized at CCPs may play a role in CCP dynamics and that impaired dephosphorylation of PI(3,4,5)P3 may contribute to the accelerated maturation of CCPs observed in SHIP2 KD cells. Type I PI 3-kinases are expected to be localized at endocytic CCPs that mediate the internalization of growth factor receptors (Cantley, 2002; Engelman et al., 2006). Furthermore, studies of type II PI 3-kinases α and β (PI3KC2-α and PI3KC2-β) have revealed links of these enzymes to CCPs. More specifically, (a) both PI3KC2-α and PI3KC2-β contain binding sites for endocytic clathrin coats (Engelman et al., 2006), (b) PI3KC2-α was detected at CCPs (Gaidarov et al., 2001), and (c) PI3KC2-β was shown to interact with SH3 domains of intersectin 1, an endocytic CCP component (Fig. S2; Das et al., 2007; Henne et al., 2010). In fact, TIRFM analysis confirmed that both PI3KC2-α and PI3KC2-β localized at CCPs in COS-7 cells (Fig. S2, E and F). Although the main in vitro product of type II PI 3-kinases is PI 3-phosphate, it was reported that binding of PI3KC2-α to clathrin modifies its substrate preference, thus enabling it to synthesize PI(3,4,5)P3 from PI(4,5)P2 (Gaidarov et al., 2001).
Figure 4.
PI(3,4,5)P3 production accelerates endocytic CCP dynamics. (A) Ratio of PI(3,4,5)P3 to total inositol phospholipids (PIs) in control and SHIP2 KD cells after serum stimulation as determined by HPLC. Error bars show mean ± SD (*, P < 0.05; t test). (B) Schematic representation of the rapamycin-inducible heterodimerization system used to recruit a PI 3-kinase at the plasma membrane. (C) COS-7 cells expressing RFP–FKBP-iSH2 or GFP-Akt-PH in the absence or presence of the rapamycin analogue (RAPA). (D) Cumulative histograms of CCP lifetime in cells expressing RFP-FKBP (n = 186; blue), RFP–FKBP-iSH2 (n = 227; red), or RFP–FKBP-iSH2 in ruffle (n = 30; green). Bar, 10 µm.
PI(3,4,5)P3 production accelerates endocytic CCP dynamics
Based on the aforementioned considerations, we tested directly the effect of PI(3,4,5)P3 on CCP dynamics. Because acute stimulation by serum or purified growth factors is expected not only to increase PI(3,4,5)P3 levels but also to affect the dynamics of proteins associated with the plasma membrane indirectly, via the stimulation of tyrosine phosphorylation, we used a protocol selectively aimed at increasing PI(3,4,5)P3. More specifically, we used the rapamycin-inducible FRB (FKBP12-rapamycin binding)-FKBP (FK506-binding protein) heterodimerization system to acutely recruit PI 3-kinase to the plasma membrane (Fig. 4 B; Suh et al., 2006; Varnai et al., 2006; Zoncu et al., 2007). Plasma membrane–targeted CFP-FRB was coexpressed in COS-7 cells together with RFP-FKBP (as a control) or with RFP-FKBP fused to the p110-binding domain (also called the inter-SH2 domain) of the p85 regulatory subunit of type I PI 3-kinase (RFP–FKBP-iSH2), which functions as a ligand for the endogenous p110 catalytic subunit of the kinase. Addition of the rapamycin analogue iRAP (indole-modified analogue of rapamycin) induced a robust translocation of RFP-FKBP (not depicted) or RFP–FKBP-iSH2 to the plasma membrane, as expected (Fig. 4 C). However, only in cells expressing RFP–FKBP-iSH2, and not in cells expressing RFP-FKBP (not depicted), were a recruitment of the PI(3,4,5)P3 reporter GFP-Akt–pleckstrin homology (PH) to the plasma membrane and formation of ruffles also observed, indicating production of PI(3,4,5)P3 in this membrane (Fig. 4 C). In cells expressing RFP–FKBP-iSH2 and treated with iRAP, the mean CCP lifetime (115 s; n = 227; Fig. 4 D, red) was ∼20% shorter than in control cells expressing RFP-FKBP (149 s; n = 186; Fig. 4 D, blue), supporting the hypothesis that higher levels of PI(3,4,5)P3 in the plasma membrane accelerate CCP growth and turnover. Furthermore, the lifetime of CCPs was even faster (92 s; n = 30; Fig. 4 D, green) at the membrane ruffles (Video 4), i.e., sites where the concentration of GFP-Akt-PH, and thus PI(3,4,5)P3, in the membrane is the highest. Collectively, these results suggest that dephosphorylation of PI(3,4,5)P3, in addition to dephosphorylation of PI(4,5)P2, is a mechanism through which SHIP2 controls CCP dynamics and increased levels of either of these phosphoinositides leads to accelerated CCP turnover.
Concluding remarks
Collectively, these results demonstrate that SHIP2 has an important function in coat dynamics and signaling at endocytic CCPs, where intersectin plays a major role in its recruitment. Based on the impact of SHIP2 depletion on total PI(4,5)P2 levels, at least some of these effects are likely to be mediated by its property to dephosphorylate PI(4,5)P2 in addition to PI(3,4,5)P3. PTEN, the other negative regulator of PI(3,4,5)P3 signaling, does not act on PI(4,5)P2 (Maehama and Dixon, 1999), which may account for some of the different physiological actions of PTEN and SHIP2.
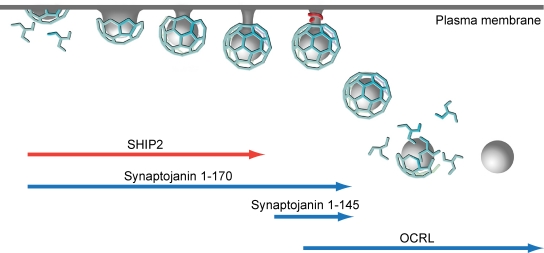
The previously unknown robust effect of SHIP2 in the regulation of PI(4,5)P2 levels raises the possibility that a reduction in the availability of this substrate for PI 3-kinases may contribute to the negative effect of SHIP2 on PI(3,4,5)P3 signaling. The faster maturation and shorter lifetime of CCPs in SHIP2 KD cells not only agrees with a role of PI(4,5)P2 and possibly PI(3,4,5)P3 in their dynamics but also supports the occurrence of phosphoinositide turnover at CCPs, as first suggested by the early recruitment at pits of another inositol 5-phosphatase, synaptojanin-170 (Fig. 5; Perera et al., 2006). Like SHIP2, synaptojanin can act both on PI(4,5)P2 and PI(3,4,5)P3 (Woscholski et al., 1997; Cremona et al., 1999; Mani et al., 2007). Because SHIP2 is recruited to CCPs from the initial stages but leaves before fission, it most likely controls only growth and/or maturation of CCPs, whereas synaptojanin-170 is also implicated in late endocytic steps, including clathrin uncoating after fission (Fig. 5; Perera et al., 2006). Interestingly, intersectin also binds synaptojanin (McPherson, 2002), although it does not appear to have a dominant role in the recruitment of this protein to CCPs. OCRL, another inositol 5-phosphatase that acts on both PI(4,5)P2 and PI(3,4,5)P3, is recruited to CCPs only at late stages and remains associated with newly formed endocytic vesicles (Fig. 5; Erdmann et al., 2007). Thus, it would appear that CCP turnover and signaling involve sequential and coordinated actions of multiple inositol 5-phosphatases. The relative importance of each inositol 5-phosphatase likely differs in different cell types, given the heterogeneous expression of these enzymes across tissues.
Figure 5.
Inositol 5-phosphatases at endocytic CCPs. Schematic cartoon showing that different inositol 5-phosphatases associate with endocytic CCPs at different stages of their maturation.
Overall, an important role in endocytic clathrin coat dynamics and signaling appears to be a shared, evolutionary conserved function of several inositol 5-phospahtases. The enzymes that synthesize the substrates of these phosphatases, i.e., PI(4,5)P2 and PI(3,4,5)P3, also have links to endocytic clathrin coats. More specifically, all three type I PIP kinases, i.e., the PI 4-phosphate 5-kinases which are responsible for the bulk of PI(4,5)P2 synthesis in mammals (Heck et al., 2007; Volpicelli-Daley et al., 2010), can interact with the endocytic clathrin adaptor AP-2 (with the γ isoform having additional high affinity binding sites for such adaptors; Bairstow et al., 2006; Krauss et al., 2006; Thieman et al., 2009). As discussed in this study, PI 3-kinases are also recruited to endocytic clathrin coats, although their contribution to the endocytic CCP dynamics at steady-state might be minimal because wortmannin, an inhibitor for PI 3-kinases, hardly affected the dynamics (Fig. S3 E; Shpetner et al., 1996).
The action of SHIP2 is important for attenuation of insulin-mediated signaling. Thus, endocytic CCPs represent major sites where the functions of phosphoinositide metabolism in membrane dynamics and signaling are integrated. Because SHIP2 is a promising target for therapeutic interventions aimed at restoring insulin sensitivity in type II diabetes (Lazar and Saltiel, 2006; Suwa et al., 2009), our findings, which define a critical site of action of SHIP2, may have implications toward the development of drugs that may help treat this disease.
Materials and methods
Plasmids and reagents
Sources of cDNAs were as follows: GFP-(mouse)SHIP2 from U. Phillippar (Massachusetts Institute of Technology, Cambridge, MA); cortactin-DsRed from M. Kaksonen (European Molecular Biology Laboratory, Heidelberg, Germany); clathrin-GFP (LCa-GFP) from J. Keen (Thomas Jefferson University, Philadelphia, PA); GFP–intersectin 1–L and –S from P. McPherson (McGill University, Montreal, Quebec, Canada); mRFP-epsin from H. Chen (De Camilli laboratory); epsin-GFP from G. Ko (De Camilli laboratory); clathrin-mRFP (LCa-mRFP) from the De Camilli laboratory (Zoncu et al., 2007); μ2-GFP and GFP-CALM from A. Sorkin (University of Pittsburgh, Pittsburgh, PA); and dynamin-GFP from M. McNiven (Mayo Clinic, Rochester, MN). SHIP2 mutants were generated by PCR, PCR-based mutagenesis (Nakatsu et al., 2000) or the QuikChange kit (Agilent Technologies). mCherry-actin was constructed by PCR. mCherry-SHIP2 was made by replacing GFP with mCherry in the GFP-SHIP2 plasmid. To generate mRFP–FKBP-iSH2, the inter-SH2 domain of the p85 regulatory subunit of human class I PI 3-kinase was amplified by PCR and inserted at the C-terminal side of the FKBP module in the mRFP-FKBP backbone vector (Zoncu et al., 2007). HuSH 29-mer SHIP2-specific shRNA and control shRNA constructs were purchased from OriGene Technologies.
Anti–intersectin 1 antibodies were provided by P. McPherson. The following antibodies were purchased: anti-GFP antibodies from Takara Bio Inc. and Invitrogen and anti–β-actin from Sigma-Aldrich. Anti-SHIP2 polyclonal antibodies were obtained by immunizing rabbits with a human SHIP2 fragment corresponding to aa 887–1019 and then affinity purified with protein G–Sepharose (GE Healthcare) by T. Itoh in the De Camilli laboratory.
GST pull-down
GST pull-downs were performed as described previously (Erdmann et al., 2007). In brief, GST, GST–SHIP2-1022–1078, or GST–SHIP2-1022–1078-F1054A recombinant proteins were purified from Escherichia coli using glutathione-Sepharose beads (GE Healthcare). Proteins immobilized on the beads were subsequently incubated with a rat brain extract generated by lysing brain tissue in lysis buffer (PBS containing 1% Triton X-100) for 2 h at 4°C, followed by extensive washes with lysis buffer and PBS. Proteins retained on beads were eluted with SDS-PAGE sample buffer and separated by SDS-PAGE. Protein identification by liquid chromatography–mass spectrometry was performed at Keck Research Facility at Yale University.
RNA interference and stable cell lines
COS-7 cell lines expressing either control shRNA vector (pRS-shGFP) or pRS vector encoding SHIP2-specific shRNA were established and maintained in the presence of puromycin. For intersectin KD, COS-7 cells were transfected with either control siRNA or stealth siRNA directed against intersectin 1 (Invitrogen) using RNAiMAX reagent (Invitrogen).
TIRFM
Image acquisition was performed as described previously (Perera et al., 2006). In brief, cells were imaged at 37°C by TIRFM using an objective-type inverted microscope (IX-70; Olympus) fitted with a 60× NA 1.45 TIRFM lens (Olympus) and controlled by iQ software (Andor Technology). Laser lines (488 and 568 nm) from argon and argon/krypton lasers (CVI Melles Griot) were coupled to the TIRFM condenser through a single optical fiber. The calculated evanescent field depth was ∼100 nm. Cells were typically imaged in two channels by sequential excitation at 0.25 Hz, without binning, with 0.2–0.5-s exposures and detected with a back-illuminated electron-multiplying charge-coupled device camera (512 × 512 pixels; 16-bit; iXon887; Andor Technology).
Rapalogue-induced PI(3,4,5)P3 production
COS-7 cells expressing optimal levels of each marker (clathrin-GFP, plasma membrane–FRB-CFP [Varnai et al., 2006; Zoncu et al., 2007], and mRFP-FKBP or mRFP–FKBP-iSH2) were preselected using wide-field epifluorescence, after which the imaging modality was switched to TIRFM. Clathrin-GFP and mRFP-FKBP or mRFP–FKBP-iSH2 were imaged by sequential illumination at 488-nm and 568-nm, respectively. After an initial imaging period of 7–10 min, rapalogue (Zoncu et al., 2007) was added to cells (5 µM iRAP or 500 nM AP021967), and imaging was continued for an additional 20 min. Successful membrane recruitment of mRFP–FKBP-iSH2 was indicated by an increase in 568-nm fluorescence by TIRFM.
Image analysis
Image analysis was performed using ImageJ (National Institutes of Health) and iQ software as described previously (Perera et al., 2006). CCPs were randomly selected, and for each time point, the fluorescence intensity was normalized to the peak fluorescence intensity of that pit. Colocalization analysis was performed as described previously (Perera et al., 2006). For graphs of fluorescence intensity, we analyzed at least 12 CCPs from at least three cells. For histograms of CCP lifetime, we analyzed at least 186 CCPs from at least three cells.
Phosphoinositide analysis
Levels of phosphoinositides were analyzed by separating glycerol-inositol phosphates with HPLC after phospholipid deacylation. For the analysis of PI(4,5)P2 and PI 4-phosphate, which represent the bulk of PIP2 and PIP, respectively, the glycerol-inositol phosphate peaks derived from the PIP2 and PIP peak were identified and quantified using a conductivity detector (Voronov et al., 2008). For the analysis of PI(3,4,5)P3, cells were labeled with [3H]myo-inositol for 3 d and then harvested after stimulation with serum for 15 min after 24-h serum starvation. The PI(3,4,5)P3 peak was identified and quantified using a radiometric detector. Error bars show mean ± SD from three independent experiments.
In vitro phosphatase assay
The in vitro phosphatase assay (malachite assay) to monitor the enzymatic activity of SHIP2 against PI(4,5)P2 and PI(3,4,5)P3 was performed as described previously (Mani et al., 2007). In brief, COS-7 cells expressing GFP, GFP-SHIP2, or GFP–SHIP2-D608A were extracted in lysis buffer, and transfected proteins were purified by immunoprecipitation with protein G–Sepharose-immobilized anti-GFP antibodies. Aliquots of the immunoprecipitates were then either analyzed by Western blotting to confirm consistent recovery of transfected proteins or incubated with a water-soluble phosphoinositide substrate (with a diC8-acyl chain; Echelon) for 30 min at 37°C. Aliquots of the bead supernatant at the end of this incubation were mixed with malachite-green solution, and released free phosphate was measured by a plate reader at 650-nm wavelength (Lee et al., 2004). Data show mean ± SD of three independent experiments.
Online supplemental material
Fig. S1 shows that GFP-SHIP2 is targeted to CCPs in different cell types and demonstrates the importance of the region of SHIP2 that contains the PPPDF motif in its targeting to CCPs in COS-7 cells. Fig. S2 demonstrates the localization of intersectin, SHIP2, PI3KC2-α, and PI3KC2-β at CCPs and shows that targeting of intersectin to CCPs is independent of SHIP2. Fig. S3 shows the time course of the fluorescence intensity of clathrin-mRFP and GFP-tagged clathrin adaptors or dynamin at CCPs and that wortmannin has no effect on clathrin lifetime. Video 1 shows the dynamics of GFP-SHIP2 and clathrin-mRFP in COS-7 cells. Videos 2 and 3 show the dynamics of clathrin-GFP in control and SHIP2 KD cells, respectively. Video 4 shows the dynamics of clathrin-GFP within a ruffle of a FKBP-iSH2–expressing cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005018/DC1.
Acknowledgments
We thank Peter McPherson, Ulrike Phillippar, and Alexander Sorkin for critical reagents, Toshiki Itoh for generation of the anti-SHIP2 antibody, Abel-Alcazar Roman for lipid analysis, Agnes Ferguson for assistance with TIRFM, Wendolyn Hill for drawing Fig. 5, and Min Wu and Shawn Ferguson for critical reading of the manuscript.
This work was supported in part by grants from the National Institutes of Health (NS36251, DK45735, DA018343 to P. De Camilli and GM58801 to F.B. Gertler), the American Diabetes Association, the Yale Center for Genomics and Proteomics, and the W.M. Keck Foundation as well as the G. Harold and Leila Y. Mathers Charitable Foundation (to P. De Camilli) and the Japanese Society for the Promotion of Science (to F. Nakatsu).
Footnotes
Abbreviations used in this paper:
- CALM
- clathrin assembly lymphoid myeloid leukemia
- CCP
- clathrin-coated pit
- FKBP
- FK506-binding protein
- FRB
- FKBP12-rapamycin binding
- iRAP
- indole-modified analogue of rapamycin
- KD
- knockdown
- OCRL
- oculocerebrorenal syndrome of Lowe
- PH
- pleckstrin homology
- PI
- phosphatidylinositol
- PI(4,5)P2
- PI 4,5-bisphosphate
- PI(3,4,5)P3
- PI 3,4,5-triphosphate
- PIP
- PI phosphate
- PIP2
- PI bisphosphate
- shRNA
- short hairpin RNA
- TIRFM
- total internal reflection fluorescence microscopy
References
- Bairstow S.F., Ling K., Su X., Firestone A.J., Carbonara C., Anderson R.A. 2006. Type Igamma661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J. Biol. Chem. 281:20632–20642 10.1074/jbc.M601465200 [DOI] [PubMed] [Google Scholar]
- Cantley L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657 10.1126/science.296.5573.1655 [DOI] [PubMed] [Google Scholar]
- Chen H., Fre S., Slepnev V.I., Capua M.R., Takei K., Butler M.H., Di Fiore P.P., De Camilli P. 1998. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 394:793–797 10.1038/28660 [DOI] [PubMed] [Google Scholar]
- Choudhury R., Noakes C.J., McKenzie E., Kox C., Lowe M. 2009. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J. Biol. Chem. 284:9965–9973 10.1074/jbc.M807442200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B.M., McCoy A.J., Kent H.M., Evans P.R., Owen D.J. 2002. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 109:523–535 10.1016/S0092-8674(02)00735-3 [DOI] [PubMed] [Google Scholar]
- Cremona O., Di Paolo G., Wenk M.R., Lüthi A., Kim W.T., Takei K., Daniell L., Nemoto Y., Shears S.B., Flavell R.A., et al. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 99:179–188 10.1016/S0092-8674(00)81649-9 [DOI] [PubMed] [Google Scholar]
- Damen J.E., Liu L., Rosten P., Humphries R.K., Jefferson A.B., Majerus P.W., Krystal G. 1996. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA. 93:1689–1693 10.1073/pnas.93.4.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Scappini E., Martin N.P., Wong K.A., Dunn S., Chen Y.J., Miller S.L., Domin J., O’Bryan J.P. 2007. Regulation of neuron survival through an intersectin-phosphoinositide 3′-kinase C2beta-AKT pathway. Mol. Cell. Biol. 27:7906–7917 10.1128/MCB.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dyson J.M., O’Malley C.J., Becanovic J., Munday A.D., Berndt M.C., Coghill I.D., Nandurkar H.H., Ooms L.M., Mitchell C.A. 2001. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J. Cell Biol. 155:1065–1079 10.1083/jcb.200104005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J.A., Luo J., Cantley L.C. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7:606–619 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- Erdmann K.S., Mao Y., McCrea H.J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., De Camilli P. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 13:377–390 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G., Pearse B.M., Higgins M.K., Vallis Y., Owen D.J., Gibson A., Hopkins C.R., Evans P.R., McMahon H.T. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 291:1051–1055 10.1126/science.291.5506.1051 [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Keen J.H. 1999. Phosphoinositide–AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol. 146:755–764 10.1083/jcb.146.4.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I., Chen Q., Falck J.R., Reddy K.K., Keen J.H. 1996. A functional phosphatidylinositol 3,4,5-trisphosphate/phosphoinositide binding domain in the clathrin adaptor AP-2 alpha subunit. Implications for the endocytic pathway. J. Biol. Chem. 271:20922–20929 10.1074/jbc.271.34.20922 [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Santini F., Warren R.A., Keen J.H. 1999. Spatial control of coated-pit dynamics in living cells. Nat. Cell Biol. 1:1–7 10.1038/8971 [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Smith M.E., Domin J., Keen J.H. 2001. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell. 7:443–449 10.1016/S1097-2765(01)00191-5 [DOI] [PubMed] [Google Scholar]
- Hao W., Tan Z., Prasad K., Reddy K.K., Chen J., Prestwich G.D., Falck J.R., Shears S.B., Lafer E.M. 1997. Regulation of AP-3 function by inositides. Identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J. Biol. Chem. 272:6393–6398 10.1074/jbc.272.46.29322 [DOI] [PubMed] [Google Scholar]
- Haucke V. 2005. Phosphoinositide regulation of clathrin-mediated endocytosis. Biochem. Soc. Trans. 33:1285–1289 10.1042/BST20051285 [DOI] [PubMed] [Google Scholar]
- Heck J.N., Mellman D.L., Ling K., Sun Y., Wagoner M.P., Schill N.J., Anderson R.A. 2007. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit. Rev. Biochem. Mol. Biol. 42:15–39 10.1080/10409230601162752 [DOI] [PubMed] [Google Scholar]
- Hejna J.A., Saito H., Merkens L.S., Tittle T.V., Jakobs P.M., Whitney M.A., Grompe M., Friedberg A.S., Moses R.E. 1995. Cloning and characterization of a human cDNA (INPPL1) sharing homology with inositol polyphosphate phosphatases. Genomics. 29:285–287 10.1006/geno.1995.1247 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Boucrot E., Meinecke M., Evergren E., Vallis Y., Mittal R., McMahon H.T. 2010. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 328:1281–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N., Diao A., McKenzie E., Skippen A., Cockcroft S., Lowe M. 2006. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 25:3750–3761 10.1038/sj.emboj.7601274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 291:1047–1051 10.1126/science.291.5506.1047 [DOI] [PubMed] [Google Scholar]
- Krause M., Dent E.W., Bear J.E., Loureiro J.J., Gertler F.B. 2003. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19:541–564 10.1146/annurev.cellbio.19.050103.103356 [DOI] [PubMed] [Google Scholar]
- Krauss M., Kukhtina V., Pechstein A., Haucke V. 2006. Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc. Natl. Acad. Sci. USA. 103:11934–11939 10.1073/pnas.0510306103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar D.F., Saltiel A.R. 2006. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat. Rev. Drug Discov. 5:333–342 10.1038/nrd2007 [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Wenk M.R., Kim Y., Nairn A.C., De Camilli P. 2004. Regulation of synaptojanin 1 by cyclin-dependent kinase 5 at synapses. Proc. Natl. Acad. Sci. USA. 101:546–551 10.1073/pnas.0307813100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Dixon J.E. 1999. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 9:125–128 10.1016/S0962-8924(99)01519-6 [DOI] [PubMed] [Google Scholar]
- Mani M., Lee S.Y., Lucast L., Cremona O., Di Paolo G., De Camilli P., Ryan T.A. 2007. The dual phosphatase activity of synaptojanin1 is required for both efficient synaptic vesicle endocytosis and reavailability at nerve terminals. Neuron. 56:1004–1018 10.1016/j.neuron.2007.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Balkin D.M., Zoncu R., Erdmann K.S., Tomasini L., Hu F., Jin M.M., Hodsdon M.E., De Camilli P. 2009. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 28:1831–1842 10.1038/emboj.2009.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson P.S. 2002. The endocytic machinery at an interface with the actin cytoskeleton: a dynamic, hip intersection. Trends Cell Biol. 12:312–315 10.1016/S0962-8924(02)02309-7 [DOI] [PubMed] [Google Scholar]
- McPherson P.S., Garcia E.P., Slepnev V.I., David C., Zhang X., Grabs D., Sossin W.S., Bauerfeind R., Nemoto Y., De Camilli P. 1996. A presynaptic inositol-5-phosphatase. Nature. 379:353–357 10.1038/379353a0 [DOI] [PubMed] [Google Scholar]
- Merrifield C.J., Perrais D., Zenisek D. 2005. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 121:593–606 10.1016/j.cell.2005.03.015 [DOI] [PubMed] [Google Scholar]
- Mongioví A.M., Romano P.R., Panni S., Mendoza M., Wong W.T., Musacchio A., Cesareni G., Di Fiore P.P. 1999. A novel peptide-SH3 interaction. EMBO J. 18:5300–5309 10.1093/emboj/18.19.5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Sakuma M., Matsuo Y., Arase H., Yamasaki S., Nakamura N., Saito T., Ohno H. 2000. A Di-leucine signal in the ubiquitin moiety. Possible involvement in ubiquitination-mediated endocytosis. J. Biol. Chem. 275:26213–26219 10.1074/jbc.M907720199 [DOI] [PubMed] [Google Scholar]
- Ooms L.M., Horan K.A., Rahman P., Seaton G., Gurung R., Kethesparan D.S., Mitchell C.A. 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419:29–49 10.1042/BJ20081673 [DOI] [PubMed] [Google Scholar]
- Perera R.M., Zoncu R., Lucast L., De Camilli P., Toomre D. 2006. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc. Natl. Acad. Sci. USA. 103:19332–19337 10.1073/pnas.0609795104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X., Moreau C., Drayer A.L., Woscholski R., Parker P., Erneux C. 1998. The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity. FEBS Lett. 437:301–303 10.1016/S0014-5793(98)01255-1 [DOI] [PubMed] [Google Scholar]
- Prasad N., Topping R.S., Decker S.J. 2001. SH2-containing inositol 5′-phosphatase SHIP2 associates with the p130(Cas) adapter protein and regulates cellular adhesion and spreading. Mol. Cell. Biol. 21:1416–1428 10.1128/MCB.21.4.1416-1428.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I., Miyazaki M., Boll W., Duckworth B., Cantley L.C., Shoelson S., Kirchhausen T. 1997. Regulatory interactions in the recognition of endocytic sorting signals by AP-2 complexes. EMBO J. 16:2240–2250 10.1093/emboj/16.9.2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M.R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., et al. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607–618 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpetner H., Joly M., Hartley D., Corvera S. 1996. Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J. Cell Biol. 132:595–605 10.1083/jcb.132.4.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh B.C., Inoue T., Meyer T., Hille B. 2006. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 314:1454–1457 10.1126/science.1131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa A., Yamamoto T., Sawada A., Minoura K., Hosogai N., Tahara A., Kurama T., Shimokawa T., Aramori I. 2009. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br. J. Pharmacol. 158:879–887 10.1111/j.1476-5381.2009.00358.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor V., Wong M., Brandts C., Reilly L., Dean N.M., Cowsert L.M., Moodie S., Stokoe D. 2000. 5′ phospholipid phosphatase SHIP-2 causes protein kinase B inactivation and cell cycle arrest in glioblastoma cells. Mol. Cell. Biol. 20:6860–6871 10.1128/MCB.20.18.6860-6871.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieman J.R., Mishra S.K., Ling K., Doray B., Anderson R.A., Traub L.M. 2009. Clathrin regulates the association of PIPKIgamma661 with the AP-2 adaptor beta2 appendage. J. Biol. Chem. 284:13924–13939 10.1074/jbc.M901017200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Thyagarajan B., Rohacs T., Balla T. 2006. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175:377–382 10.1083/jcb.200607116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M., D’Angelo G., Di Campli A., De Matteis M.A. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27:2457–2470 10.1038/emboj.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley L.A., Lucast L., Gong L.W., Liu L., Sasaki J., Sasaki T., Abrams C.S., Kanaho Y., De Camilli P. 2010. Phosphatidylinositol 4-phosphate 5-kinases (PIPKIs) and phosphatidylinositol 4,5 bisphosphate [PI(4,5)P2] synthesis in the brain. J. Biol. Chem. 10.1074/jbc.M110.132191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronov S.V., Frere S.G., Giovedi S., Pollina E.A., Borel C., Zhang H., Schmidt C., Akeson E.C., Wenk M.R., Cimasoni L., et al. 2008. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc. Natl. Acad. Sci. USA. 105:9415–9420 10.1073/pnas.0803756105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woscholski R., Finan P.M., Radley E., Totty N.F., Sterling A.E., Hsuan J.J., Waterfield M.D., Parker P.J. 1997. Synaptojanin is the major constitutively active phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase in rodent brain. J. Biol. Chem. 272:9625–9628 10.1074/jbc.272.15.9625 [DOI] [PubMed] [Google Scholar]
- Xie J., Vandenbroere I., Pirson I. 2008. SHIP2 associates with intersectin and recruits it to the plasma membrane in response to EGF. FEBS Lett. 582:3011–3017 10.1016/j.febslet.2008.07.048 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hartz P.A., Philip E., Racusen L.C., Majerus P.W. 1998. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 273:1574–1582 10.1074/jbc.273.3.1574 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Sebastian R., Nakatsu F., Chen H., Balla T., Ayala G., Toomre D., De Camilli P.V. 2007. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 104:3793–3798 10.1073/pnas.0611733104 [DOI] [PMC free article] [PubMed] [Google Scholar]