A systems biology–based approach shows that life and death decisions for cells depend on the stoichiometry of c-FLIP isoforms.
Abstract
Cellular FADD-like interleukin-1β–converting enzyme inhibitory proteins (c-FLIPs; isoforms c-FLIP long [c-FLIPL], c-FLIP short [c-FLIPS], and c-FLIP Raji [c-FLIPR]) regulate caspase-8 activation and death receptor (DR)–induced apoptosis. In this study, using a combination of mathematical modeling, imaging, and quantitative Western blots, we present a new mathematical model describing caspase-8 activation in quantitative terms, which highlights the influence of c-FLIP proteins on this process directly at the CD95 death-inducing signaling complex. We quantitatively define how the stoichiometry of c-FLIP proteins determines sensitivity toward CD95-induced apoptosis. We show that c-FLIPL has a proapoptotic role only upon moderate expression in combination with strong receptor stimulation or in the presence of high amounts of one of the short c-FLIP isoforms, c-FLIPS or c-FLIPR. Our findings resolve the present controversial discussion on the function of c-FLIPL as a pro- or antiapoptotic protein in DR-mediated apoptosis and are important for understanding the regulation of CD95-induced apoptosis, where subtle differences in c-FLIP concentrations determine life or death of the cells.
Introduction
CD95 (APO-1/Fas) is a member of the death receptor (DR) family, a subfamily of the TNF-R superfamily (Lavrik et al., 2005a). Cross-linking of CD95 with its natural ligand CD95L (CD178; Suda et al., 1993) or with agonistic antibodies such as anti–APO-1 induces apoptosis in sensitive cells (Trauth et al., 1989). Stimulation of CD95 has also been reported to induce nonapoptotic pathways such as NF-κB, Akt, Erk, and others (Peter et al., 2007). The death-inducing signaling complex (DISC) is formed within seconds after CD95 stimulation (Kischkel et al., 1995). The DISC comprises oligomerized, and probably trimerized, CD95 and the adaptor protein FADD, two isoforms of procaspase-8 (procaspase-8a and procaspase-8b), procaspase-10, and cellular FADD-like interleukin-1β–converting enzyme inhibitory proteins (c-FLIPs) long/short/Raji (c-FLIPL/S/R; Muzio et al., 1996; Scaffidi et al., 1999; Sprick et al., 2002; Krammer et al., 2007). The interactions between the molecules at the DISC are based on homotypic contacts. The death domain of CD95 interacts with the death domain of FADD, whereas the death effector domain (DED) of FADD interacts with the N-terminal tandem DEDs of procaspase-8, procaspase-10, and the c-FLIP isoforms.
After binding to the DISC, procaspase-8a/b (p55/p53) undergoes autocatalytic processing, resulting in the generation of active caspase-8 (Muzio et al., 1996; Medema et al., 1997; Lavrik et al., 2005b). This processing involves dimerization of two procaspase-8 molecules followed by a conformational change, leading to autoactivation of procaspase-8 homodimers (Muzio et al., 1998; Chang et al., 2003; Fuentes-Prior and Salvesen, 2004). During subsequent procaspase-8–processing steps at the DISC, cleavage occurs at several Asp (D) residues between the prodomain and the small and large catalytic subunits (Fig. 1 A). This results in the formation of the N-terminal cleavage product p43/p41, the prodomain p26/p24, and the C-terminal cleavage products p30, p18, and p10 (Medema et al., 1997; Hoffmann et al., 2009; Hughes et al., 2009). Active caspase-8 heterotetramers p102-p182 generated at the DISC trigger the apoptotic signal. Recently, it has been reported that the cleavage products p30 and p43/p41 also possess catalytic activity, which leads to apoptosis initiation (Hoffmann et al., 2009; Hughes et al., 2009). Thus, procaspase-8 processing at the DISC initiates apoptosis through generation of several catalytically active cleavage products.
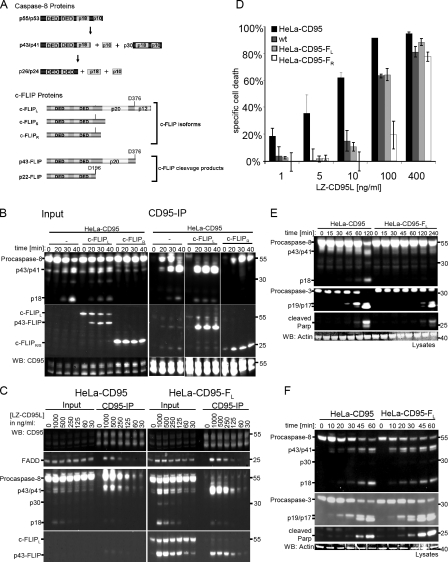
Figure 1.
c-FLIP isoforms have different effects on caspase-8 activation at the DISC. (A) Scheme of the DED proteins of the DISC, procaspase-8, and c-FLIP and their cleavage products. Procaspase-8 proteins comprise two isoforms (procaspase-8a [p55] and procaspase-8b [p53]) and their cleavage products, p43/p41, p30, and p18. c-FLIP proteins comprise three isoforms, namely c-FLIPL, c-FLIPS, and c-FLIPR, and two c-FLIP cleavage products (p43-FLIP and p22-FLIP). (B) HeLa-CD95, HeLa-CD95–FL, and HeLa-CD95–FR cells were stimulated with 1 µg/ml LZ-CD95L for the indicated time points. CD95 DISCs were immunoprecipitated using anti–APO-1 antibodies and analyzed along with total cellular lysates using Western blotting (WB) with antibodies against caspase-8 (C15), c-FLIP (NF6), FADD (1C4), and CD95 (C20). (C) HeLa-CD95 and HeLa-CD95–FL cells were stimulated with the indicated amounts of LZ-CD95L. CD95 DISCs were immunoprecipitated 20 min after addition of LZ-CD95L using anti–APO-1 antibodies and analyzed along with total cellular lysates using Western blotting with antibodies against caspase-8 (C15), c-FLIP (NF6), FADD (1C4), and CD95 (C20). (D) HeLa wt (dark gray bars), HeLa-CD95 (black bars), HeLa-CD95–FL (light gray bars), and HeLa-CD95–FR (white bars) cells were stimulated with the indicated amounts of LZ-CD95L. Specific cell death was determined after 24 h of CD95 stimulation with PI stain and flow cytometry. Mean and SEM of three independent experiments are shown. (E) HeLa-CD95 and HeLa-CD95–FL cells were stimulated with 200 ng/ml LZ-CD95L for the indicated time points. Total cellular lysates were analyzed using Western blotting with antibodies against caspase-8, caspase-3, cleaved PARP, and actin. (F) HeLa-CD95 and HeLa-CD95–FL cells were stimulated with 3 µg/ml LZ-CD95L for the indicated time points. Total cellular lysates were analyzed using Western blotting with antibodies against caspase-8, caspase-3, cleaved PARP, and actin. (B, C, E, and F) One representative experiment out of three is shown. White lines indicate that intervening lanes have been spliced out.
Procaspase-10 is also activated at the DISC. However, it was reported to be unable to induce apoptosis in the absence of procaspase-8 (Sprick et al., 2002). Therefore, the role of procaspase-10 in regulation of apoptosis remains unclear.
c-FLIP is a well-described inhibitor of DR-mediated apoptosis (Scaffidi et al., 1999; Krueger et al., 2001; Golks et al., 2005). At the mRNA level, it can be found in multiple splice variants, whereas at the protein level, only three isoforms, c-FLIPL, c-FLIPS, and c-FLIPR, have been detected so far (Fig. 1 A). All three c-FLIP isoforms contain two DEDs structurally similar to the N-terminal part of procaspase-8. c-FLIPL also contains catalytically inactive caspase-like domains (p20 and p12). In addition to their antiapoptotic role in DR-induced apoptosis, c-FLIP proteins were demonstrated to play a prominent role in engagement of NF-κB signaling (Budd et al., 2006; Neumann et al., 2010). Remarkably, two cleavage products of c-FLIP, p43-FLIP and p22-FLIP (Fig. 1 A), were shown to play an important role in NF-κB induction (Kataoka and Tschopp, 2004; Dohrman et al., 2005; Golks et al., 2006).
c-FLIP proteins are recruited to the DISC by DED interactions. Both short c-FLIP isoforms, c-FLIPS and c-FLIPR, block DR-induced apoptosis by inhibiting procaspase-8 activation at the DISC (Krueger et al., 2001; Golks et al., 2005; Ueffing et al., 2008). This has been suggested to occur via formation of catalytically inactive procaspase-8/c-FLIPR/S heterodimers. The role of c-FLIPL at the DISC is still a matter of controversy (Yu and Shi, 2008). It has been shown that c-FLIPL can act as an antiapoptotic molecule, functioning in a way analogous to c-FLIPS when expressed at high concentrations in the cell (Krueger et al., 2001; Chang et al., 2002). It has also been reported that c-FLIPL can act as a proapoptotic molecule when expressed at low concentrations, facilitating the activation of procaspase-8 at the DISC (Chang et al., 2002). This has been reported to be mediated by formation of catalytically active procaspase-8/c-FLIPL heterodimers in which the procaspase-8 active loop is stabilized by c-FLIPL (Micheau et al., 2002; Yu et al., 2009), thereby allowing cleavage of procaspase-8 into p43/41. However, this proapoptotic role of c-FLIPL contradicts the findings of Sharp et al. (2005), who have demonstrated that the selective knockdown of c-FLIPL sensitizes the cells toward DR-induced apoptosis. These findings motivated us to apply a more systematic approach to understand how different quantities of c-FLIP proteins at the DISC influence caspase-8 activation at the DISC and subsequent apoptosis induction.
Studies of apoptosis using systems biology have recently provided significant insight in cell death. There are several models describing the regulation of caspase activation (Eissing et al., 2004; Legewie et al., 2006), the intrinsic pathway (Rehm et al., 2006), and death pathway regulation (Bentele et al., 2004; Spencer et al., 2009). However, a detailed model considering the dynamics of all DED proteins, e.g., procaspase-8 and c-FLIP, at the DISC was still missing.
Based on quantitative Western blot data, we built a mathematical model of CD95 DISC signaling. The model made several important predictions, which were confirmed experimentally. In this way, we show that the same amount of c-FLIPL can accelerate cell death at high CD95L concentrations, whereas it slows down induction of cell death at low CD95L concentrations. We demonstrate that c-FLIPL acts proapoptotic in the presence of high amounts of one of the short c-FLIP isoforms. Our data show that c-FLIPL can abrogate the c-FLIPR/S–mediated inhibition of procaspase-8 processing. Thus, we found a novel function of the short c-FLIP isoforms: they modulate the sensitizing effect of c-FLIPL.
Results
c-FLIPL can accelerate caspase-8 activation, whereas c-FLIPR/S inhibits it
To understand the influence of the three c-FLIP isoforms on procaspase-8 activation (Fig. 1 A), we generated HeLa cell lines stably overexpressing CD95 along with different c-FLIP isoforms, c-FLIPL, c-FLIPS, or c-FLIPR (HeLa-CD95–FL, HeLa-CD95–FS, or HeLa-CD95–FR, respectively). The expression of all other main components of the CD95 DISC (CD95, FADD, and procaspase-8) was similar in these cell lines (Fig. S1 A). Surface expression of CD95 was also uniform in the different HeLa-CD95 cell lines (Fig. S1 B).
To analyze procaspase-8 activation, HeLa-CD95–FL, HeLa-CD95–FR, and HeLa-CD95 cells were stimulated for different time intervals with 1 µg/ml LZ-CD95L. This was followed by analysis of the CD95 DISCs and total cellular lysates on Western blots (Fig. 1 B). In HeLa-CD95 cells, procaspase-8a/b processing at the DISC occurred as described in a previous study with generation of the cleavage products p43/p41 and p18 (Lavrik et al., 2007). Overexpression of a short c-FLIP isoform, c-FLIPR (HeLa-CD95–FR cells), efficiently blocked procaspase-8a/b cleavage at the DISC. In these cells, only unprocessed procaspase-8a/b (p55/p53) was detected at the DISC. Even after several hours of stimulation, procaspase-8 at the DISC was only processed to a minor extent (Fig. S1 C). Another short isoform of c-FLIP, c-FLIPS, blocked procaspase-8 activation in a similar way (unpublished data). Therefore, we considered that c-FLIPS and c-FLIPR act in a uniform manner on caspase-8 activation and did not distinguish between the two short isoforms in our further work. Strikingly, overexpression of c-FLIPL (HeLa-CD95–FL cells), accelerated the first cleavage step of procaspase-8a/b processing at the DISC. The entire pool of procaspase-8a/b at the DISC was processed to p43/p41 already after 20 min of stimulation (Fig. 1 B). These data were in accordance with previous studies that demonstrated similar effects on procaspase-8 activation at the DISC upon overexpression of the long or short c-FLIP isoforms (Krueger et al., 2001; Micheau et al., 2002; Golks et al., 2005).
A remarkably similar activating effect of c-FLIPL on procaspase-8 cleavage at the DISC was observed upon addition of different doses of LZ-CD95L (Fig. 1 C). Independent of the amount of LZ-CD95L, procaspase-8a/b was completely cleaved to p43/p41 after 20 min of stimulation at the DISC of HeLa-CD95–FL cells. In contrast, in HeLa-CD95 cells, most of the procaspase-8 at the DISC had not been processed at this time point. We also observed an increase of procaspase-8 cleavage products in the lysates of HeLa-CD95–FL cells compared with HeLa-CD95 cells after 20 min of stimulation at all ligand concentrations used (Fig. 1 C). The same results were obtained for independent clones of HeLa-CD95–FL cells (Fig. S1 D).
Although c-FLIPL accelerated procaspase-8 cleavage at the DISC, the sensitivity of HeLa-CD95–FL cells toward CD95-induced death was reduced in comparison with HeLa-CD95 cells (Fig. 1 D). However, HeLa-CD95–FL cells were more sensitive than HeLa-CD95–FR cells (Fig. 1 D), which is in accordance with Western blot data on procaspase-8 processing in these cell lines (Fig. 1 B).
Because there was a decrease in CD95-mediated cell death after c-FLIPL overexpression, although initial procaspase-8 cleavage was accelerated, we aimed to get more insight into this phenomenon. We analyzed the kinetics of procaspase-8 processing in the lysate for longer time periods. Upon addition of 200 ng/ml LZ-CD95L, procaspase-8 cleavage occurred in HeLa-CD95 cells after 120 min but was almost completely blocked in HeLa-CD95–FL cells (Fig. 1 E). Caspase-3 and poly(ADP-ribose) polymerase (PARP) cleavage were also significantly impaired. This was in accordance with the decreased cell death in HeLa-CD95–FL cells for low concentrations of CD95L (Fig. 1 D).
However, upon stimulation with a high amount of 3 µg/ml LZ-CD95L, not only the formation of the procaspase-8 cleavage products p43/p41, p30, and p18 was accelerated in total cellular lysates of HeLa-CD95–FL cells (Fig. 1 F), but also the amount of caspase-3 and PARP cleavage products was slightly higher in HeLa-CD95–FL cells. To test whether this increase in procaspase-8 and procaspase-3 processing led to an acceleration of the induction of cell death, we measured cell death upon stimulation with 3 µg/ml or 200 ng/ml LZ-CD95L in HeLa-CD95 and HeLa-CD95–FL cells at various time points (Fig. S2 A). Induction of cell death was clearly slowed down after addition of 200 ng/ml LZ-CD95L in HeLa-CD95–FL cells. Even upon addition of 3 µg/ml LZ-CD95L, cell death occurred slightly slower in HeLa-CD95–FL cells, although there was more procaspase-8 processing initially (Fig. 1 F and Fig. S2 A).
Collectively, we observed nonlinear effects of c-FLIP overexpression on CD95-mediated procaspase-8 processing and cell death. To explain these differential effects of c-FLIP on caspase-8 cleavage, we built a mathematical model of caspase-8 processing at the CD95 DISC.
A mathematical model of procaspase-8 activation at the DISC
The observation that c-FLIPL may either block or accelerate procaspase-8 cleavage depending on its concentration at the DISC and the strength of CD95 stimulation prompted us to explain these effects using a mathematical model. To generate a systems biology model of procaspase-8 activation at the DISC, we developed a mathematical model based on ordinary differential equations. The topology of the model is shown in Fig. 2 A. It involves binding of CD95L to CD95, followed by FADD recruitment, which leads to binding of DED proteins c-FLIP and procaspase-8.
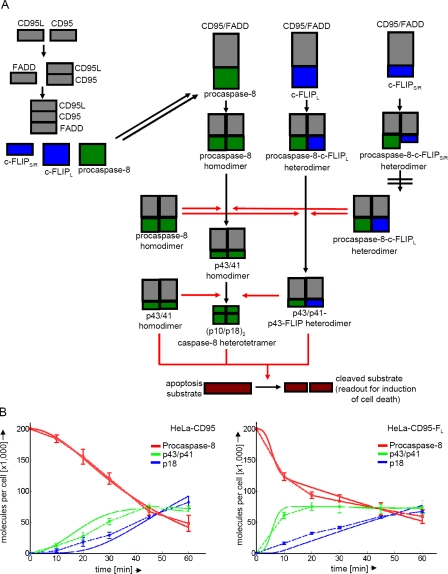
Figure 2.
A mathematical model of CD95 signaling. (A) Scheme showing the topology of the mathematical model. CD95, CD95L, and FADD are depicted in gray, the long and short isoforms of c-FLIP in blue, and caspase-8 in green. At the DISC procaspase-8 homodimers, procaspase-8/c-FLIPL dimers and procaspase-8/c-FLIPS/R dimers are formed. Red arrows indicate catalytic processing. (B) Fitting of simulated data (solid lines) to measurement data (dashed lines) in HeLa-CD95 and HeLa-CD95–FL cells for procaspase-8 processing upon stimulation with 3 µg/ml LZ-CD95L. Numbers are given in thousands of molecules per cell. Procaspase-8 is shown in red, p43/p41 in green, and p18 in blue. Mean and SEM of three independent experiments are shown.
We considered that procaspase-8 forms three types of dimers at the DISC: procaspase-8 homodimers, procaspase-8/c-FLIPL heterodimers, and procaspase-8/c-FLIPS/R heterodimers. Generation of procaspase-8 homodimers results in full processing of procaspase-8 to its cleavage products p43/p41 and p18. Procaspase-8/c-FLIPL heterodimers are only processed to p43/p41 and p43-FLIP in our model. This consideration is based on a previous study (Krueger et al., 2001) as well as our own experimental data. To this point, we have generated HeLa-CD95–FL/R cells, which overexpress both the short (c-FLIPR) and the long c-FLIP isoforms. Therefore, the amount of procaspase-8 at the DISC is reduced, and mainly procaspase-8/c-FLIPR and procaspase-8/c-FLIPL heterodimers are formed. In these cells, we observed processing of procaspase-8 at the DISC only to p43/p41 but not to p18 (Fig. S2 B). In our model, procaspase-8 homodimers and procaspase-8/c-FLIPL heterodimers were catalytically active and could process other procaspase-8 dimers. The third type of procaspase-8 dimers, procaspase-8/c-FLIPS/R heterodimers, did not possess any catalytic activity and were not processed. Additionally, we introduced a turnover of procaspase-8 at the DISC in our mathematical model. Our assumption was that once one procaspase-8 molecule had been completely cleaved, it could be replaced at the DISC by new procaspase-8 molecules recruited to this complex. This was necessary to explain the observed data.
Finally, in our model, all active procaspase-8 cleavage products (p41/p43–procaspase-8 homodimers, p41/p43–procaspase-8/p43-FLIP heterodimers, and (p10-p18)2 caspase-8 heterotetramers) cleaved a universal apoptosis substrate (Fig. 2 A). Procaspase-8 homodimers were unable to cleave the apoptosis substrate, as it was reported that procaspase-8 activity itself is not sufficient to induce cell death (Kang et al., 2008). Cleavage of this substrate was assumed to lead directly to cell death and, consequently, was taken as readout for cell death.
Our mathematical model includes several simplifications to reduce the complexity of the model. It has been previously reported by Hughes et al. (2009) that procaspase-8a and procaspase-8b possess similar catalytic properties at the DISC. Therefore, procaspase-8a and procaspase-8b were considered as one entity. As the first cleavage step, we modeled only processing to p43/p41. The processing of p55/p53 to p30 was neglected in our modeling, as the amount of p30 generated is ∼20 times lower than p43/p41 (Fig. 1 C; Hoffmann et al., 2009). As previously mentioned, c-FLIPS and c-FLIPR were considered as one entity, as they inhibit caspase-8 activation in a similar way (Golks et al., 2005). Procaspase-10 activation was not considered, as it was reported to not contribute to the onset of apoptosis (Sprick et al., 2002).
To estimate the parameters of the model, we measured the absolute concentrations of all relevant proteins. We applied quantitative Western blotting using recombinant proteins as concentration standards (Fig. S3 A). The resulting numbers of molecules are presented in Table S1. Strikingly, the number of c-FLIP proteins in HeLa-CD95 cells was very low, suggesting a high affinity of c-FLIP proteins to the DISC, leading to high concentration of c-FLIP proteins at the DISC as reported in previous studies (Chang et al., 2002; Lavrik et al., 2007).
The model was fitted to quantitative Western blot data of procaspase-8 processing in HeLa-CD95, HeLa-CD95–FL (Fig. 1 F), and HeLa-CD95–c-FLIP–deficient cells, which were generated using shRNA (Fig. S3 B). A good fit between model simulation (Fig. 2 B, solid lines) and experimental data (Fig. 2 B, dashed lines) could be achieved reproducing the cleavage of procaspase-8 into p43/p41 and p18. We concluded that the mathematical model was well suited to quantitatively describe the processing of procaspase-8 at the DISC.
The model predicts CD95L dose-dependent effects of c-FLIPL
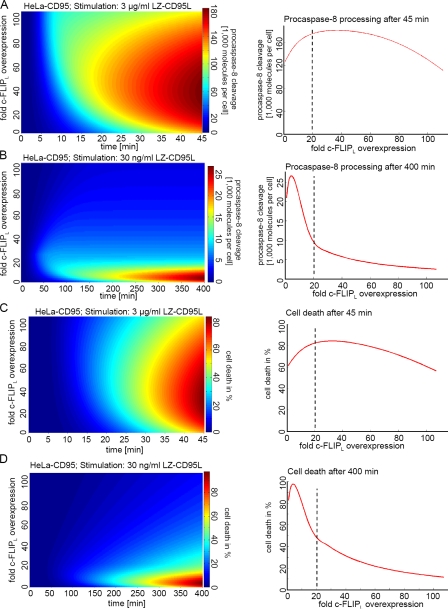
Our experiments (Fig. 1) as well as data in previous literature (Bentele et al., 2004; Neumann et al., 2010) suggest nonlinear effects on procaspase-8 processing by c-FLIP proteins. Therefore, the question arose whether our model could describe how changes in the amount of c-FLIPL influenced processing of procaspase-8 and CD95-mediated cell death. Fig. 3 shows procaspase-8 cleavage and cell death depending on the time and amount of c-FLIPL upon stimulation with 3 µg/ml (Fig. 3, A and C) and 30 ng/ml (Fig. 3, B and D) LZ-CD95L. Interestingly, the model could reproduce the CD95L dose-dependent effect of c-FLIPL on procaspase-8 cleavage. Our model predicted that a 20-fold c-FLIPL overexpression would reduce procaspase-8 cleavage and cell death at a low concentration of LZ-CD95L but would accelerate procaspase-8 cleavage and cell death upon a high concentration. There are two reasons for this phenomenon: the first is that the ratio of c-FLIPL to caspase-8 at the DISC is dependent on the strength of the receptor signal. Because the total amount of c-FLIP in HeLa cells is very low (Table S1), the model predicted a depletion of c-FLIP in the cytosol upon high CD95L stimulation because all c-FLIP had already been recruited to the DISC (Bentele et al., 2004; Lavrik et al., 2007). As a consequence, the relative amount of c-FLIP at the DISC is higher upon weak CD95 stimulation than upon strong CD95 stimulation. The second reason is that because of the high affinity of c-FLIP to FADD, it would remain bound to the DISC and could reduce a possible turnover of caspase-8 at the DISC. Therefore, although initial procaspase-8 processing upon c-FLIPL overexpression is increased, c-FLIPL would block further caspase-8 activation. Two opposite effects of c-FLIPL, activation of procaspase-8 versus blockage of its turnover, result in nonlinear effects and lead to a narrow range of c-FLIPL concentrations, which result in sensitization toward CD95-induced apoptosis (Fig. 3, C and D).
Figure 3.
The model predicts a CD95L dose-dependent sensitization of c-FLIPL. (A, left) Prediction of the amount of procaspase-8 processing depending on time and the concentration of c-FLIPL after stimulation with 3 µg/ml CD95L. Blue indicates low amounts of procaspase-8 processing, and red indicates high amounts of procaspase-8 cleavage. (right) Prediction of the amount of procaspase-8 processing depending on the concentration of c-FLIPL after 45 min of stimulation with 3 µg/ml CD95L. (B, left) Prediction of the amount of procaspase-8 processing depending on time and the concentration of c-FLIPL after stimulation with 30 ng/ml CD95L. (right) Prediction of the amount of procaspase-8 processing depending on the concentration of c-FLIPL after 400 min of stimulation with 30 ng/ml CD95L. (C, left) Simulation of the amount of cell death depending on time and the concentration of c-FLIPL after stimulation with 3 µg/ml CD95L in HeLa-CD95 cells. (right) Predicted amount of cell death depending on the concentration of c-FLIPL after 45 min of stimulation with 3 µg/ml CD95L in HeLa-CD95 cells. (D, left) Simulation of the amount of cell death depending on time and the concentration of c-FLIPL after stimulation with 30 ng/ml CD95L in HeLa-CD95 cells. (right) Prediction of the amount of cell death depending on the concentration of c-FLIPL after 400 min of stimulation with 30 ng/ml CD95L in HeLa-CD95 cells. (A and B) The number of cleaved procaspase-8 molecules is given in thousands of molecules per cell. Dashed lines indicate the consequence of a 20-fold c-FLIPL overexpression, which is achieved with the H2 GFP-IRES–c-FLIPL plasmid.
Collectively, our model explained the CD95L dose-dependent effects of c-FLIPL on procaspase-8 processing as well as on the onset of CD95-induced apoptosis. The next challenge was to test the predictions of the model in our cellular systems.
Experimental verification of model predictions: c-FLIPL both accelerated and slowed down cell death in a CD95L and c-FLIPL dose-dependent manner
Next, we tested whether we could confirm the predictions made by our model experimentally. First, we analyzed the optimal concentration of c-FLIPL, leading to an increase of caspase-8 activity and apoptosis induction. The model predicted the highest amount of procaspase-8 cleavage and cell death to occur at a 20–40-fold c-FLIPL overexpression at a concentration of 3 µg/ml LZ-CD95L and a 45-min time of stimulation (Fig. 3, A and C, right). Because the endogenous amount of c-FLIPL is very low (∼320 molecules per cell; Table S1), a 20–40-fold overexpression corresponds to ∼6,500–13,000 molecules per cell. This moderate overexpression could be achieved after transient transfection with a plasmid containing a histone H2 promoter followed by the coding sequence of GFP-IRES–c-FLIPL. The plasmid has a weak promoter, and the c-FLIPL cDNA was inserted behind and not in front of the IRES sequence. This construction results in a low overexpression of c-FLIPL. The resulting level of expression of c-FLIPL was ∼20 times higher than in the control-transfected HeLa-CD95 cells (Fig. S4 A). The percentage of transfected cells was determined counting the GFP-positive cells with flow cytometry. The fold overexpression of c-FLIPL was estimated by quantitative Western blotting. Dividing the total fold c-FLIPL overexpression by the percentage of transfected cells yielded the mean overexpression per transfected cell. The expression of the other main components of the CD95 system in these cells remained unaltered (Fig. S4 B).
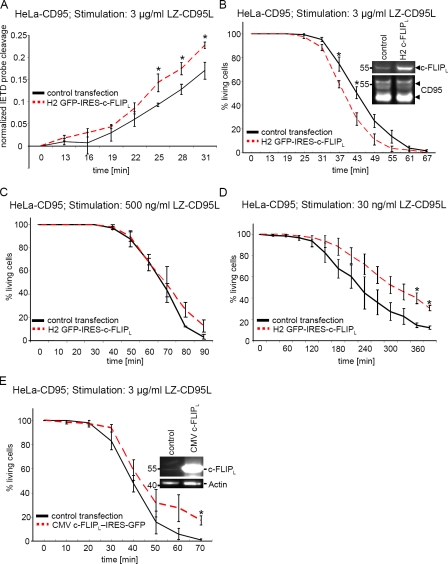
As the transfection efficiency was only ∼10%, we could not analyze the effect of moderate c-FLIPL overexpression using Western blot analysis. Therefore, we decided to perform a single-cell analysis with time-lapse microscopy. To determine caspase-8 activity in the cells, we transfected them with a plasmid encoding a nuclear export signal (NES)–IETD-mCherry fusion protein. This protein consists of an NES followed by the peptide sequence IETD fused to mCherry via a linker and thereby served as a caspase-8 activity probe (Fig. S4 D). In unstimulated cells, this probe resided in the cytosol. Upon stimulation with LZ-CD95L, procaspase-8 was activated and cleaved the probe after the IETD sequence, allowing mCherry to enter into the nucleus (Fig. S4 D). We cotransfected HeLa-CD95 cells either with the NES-IETD-mCherry probe and the H2 GFP-IRES–c-FLIPL plasmid or the NES-IETD-mCherry probe and an empty control vector. GFP- (GFP+) and mCherry-positive cells (mCherry+) were analyzed in time-lapse microscopy, and the amount of mCherry translocating into the nucleus after CD95 stimulation was measured. In GFP+mCherry+ cells, we found a moderate increase in caspase-8 activity in comparison with GFP−mCherry+ cells, as determined by cleavage of the caspase-8 activity probe in live cell imaging (Fig. 4 A).
Figure 4.
c-FLIPL accelerates or slows down CD95-induced cell death in a CD95L dose-dependent manner. (A) HeLa-CD95 cells were transfected with a plasmid encoding the NES-IETD-mCherry probe along with the H2 GFP-IRES–c-FLIPL plasmid or an empty control plasmid. 48 h after transfection, cells were stimulated with LZ-CD95L. The graph shows caspase-8 activity probe cleavage of GFP+mCherry+ and GFP−mCherry+ cells after addition of 3 µg/ml LZ-CD95L. (B–D) HeLa-CD95 cells were transfected with the H2 GFP-IRES–c-FLIPL (B–D), the CMV c-FLIPL–IRES-GFP (E), or an mCherry-encoding control plasmid. 48 h after transfection, LZ-CD95L was added to the cells. The graphs in B and E show cell death of GFP+ and mCherry+ cells after addition of 3 µg/ml LZ-CD95L. The graphs in C and D show cell death of GFP+ and mCherry+ cells after addition of 500 ng/ml and 30 ng/ml LZ-CD95L, respectively. Mean and SEM of four (A and B) or three (C–E) independent experiments are shown. *, P < 0.05.
To verify that this increase of caspase-8 activity led to an acceleration of cell death, we investigated death of the H2 GFP-IRES–c-FLIPL–transfected cells versus the mCherry control–transfected cells upon stimulation with 3 µg/ml LZ-CD95L. GFP+ and mCherry+ cells were determined with confocal microscopy followed by live cell imaging, and cell death was monitored by cell shrinkage and formation of apoptotic bodies (Fig. S4 C). In accordance with the model predictions, there was a sensitization toward CD95-induced apoptosis upon moderate c-FLIPL overexpression (Fig. 4 B).
Furthermore, we wanted to verify the predictions made by the model for a weak stimulation with 30 ng/ml LZ-CD95L. Already, a 20-fold up-regulation of c-FLIPL was predicted to lead to a reduction of both procaspase-8 processing and cell death after 400 min of stimulation (Fig. 3, B and D, right). Indeed, upon stimulation with 30 ng/ml LZ-CD95L, induction of cell death was slowed down in cells transfected with the H2 GFP-IRES–c-FLIPL plasmid, as determined with live cell imaging (Fig. 4 D). The same amount of c-FLIPL that sensitized HeLa-CD95 cells upon a stimulation with 3 µg/ml LZ-CD95L slowed down cell death upon stimulation with 30 ng/ml LZ-CD95L. Upon addition of 500 ng/ml LZ-CD95L, almost no differences in cell death between control-transfected and H2 GFP-IRES–c-FLIPL–transfected cells were observed (Fig. 4 C). We concluded that upon c-FLIPL overexpression, there is a continuum from blocking cell death upon weak stimulation to accelerating induction of cell death upon strong stimulation.
For high overexpression levels of c-FLIPL, the model predicted a significant reduction of procaspase-8 processing and sensitivity toward CD95-induced apoptosis (Fig. 3, A and C). To test this prediction, we overexpressed c-FLIPL in high amounts. This was achieved after transient transfection of a plasmid encoding c-FLIPL–IRES-GFP under control of a cytomegalovirus (CMV) promoter. 15% of GFP+ cells were still alive after 70 min of stimulation, whereas all control-transfected mCherry+ cells were already dead at this time point (Fig. 4 E). This was in accordance with the model-based prediction that a drop of procaspase-8 cleavage and cell death upon very high c-FLIPL expression would be observed (Fig. 3, A and C). Collectively, we could confirm the predictions made by the model and observed that the same amount of c-FLIPL can accelerate or slow down CD95-mediated cell death depending on the stimulating dose of CD95L.
Moderate c-FLIPL up-regulation slows down cell death in HeLa wild-type (wt) cells
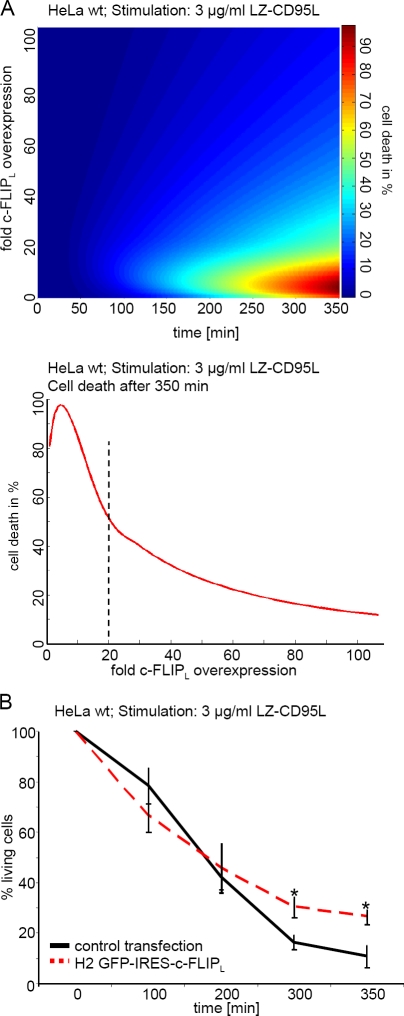
So far, we have shown that the effect of c-FLIPL depends on the strength of CD95 stimulation and the amount of c-FLIPL. To further verify the proposed mechanism by testing the model predictions for yet another scenario, we investigated the effect of c-FLIPL in cells that express reduced amounts of CD95. The model predicted that in HeLa wt cells, which have an ∼10 times lower CD95 expression than HeLa-CD95 cells, a 20-fold c-FLIPL overexpression led to a reduction in cell death after 350 min of stimulation with 3 µg/ml LZ-CD95L (Fig. 5 A). After 350 min of stimulation with 3 µg/ml LZ-CD95L, 30% of H2 GFP-IRES–c-FLIPL–transfected cells were still alive, as determined with live cell imaging, in contrast to only 10% survival of control-transfected cells (Fig. 5 B). This is in agreement with the model prediction and with previous results (Sharp et al., 2005). Therefore, moderate c-FLIPL overexpression in HeLa wt reduced CD95-induced cell death.
Figure 5.
Moderate c-FLIPL overexpression blocks cell death in HeLa wt cells. (A, top) Prediction of the amount of cell death depending on time and concentration of c-FLIPL after stimulation with 3 µg/ml CD95L in HeLa wt cells. (bottom) Prediction of the amount of cell death depending on the concentration of c-FLIPL after 350 min of stimulation with 3 µg/ml CD95L in HeLa wt cells. (B) HeLa wt cells were transfected with the H2 GFP-IRES–c-FLIPL or an mCherry-encoding control plasmid. 48 h after transfection, LZ-CD95L was added to the cells. The graph shows cell death of GFP+ and mCherry+ cells after addition of 3 µg/ml LZ-CD95L. Mean and SEM of three independent experiments are shown. *, P value < 0.05.
c-FLIPS/R promotes the c-FLIPL–sensitizing effect on CD95-induced cell death
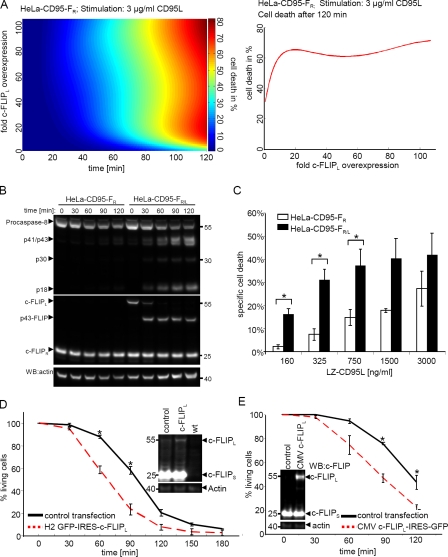
We observed that we could sensitize HeLa-CD95 cells to CD95-induced death with overexpression of c-FLIPL only to a minor extent (Fig. 4). To explain this phenomenon, we suggest that processing of procaspase-8 at the DISC in HeLa-CD95 cells is relatively fast. Therefore, accelerating procaspase-8 cleavage at the DISC with c-FLIPL overexpression has only marginal effects. To test this hypothesis, we searched for a system in which procaspase-8 processing at the DISC would be slowed down. HeLa-CD95–FS and HeLa-CD95–FR cells with a high amount of short c-FLIP isoforms ideally suited this scenario, as processing of procaspase-8 in these cells occurs much slower than in the absence of c-FLIPS/R (Fig. 1 B). The model predicted that in HeLa-CD95–FR and HeLa-CD95–FS cells, c-FLIPL overexpression would lead to a strong increase in both procaspase-8 processing and cell death upon addition of 3 µg/ml LZ-CD95L (Fig. 6 A and Fig. S5). To verify this prediction, we compared procaspase-8 cleavage in stable HeLa-CD95–FR/L cells with HeLa-CD95–FR cells on Western blots. Indeed, procaspase-8 processing to p43/p41 and p18 was visible in HeLa-CD95–FR/L cells already 30 min after addition of 3 µg/ml LZ-CD95L but was absent in HeLa-CD95–FR cells at this time point (Fig. 6 B). We could also confirm an increase in sensitivity of HeLa-CD95–FR/L as compared with HeLa-CD95–FR cells toward CD95-induced cell death, as measured with flow cytometry (Fig. 6 C).
Figure 6.
c-FLIPL sensitizes cells with high c-FLIPS/R levels. (A, left) Prediction of the amount of cell death depending on time and the concentration of c-FLIPL after stimulation with 3 µg/ml CD95L in HeLa-CD95–FR cells. (right) Prediction of the amount of cell death depending on the concentration of c-FLIPL after 120 min of stimulation with 3 µg/ml CD95L in HeLa-CD95–FR cells. (B) HeLa-CD95–FR/L and HeLa-CD95–FR cells were stimulated with 3 µg/ml LZ-CD95L for the indicated time points. Total cellular lysates were analyzed using Western blotting (WB) with antibodies against caspase-8, c-FLIP, and actin. One representative experiment out of three is shown. White lines indicate that intervening lanes have been spliced out. (C) HeLa-CD95–FR/L (black bars) and HeLa-CD95–FR (white bars) cells were stimulated with the indicated amounts of LZ-CD95L. Specific cell death was determined after 5 h of CD95 stimulation with PI stain and flow cytometry. (D and E) HeLa-CD95–FS cells were transfected with the H2 GFP-IRES–c-FLIPL (D), CMV c-FLIPL–IRES-GFP (E), or an mCherry-encoding control plasmid. 48 h after transfection, CD95L was added to the cells. The graph shows cell death of GFP+ and mCherry+ cells after addition of 3 µg/ml LZ-CD95L. Mean and SEM of three independent experiments are shown. *, P < 0.05.
Next, we analyzed CD95-induced death in HeLa-CD95–FS cells upon c-FLIPL overexpression using live cell imaging. HeLa-CD95–FS cells were transfected transiently with c-FLIPL using both promoters CMV and H2, respectively. For both scenarios, we observed significant sensitization toward CD95-induced apoptosis (Fig. 6, D and E). 90 min after addition of LZ-CD95L, only 40% of the cells transfected with a control plasmid were dead. In contrast, ∼75% of the c-FLIPL-transfected cells had already undergone apoptosis at this time point. Thus, we confirmed the model predictions and demonstrated that the sensitizing effect of c-FLIPL toward CD95-induced cell death is strongly increased in the presence of c-FLIPS/R.
Discussion
So far, the effect of c-FLIPL on DR-induced cell death has been unclear. Although several studies showed that c-FLIPL accelerates procaspase-8 processing at the DISC (Chang et al., 2002; Micheau et al., 2002), this did not necessarily lead to a sensitization of cells toward DR-induced apoptosis. Chang et al. (2002) proposed that small amounts of c-FLIPL sensitize cells, whereas high amounts block CD95-induced cell death. However, Sharp et al. (2005) showed in quite a few cell lines that down-regulation of c-FLIPL leads to sensitization toward DR-induced apoptosis. The authors concluded that c-FLIPL exclusively blocks cell death. Our results demonstrate that the effect of c-FLIPL on induction of cell death is CD95L dose dependent. The same amount of c-FLIPL can slow down induction of cell death at weak receptor stimulation but accelerates cell death upon stimulation with high doses of CD95L. This complex behavior shows that understanding of the effects of c-FLIPL is possible only considering the exact concentration of the DISC components, e.g., amounts of CD95L and active DISCs as well as concentration of c-FLIPL at the DISC, and also provides an explanation for different results in different cellular systems (Chang et al., 2002; Sharp et al., 2005).
Our model can also explain two opposite effects of c-FLIPL on procaspase-8 activation and CD95-induced apoptosis. c-FLIPL can either promote the activation of procaspase-8 or block the binding of procaspase-8 and, furthermore, procaspase-8 turnover at the DISC. c-FLIPL has a high affinity to the DISC (Chang et al., 2002), and after recruitment to the DISC, it might remain bound, blocking turnover of procaspase-8 at the DISC. These two opposite effects result in the nonlinearity and lead to a narrow range of c-FLIPL concentrations, which result in sensitization toward CD95-induced apoptosis.
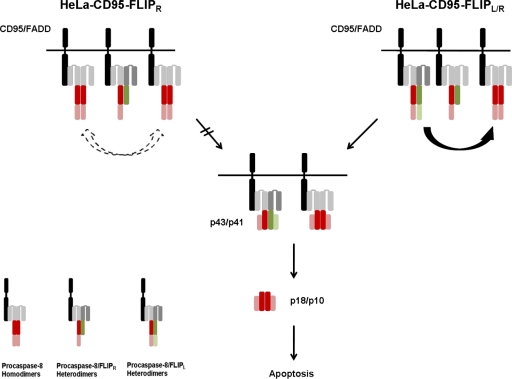
Furthermore, we show that c-FLIPR/S has the potential to modulate the strength of the sensitizing function of c-FLIPL (Fig. 7). In cells with high c-FLIPR/S levels, c-FLIPL has a strong sensitizing effect. So far, c-FLIPR/S have always been considered as molecules, which simply block procaspase-8 activation (Krueger et al., 2001; Golks et al., 2005; Ueffing et al., 2008; Kaunisto et al., 2009). The newly discovered function of c-FLIPR/S offers a novel possibility to further modulate CD95 signaling. This is especially important given that a lot of tumor cells regulate CD95 signaling by changing c-FLIP amounts and the ratio of c-FLIPS to c-FLIPL. Therefore, subtle changes in the stoichiometry of c-FLIP proteins at the DISC might significantly change the phenotype of the cells from resistant to sensitive. Moreover, we show that considering c-FLIPR/S as molecules that just block procaspase-8 processing might be too simplistic. In addition, our work shows that looking at the effects of c-FLIPS/R and c-FLIPL as if they were components of separate signaling pathways can be misleading, as this approach ignores the interplay of those two molecules.
Figure 7.
c-FLIPL sensitizes cells with high c-FLIPS/R levels. The scheme shows the effect of c-FLIPL in cells with high c-FLIPS/R levels. Caspase-8 is shown in red, and c-FLIP is shown in green. (left) Procaspase-8 processing occurs slowly in cells with high c-FLIPS/R amounts. (right) Additional expression of c-FLIPL leads to formation of procaspase-8/c-FLIPL heterodimers, which have a higher activity than homodimers and are able to cleave and activate procaspase-8, leading to cell death.
For the first time, our model has described the activation of procaspase-8 at the DISC and apoptosis with the generation of different homo- and heterodimers of procaspase-8 at the DISC leading to the cell death. Our model also introduced a direct readout for the rate of cell death. The active procaspase-8 cleavage products p43/p41 procaspase-8 homodimer, p43/p41-p43-FLIP heterodimer, and the (p10/p18)2 heterotetramer could cleave an apoptosis substrate leading to cell death. Interestingly, in our model, the activity of the p43/p41 procaspase-8 homodimer toward the apoptosis substrate was estimated to be roughly five times as high as that of the p43/p41-p43-FLIP heterodimer. A similar decrease in activity of the p43/p41 homodimer compared with the heterodimer was also proposed by Yu et al. (2009). The activity of the caspase-8 heterotetramer was much less than the activity of one of the homodimers. This is in accordance with the finding of Hughes et al. (2009). They proposed that the second cleavage step of procaspase-8 processing might reduce the activity of the molecule. Importantly, setting the activities of the procaspase-8 proteins in this way, we could reproduce the experimental data well (Figs. 4–6).
Another important role in the regulation of life/death decisions at the DISC is the induction of other nonapoptotic pathways via CD95, such as NF-κB (Budd et al., 2006; Neumann et al., 2010). c-FLIP proteins have been reported to be crucial regulators of NF-κB activity (Kataoka et al., 2000; Kataoka and Tschopp, 2004; Dohrman et al., 2005; Budd et al., 2006; Golks et al., 2006). Therefore, understanding of the quantitative stoichiometry of c-FLIP proteins and generation of c-FLIP cleavage products at the DISC might potentially lead to new insights in the understanding of life/death decisions at CD95.
Our model explains the regulation of procaspase-8 activation by c-FLIP proteins in HeLa cells, which overexpress CD95. With exception of the first model of the CD95-induced apoptosis (Bentele et al., 2004), which was performed using B lymphoblastoid cells, most systems biology models of apoptosis currently published use HeLa cells as a cellular system (Lavrik et al., 2009). Previous studies defined the mitochondrial outer membrane permeabilization regulation using HeLa cells (Rehm et al., 2006, 2009). Several sophisticated models of TRAIL-induced apoptosis using HeLa cells were also previously developed (Albeck et al., 2008a,b; Spencer et al., 2009). Therefore, up to now, using HeLa cells is a well-defined experimental setup for systems biology studies. The future challenge will be to develop systems biology models toward understanding of apoptosis regulation in various other cell types. Especially important will be the understanding of primary cancer cells.
We have discovered a new view of caspase-8 activation by c-FLIP proteins. Another question to address in the future will be the contribution of c-FLIP proteins to procaspase-10 activation at the DISC. Procaspase-10 has been reported to lack a major impact on apoptosis induction (Sprick et al., 2002). However, these findings might only apply to certain cell types and low levels of procaspase-10 in these cells. Therefore, the investigation of all DED proteins of the DISC, procaspase-8, -10, and c-FLIP should be the focus of future studies.
Collectively, whether c-FLIPL accelerates or slows down DR-induced cell death does not only depend on its amount but also on the cell type and the strength of receptor stimulation. These findings explain why different investigators came to completely different conclusions and can resolve the contradictions reported in recent literature (Chang et al., 2002; Sharp et al., 2005).
Materials and methods
Cell lines
HeLa-CD95 was generated by selection with 20 ng/ml puromycin according to standard protocols. Stable c-FLIPL and c-FLIPR cell lines were selected with 100 µg/ml G418 and 100 µg/ml zeocin, respectively. HeLa-CD95 cells were maintained in DME (Life Technologies), 10 mM Hepes (Life Technologies), 50 µg/ml gentamycin (Life Technologies), and 10% FCS (Life Technologies) in 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen).
DNA constructs
For stable CD95 overexpression, CD95 cDNA was cloned into a pIRESpuro3 plasmid (Takara Bio Inc.). To generate cells stably expressing c-FLIPL, the coding sequence of c-FLIPL was cloned in the pIRESneo3 vector (Takara Bio Inc.). For stable c-FLIPR expression, we used a pEF4 plasmid (Invitrogen). To generate the c-FLIPL–IRES-GFP plasmid, the neomycin-resistance sequence of the pIRESneo3 plasmid was replaced by GFP. For the c-FLIP kinase-dead plasmid, the following sequence was cloned into a vector (pSilencer 3.1-H1; Applied Biosystems): 5′-GATCCGCAAGGAGAAGAGTTTCTTCTCAAGAGAAAGAAACTCTTCTCCTTGCTTTTTTGGAAA-3′. The c-FLIP targeting sequence was 5′-GCAAGGAGAAGAGTTTCTT-3′. To construct the NES-IETD-mCherry caspase-8 activity probe, an NES was fused to the caspase-8 cleavage sequence IETD followed by a linker, leading to the sequence MNLVDLQKKLEELELDEQQTGGPIETDSGGGGG. This sequence was then fused to mCherry. To obtain a plasmid encoding for GFP-IRES–c-FLIPL under a histone H2 promoter, we amplified a GFP-IRES sequence with the following primers: (top) 5′-GCGGCCTAGCTAGCGCTTAAGGCCTG-3′ and (bottom) 5′-GGGTTGCTAGCGGGTTGTGGCAAGCTTA-3′. Additionally, we used a c-FLIPL coding sequence that was flanked at 3′ with an NheI sticky end and at 5′ with an NotI sticky end. The empty plasmid containing an H2 promoter was digested with NheI and NotI in the multiple-cloning site. After digestion with NheI, the PCR fragment was ligated together with the c-FLIPL–encoding fragment into the plasmid containing an H2 promoter. The coding sequence of LZ-CD95L (Walczak et al., 1999) was cloned into a pIRESpuro3 plasmid (Takara Bio Inc.).
Antibodies and reagents
Anti–caspase-8 monoclonal antibody C15 (mouse IgG2b) recognizes the p18 subunit of caspase-8 (Scaffidi et al., 1997). Anti-FLIP monoclonal antibody NF6 (mouse IgG1) recognizes the N-terminal part of c-FLIP (Scaffidi et al., 1999). Anti–APO-1 (anti-CD95) is an agonistic monoclonal antibody (IgG3) recognizing an epitope on the extracellular part of CD95 (APO-1/Fas; Trauth et al., 1989). Additionally, we used a polyclonal C3 antibody (Cell Signaling Technology), C2-10 anti-PARP antibody (BD), and the C20 monoclonal CD95 antibody (Santa Cruz Biotechnology, Inc.). Horseradish peroxidase–conjugated goat anti–mouse IgG1, -2a, and -2b were obtained from SouthernBiotech. The coding sequence of LZ-CD95L (Walczak et al., 1999) was cloned into a pIRESpuro3 plasmid (Takara Bio Inc.). Recombinant LZ-CD95L was produced using 293T cells stably transfected with this vector. All chemicals used were of analytical grade and purchased from Merck or Sigma-Aldrich.
Analysis of total cellular lysates
3 × 105 cells were either treated with indicated amounts of LZ-CD95L for indicated periods of time at 37°C or left untreated, washed twice in 1x PBS, and lysed subsequently in lysis buffer A (30 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [Sigma-Aldrich], protease inhibitor cocktail, 1% Triton X-100 [Serva], and 10% glycerol). Total cellular lysates were analyzed using SDS-PAGE (Invitrogen). Proteins were transferred to Hybond nitrocellulose membrane using IBlot technology (Invitrogen) blocked with 5% nonfat dry milk in PBS/Tween (PBS plus 0.05% Tween 20) for 1 h, washed with PBS/Tween, and incubated with the primary antibody in PBS/Tween overnight at 4°C. Blots were developed with a chemoluminescence method according to the manufacturer’s protocol (PerkinElmer).
DISC immunoprecipitation
107 HeLa-CD95 cells were lysed in lysis buffer A with or without stimulation, divided into two equal parts, and DISC immunoprecipitation was performed using 5 µg anti-CD95 antibody (anti–APO-1) with 30 µl protein A–Sepharose. Immunoprecipitations were performed overnight at 4°C, then beads were washed five times with 20 vol of lysis buffer and subjected to Western blot analysis as described in the previous paragraph.
Microscope image acquisition
Cells were cultured in 8-well Laboratory-Tek–chambered cover glasses (Thermo Fisher Scientific). During imaging, cells were maintained at 37°C in DME without phenol red and complemented with 20 mM Hepes buffer and 10% FCS. We used a laser-scanning confocal microscope (TCS SP2; Leica) with a 63× 1.4 NA oil immersion lens and a camera (DC300FX; Leica) for image acquisition. For excitation, the laser wavelength was set to 488 nm for GFP and 594 nm for mCherry. The recording spectrum was 498–535 nm for GFP and 604–686 nm for mCherry. Image acquisition was performed using confocal software (Leica). Image analysis was performed using ImageJ (National Institutes of Health). To measure cell death of GFP-IRES–c-FLIPL– versus mCherry control–transfected cells, we determined which cells were GFP+ or mCherry+ with fluorescence microscopy. Then, CD95L was added to the cells. Cell fate of mCherry+ and GFP+ cells was measured with live cell imaging. To measure cleavage of the caspase-8 activity, probe cells were cotransfected with a GFP-IRES–c-FLIPL plasmid plus the NES-IETD-mCherry probe or the NES-IETD-mCherry probe plus an empty control vector. Cleavage of the caspase-8 activity probe was determined by translocation of mCherry to the nucleus in confocal microscopy. To measure mCherry translocation to the nucleus, the mean fluorescence intensity (MFI) of the nucleus and cytoplasm was determined using ImageJ. The following formula was applied to calculate the translocation R(t): R(t) = [MFI (nucleus) − MFI (cytoplasm)]/[MFI (nucleus) + MFI (cytoplasm)] − R(0).
Cell death assay
Cells were plated and treated as indicated with LZ-CD95L for indicated periods of time and 1 µg/ml propidium iodide (PI) for 30 min. Cell death was assessed by PI uptake and quantified with a flow cytometer (Cytomics FC 500 MPL; Beckman Coulter).
Statistical methods
Student’s t test was applied to test for significance of differences in cell death between cell lines as measured by live cell imaging.
Mathematical modeling
Modeling was performed using the SBtoolbox (Schmidt and Jirstrand, 2006) in combination with Matlab (MathWorks). For local parameter fitting, a simplex algorithm was used. For parameter fitting, we used data from quantitative Western blots for processing of procaspase-8. Additionally, proteins amounts were set in the model according to measured data shown in Table S1 with acceptance of a 20% deviation. After 1,000 times of local parameter estimation with random start values, best fits were analyzed. The model is available in SBtoolbox model format as Supplemental data.
Online supplemental material
Fig. S1 shows characterization of stable cell lines overexpressing different c-FLIP isoforms. Fig. S2 shows the effect of c-FLIPL on procaspase-8 processing and cell death. Fig. S3 shows protein quantification with Western blots and the effect of c-FLIP down-regulation in HeLa-CD95 cells. Fig. S4 shows live cell imaging of HeLa-CD95 cells. Fig. S5 shows that the model predicts an increase in procaspase-8 processing upon c-FLIPL overexpression in HeLa-CD95–FR cells. Table S1 shows mean protein numbers per cell in HeLa-CD95 and c-FLIP–overexpressing HeLa-CD95 cells. Additional supplementary data shows the CD95 signaling model as an SBtoolbox file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002060/DC1.
Acknowledgments
We thank Julia Schwuchow, Alexander Pappa, Julia Hoffmann, Stefan Legewie, Karsten Gülow, Carina Pforr, and Olga Shatnyeva for critical comments on the manuscript and Michaela Reichenzeller for providing a plasmid with histone H2 promoter.
We acknowledge support by the Helmholtz Alliance on Systems Biology (NW1SBCancer/UniHD) and The German Federal Ministry of Education and Research (BMBF)–funded FORSYS center ViroQuant (grant 0313923). We also thank the Wilhelm Sander Stiftung, Helmholtz-Russia Joint Research Groups (2008-2, SFB 405), and the Tumorzentrum Heidelberg/Mannheim for supporting our work.
Footnotes
Abbreviations used in this paper:
- c-FLIP
- cellular FADD-like interleukin-1β–converting enzyme inhibitory protein
- c-FLIPL
- c-FLIP long
- c-FLIPR
- c-FLIP Raji
- c-FLIPS
- c-FLIP short
- CMV
- cytomegalovirus
- DED
- death effector domain
- DISC
- death-inducing signaling complex
- DR
- death receptor
- MFI
- mean fluorescence intensity
- NES
- nuclear export signal
- PARP
- poly(ADP-ribose) polymerase
- PI
- propidium iodide
- wt
- wild type
References
- Albeck J.G., Burke J.M., Aldridge B.B., Zhang M., Lauffenburger D.A., Sorger P.K. 2008a. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 30:11–25 10.1016/j.molcel.2008.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck J.G., Burke J.M., Spencer S.L., Lauffenburger D.A., Sorger P.K. 2008b. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 6:2831–2852 10.1371/journal.pbio.0060299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele M., Lavrik I., Ulrich M., Stösser S., Heermann D.W., Kalthoff H., Krammer P.H., Eils R. 2004. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 166:839–851 10.1083/jcb.200404158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd R.C., Yeh W.C., Tschopp J. 2006. cFLIP regulation of lymphocyte activation and development. Nat. Rev. Immunol. 6:196–204 10.1038/nri1787 [DOI] [PubMed] [Google Scholar]
- Chang D.W., Xing Z., Pan Y., Algeciras-Schimnich A., Barnhart B.C., Yaish-Ohad S., Peter M.E., Yang X. 2002. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 21:3704–3714 10.1093/emboj/cdf356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.W., Xing Z., Capacio V.L., Peter M.E., Yang X. 2003. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 22:4132–4142 10.1093/emboj/cdg414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrman A., Kataoka T., Cuenin S., Russell J.Q., Tschopp J., Budd R.C. 2005. Cellular FLIP (long form) regulates CD8+ T cell activation through caspase-8-dependent NF-kappa B activation. J. Immunol. 174:5270–5278 [DOI] [PubMed] [Google Scholar]
- Eissing T., Conzelmann H., Gilles E.D., Allgöwer F., Bullinger E., Scheurich P. 2004. Bistability analyses of a caspase activation model for receptor-induced apoptosis. J. Biol. Chem. 279:36892–36897 10.1074/jbc.M404893200 [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P., Salvesen G.S. 2004. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384:201–232 10.1042/BJ20041142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golks A., Brenner D., Fritsch C., Krammer P.H., Lavrik I.N. 2005. c-FLIPR, a new regulator of death receptor-induced apoptosis. J. Biol. Chem. 280:14507–14513 10.1074/jbc.M414425200 [DOI] [PubMed] [Google Scholar]
- Golks A., Brenner D., Krammer P.H., Lavrik I.N. 2006. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappaB activation. J. Exp. Med. 203:1295–1305 10.1084/jem.20051556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J.C., Pappa A., Krammer P.H., Lavrik I.N. 2009. A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol. Cell. Biol. 29:4431–4440 10.1128/MCB.02261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M.A., Harper N., Butterworth M., Cain K., Cohen G.M., MacFarlane M. 2009. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol. Cell. 35:265–279 10.1016/j.molcel.2009.06.012 [DOI] [PubMed] [Google Scholar]
- Kang T.B., Oh G.S., Scandella E., Bolinger B., Ludewig B., Kovalenko A., Wallach D. 2008. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J. Immunol. 181:2522–2532 [DOI] [PubMed] [Google Scholar]
- Kataoka T., Tschopp J. 2004. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol. Cell. Biol. 24:2627–2636 10.1128/MCB.24.7.2627-2636.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Budd R.C., Holler N., Thome M., Martinon F., Irmler M., Burns K., Hahne M., Kennedy N., Kovacsovics M., Tschopp J. 2000. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr. Biol. 10:640–648 10.1016/S0960-9822(00)00512-1 [DOI] [PubMed] [Google Scholar]
- Kaunisto A., Kochin V., Asaoka T., Mikhailov A., Poukkula M., Meinander A., Eriksson J.E. 2009. PKC-mediated phosphorylation regulates c-FLIP ubiquitylation and stability. Cell Death Differ. 16:1215–1226 10.1038/cdd.2009.35 [DOI] [PubMed] [Google Scholar]
- Kischkel F.C., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P.H., Peter M.E. 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14:5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer P.H., Arnold R., Lavrik I.N. 2007. Life and death in peripheral T cells. Nat. Rev. Immunol. 7:532–542 10.1038/nri2115 [DOI] [PubMed] [Google Scholar]
- Krueger A., Schmitz I., Baumann S., Krammer P.H., Kirchhoff S. 2001. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 276:20633–20640 10.1074/jbc.M101780200 [DOI] [PubMed] [Google Scholar]
- Lavrik I.N., Golks A., Krammer P.H. 2005a. Death receptor signaling. J. Cell Sci. 118:265–267 10.1242/jcs.01610 [DOI] [PubMed] [Google Scholar]
- Lavrik I.N., Golks A., Krammer P.H. 2005b. Caspases: pharmacological manipulation of cell death. J. Clin. Invest. 115:2665–2672 10.1172/JCI26252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrik I.N., Golks A., Riess D., Bentele M., Eils R., Krammer P.H. 2007. Analysis of CD95 threshold signaling: triggering of CD95 (FAS/APO-1) at low concentrations primarily results in survival signaling. J. Biol. Chem. 282:13664–13671 10.1074/jbc.M700434200 [DOI] [PubMed] [Google Scholar]
- Lavrik I.N., Eils R., Fricker N., Pforr C., Krammer P.H. 2009. Understanding apoptosis by systems biology approaches. Mol. Biosyst. 5:1105–1111 10.1039/b905129p [DOI] [PubMed] [Google Scholar]
- Legewie S., Blüthgen N., Herzel H. 2006. Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLOS Comput. Biol. 2:e120 10.1371/journal.pcbi.0020120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema J.P., Scaffidi C., Kischkel F.C., Shevchenko A., Mann M., Krammer P.H., Peter M.E. 1997. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J. 16:2794–2804 10.1093/emboj/16.10.2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O., Thome M., Schneider P., Holler N., Tschopp J., Nicholson D.W., Briand C., Grütter M.G. 2002. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 277:45162–45171 10.1074/jbc.M206882200 [DOI] [PubMed] [Google Scholar]
- Muzio M., Chinnaiyan A.M., Kischkel F.C., O’Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J.D., Zhang M., Gentz R., et al. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death—inducing signaling complex. Cell. 85:817–827 10.1016/S0092-8674(00)81266-0 [DOI] [PubMed] [Google Scholar]
- Muzio M., Stockwell B.R., Stennicke H.R., Salvesen G.S., Dixit V.M. 1998. An induced proximity model for caspase-8 activation. J. Biol. Chem. 273:2926–2930 10.1074/jbc.273.5.2926 [DOI] [PubMed] [Google Scholar]
- Neumann L., Pforr C., Beaudouin J., Pappa A., Fricker N., Krammer P.H., Lavrik I.N., Eils R. 2010. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol. Syst. Biol. 6:352 10.1038/msb.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M.E., Budd R.C., Desbarats J., Hedrick S.M., Hueber A.O., Newell M.K., Owen L.B., Pope R.M., Tschopp J., Wajant H., et al. 2007. The CD95 receptor: apoptosis revisited. Cell. 129:447–450 10.1016/j.cell.2007.04.031 [DOI] [PubMed] [Google Scholar]
- Rehm M., Huber H.J., Dussmann H., Prehn J.H. 2006. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 25:4338–4349 10.1038/sj.emboj.7601295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M., Huber H.J., Hellwig C.T., Anguissola S., Dussmann H., Prehn J.H. 2009. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ. 16:613–623 10.1038/cdd.2008.187 [DOI] [PubMed] [Google Scholar]
- Scaffidi C., Medema J.P., Krammer P.H., Peter M.E. 1997. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J. Biol. Chem. 272:26953–26958 10.1074/jbc.272.43.26953 [DOI] [PubMed] [Google Scholar]
- Scaffidi C., Schmitz I., Krammer P.H., Peter M.E. 1999. The role of c-FLIP in modulation of CD95-induced apoptosis. J. Biol. Chem. 274:1541–1548 10.1074/jbc.274.3.1541 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Jirstrand M. 2006. Systems biology toolbox for MATLAB: a computational platform for research in systems biology. Bioinformatics. 22:514–515 10.1093/bioinformatics/bti799 [DOI] [PubMed] [Google Scholar]
- Sharp D.A., Lawrence D.A., Ashkenazi A. 2005. Selective knockdown of the long variant of cellular FLICE inhibitory protein augments death receptor-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 280:19401–19409 10.1074/jbc.M413962200 [DOI] [PubMed] [Google Scholar]
- Spencer S.L., Gaudet S., Albeck J.G., Burke J.M., Sorger P.K. 2009. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 459:428–432 10.1038/nature08012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprick M.R., Rieser E., Stahl H., Grosse-Wilde A., Weigand M.A., Walczak H. 2002. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 21:4520–4530 10.1093/emboj/cdf441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T., Takahashi T., Golstein P., Nagata S. 1993. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 75:1169–1178 10.1016/0092-8674(93)90326-L [DOI] [PubMed] [Google Scholar]
- Trauth B.C., Klas C., Peters A.M., Matzku S., Möller P., Falk W., Debatin K.M., Krammer P.H. 1989. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 245:301–305 10.1126/science.2787530 [DOI] [PubMed] [Google Scholar]
- Ueffing N., Keil E., Freund C., Kühne R., Schulze-Osthoff K., Schmitz I. 2008. Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ. 15:773–782 10.1038/sj.cdd.4402314 [DOI] [PubMed] [Google Scholar]
- Walczak H., Miller R.E., Ariail K., Gliniak B., Griffith T.S., Kubin M., Chin W., Jones J., Woodward A., Le T., et al. 1999. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5:157–163 10.1038/5517 [DOI] [PubMed] [Google Scholar]
- Yu J.W., Shi Y. 2008. FLIP and the death effector domain family. Oncogene. 27:6216–6227 10.1038/onc.2008.299 [DOI] [PubMed] [Google Scholar]
- Yu J.W., Jeffrey P.D., Shi Y. 2009. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. USA. 106:8169–8174 10.1073/pnas.0812453106 [DOI] [PMC free article] [PubMed] [Google Scholar]