Syndecan-3 is required for Notch processing by ADAM17/TACe and therefore regulates proliferation and viability of muscle satellite cells.
Abstract
Skeletal muscle postnatal growth and repair depend on satellite cells and are regulated by molecular signals within the satellite cell niche. We investigated the molecular and cellular events that lead to altered myogenesis upon genetic ablation of Syndecan-3, a component of the satellite cell niche. In the absence of Syndecan-3, satellite cells stall in S phase, leading to reduced proliferation, increased cell death, delayed onset of differentiation, and markedly reduced numbers of Pax7+ satellite cells accompanied by myofiber hypertrophy and an increased number of centrally nucleated myofibers. We show that the aberrant cell cycle and impaired self-renewal of explanted Syndecan-3–null satellite cells are rescued by ectopic expression of the constitutively active Notch intracellular domain. Furthermore, we show that Syndecan-3 interacts with Notch and is required for Notch processing by ADAM17/tumor necrosis factor-α–converting enzyme (TACE) and signal transduction. Together, our data support the conclusion that Syndecan-3 and Notch cooperate in regulating homeostasis of the satellite cell population and myofiber size.
Introduction
Adult stem cells are rare cells characterized by the ability to self-renew and to generate specialized progeny committed to tissue maintenance and repair (Schultz and McCormick, 1994; Kuang et al., 2008). The discovery in recent years of tissue-specific adult stem cells residing in anatomically and functionally defined niches has strengthened the importance of the niche in regulating stem cell fate and function (Blanpain et al., 2007; Blank et al., 2008; Discher et al., 2009). Satellite cells (SCs), the adult skeletal muscle progenitors, reside in a niche that was anatomically defined by electron microscopy (Mauro, 1961). SCs reside in a pocket within the surface of myofibers, where the basolateral side of the SC is exposed to the myofiber plasma membrane and the apical side is exposed to the basal lamina. The SC niche is thought to play a critical role in defining SC homeostasis, as disruption of the SC niche often leads to impaired muscle maintenance and impaired regeneration (Cornelison et al., 2004; Collins et al., 2005; Boonen and Post, 2008). However, the molecular mechanisms involved in signaling integration within the satellite niche are still poorly understood.
Recently, we identified Syndecan-3 as a component of the SC niche. Syndecan-3 is a transmembrane heparan sulfate proteoglycan (HSPG) expressed in SCs that interacts with extracellular matrix proteins and growth factors through its ectodomain, and with cytoskeletal proteins and intracellular signaling molecules through its intracellular domain. Syndecan-3 plays a role in SC maintenance, proliferation, and differentiation (Fuentealba et al., 1999). Syndecan-3–null (sdc3−/−) muscles regenerate but uninjured sdc3−/− muscles exhibit aberrant phenotypes, including an increase of myonuclei in myofibers (myofiber hyperplasia) and an increase of centrally nucleated myofibers (Cornelison et al., 2004). Furthermore, explanted sdc3−/− SCs show impaired proliferation and differentiation in culture, accompanied by altered response to growth factors (Cornelison et al., 2004).
Another key component of the SC niche is Notch. In adult muscle, Notch regulates proliferation and myogenic commitment of activated SCs (Nye et al., 1994; Kopan et al., 1994; Kuroda et al., 1999; Conboy and Rando, 2002; Luo et al., 2005; Holterman et al., 2007). Recent data have shown a role for Notch in asymmetric SC division (Shinin et al., 2006; Kuang et al., 2007) and myoblast progression through the cell cycle in culture (Carlson et al., 2008), as well as a requirement for maintaining a population of undifferentiated “reserve” cells in culture (Kitzmann et al., 2006).
Notch receptors are intramolecular heterodimers composed of an ectodomain disulfide linked to a transmembrane subunit (Shimizu et al., 2002; Fiúza and Arias, 2007). Four Notch receptors (Notch1–4) and six Notch ligands (Jagged1 and -2, Delta1, Delta3 and -4, and X-Delta2) are present in mammals (D’Souza et al., 2008). When a Notch receptor interacts with an adjacent Notch ligand or one present on an opposing cell, the Notch receptor undergoes a sequence of proteolytic cleavages that release the Notch intracellular domain (NICD). The soluble NICD then translocates to the nucleus, where it binds and activates the transcription factor CBF1/suppressor of hairless/Lag-1 (CSL), inducing expression of members of the Hes and Hey families of basic helix-loop-helix transcription factors (Jarriault et al., 1995; Iso et al., 2003). A role for heparan sulfates in Drosophila Notch has been postulated (Kamimura et al., 2004), but the mechanisms involved are not known, and similar results have not been described in mammals. Here, we used Ingenuity Pathway Analysis (IPA) of global gene expression data obtained from Affymetrix arrays comparing changes occurring in uninjured versus injured wild-type and sdc3−/− SCs to investigate candidate mechanisms responsible for the observed sdc3−/− phenotypes. The IPA analysis revealed a reduction of Notch target gene expression in sdc3−/− SCs compared with wild-type SCs. We then show an interaction between Notch and Syndecan-3 and a requirement for Syndecan-3 in Notch processing and signaling. The absence of Syndecan-3 impairs Notch signaling, altering SC homeostasis and affecting skeletal muscle regeneration.
Results
Syndecan-3 loss affects SC proliferation, differentiation, and self-renewal in culture
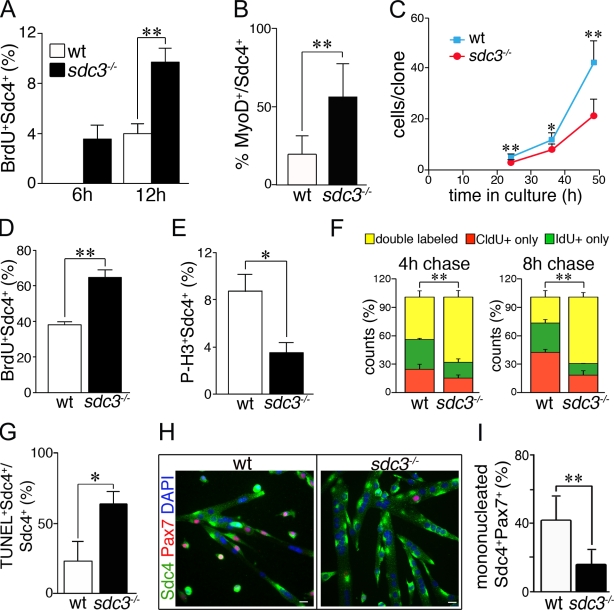
Loss of Syndecan-3 affects SC proliferation, commitment to myogenesis, and differentiation (Cornelison et al., 2004), which implicates this HSPG as an important component of the SC niche. To better understand the effects of Syndecan-3 loss on SC proliferation and differentiation, we examined the behavior of myofiber-associated SCs in culture explanted from wild-type and sdc3−/− mouse hindlimb muscles. Sdc3−/− SCs activate more rapidly after the myofiber explant, as a greater number of BrdU+ and MyoD+ SCs are found on sdc3−/− myofibers compared with the wild-type controls, (Fig. 1, A and B). Despite their rapid activation, sdc3−/− SCs expand less than wild-type cells in culture (Fig. 1 C). This difference is not caused by a difference in the numbers of proliferating cells, as demonstrated by a 4-h BrdU pulse label (unpublished data). However, a short 30-min BrdU pulse label revealed a greater number of sdc3−/− cells in S phase compared with wild-type cells (Fig. 1 D), which is inconsistent with the reduction in sdc3−/− cell expansion. When scored for the number of cells in mitosis, we found fewer phospho–histone H3+ (p-H3+) sdc3−/− cells than wild-type cells (Fig. 1 E). Together, these data suggest that the cell cycle kinetics of sdc3−/− and wild-type SCs differ in culture. We tested this idea by performing a double-label pulse-chase experiment using 5-chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU) to determine the time that cells spend in S phase. We pulse-labeled wild-type and sdc3−/− SC cultures for 30 min with CldU followed by either a 4 h or 8 h chase (Fig. S1 A). At the end of each chase period, IdU was added for 30 min (Fig. S1 A). Cells with an extended S phase will label with both CldU and IdU, whereas cells with a shorter S phase will label predominately with either CldU or IdU, depending on the chase time. Indeed, more sdc3−/− cells than wild-type cells were double-labeled with either the 4 h or 8 h chase, which indicates that loss of Syndecan-3 lengthens the S-phase of these cells (Fig. 1 F). The double-labeled cells clearly show a distinct pattern of CldU and IdU staining, which indicates that the two thymidine analogues were incorporated in different portions of the replicating genome (Fig. S1 B). This result also rules out the possibility of staining artifacts due to anti-CldU and anti-IdU antibody cross-reactivity.
Figure 1.
Cell cycle progression and cell fate decisions are dysregulated in cultured sdc3−/− SCs. (A and B) Myofiber-associated sdc3−/− SCs exit from quiescence and enter the cell cycle faster than wild-type cells. The percentage of BrdU+ (A) and MyoD+ (B) SCs is increased on sdc3−/− myofibers at 6 h and 12 h after explant compared with wild-type myofibers. (C) Sdc3−/− SCs expand less than wild-type controls during the first 2.5 d in culture (means from one representative experiment of four; ≥20 clones/plate scored). (D and E) Explanted and cultured sdc3−/− SCs were pulse-labeled for 30 min with 10 µM BrdU before fixation and immunostaining to detect BrdU, Syndecan-4 (Sdc4) as a SC marker, and P-H3 to mark mitotic cells. The percentage of Sdc4+BrdU+ cells normalized to the total number of Sdc4+ cells is higher (D, n = 5) and the percentage of Sdc4+P-H3+ cells is lower (E, n = 3) in sdc3−/− SC cultures compared with wild-type controls. (F) Wild-type and sdc3−/− SCs in culture were pulse-labeled with CldU followed by IdU after either a 4-h or 8-h chase. A higher percentage of sdc3−/− cells were in S phase during both the CldU and the IdU pulses, and were therefore double labeled, compared with wild-type cells. (G) Increased cell death is detected by a TUNEL assay in explanted sdc3−/− SC cultures compared with wild-type cultures (n = 3). (H) Explanted sdc3−/− SCs fuse into hypertrophic myotubes and generate fewer Sdc4+/Pax7+ reserve cells compared with wild-type cultures 48 h after differentiation induction. Bars, 10 µm. (I) Quantification of G. Pax7+Sdc4+ mononuclear cells normalized to total Sdc4+ mononuclear cells (15 clones/sample; n = 3). Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. **, P < 0.01; *, P < 0.05.
In addition to impaired proliferation, Syndecan-3 loss leads to a delay in the onset of differentiation, dysregulated myoblast terminal differentiation, and aberrant fusion (Cornelison et al., 2004). Proliferating myoblasts in G1 phase decide whether to reenter S phase and progress through another cell cycle or exit the cell cycle. Once they are in G0, they either differentiate or self-renew (Olguin and Olwin, 2004; Zammit et al., 2004). Thus, the delay in differentiation onset observed in sdc3−/− SC cultures (Cornelison et al., 2004) could be a direct consequence of the impaired cell cycle progression because cells that have a prolonged S phase also delay G1 phase entry. As predicted, we found that fewer sdc3−/− than wild-type cells were found in G1 phase when the cell cycle distribution was analyzed by flow cytometry (Fig. S1 C). Because fewer cells in sdc3−/− satellite cultures are in G1 at any time, cell cycle exit and Myogenin expression is delayed in sdc3−/− cultures compared with wild-type controls (Fig. S1 D). Cells that stall in S phase are also more prone to undergo apoptosis (Maddika et al., 2007), therefore we asked whether cell survival was impaired in sdc3−/− SCs. When tested for cell death by a TUNEL assay, sdc3−/− cultures had more TUNEL+ cells than wild-type cultures (Fig. 1 G).
Upon cell cycle exit, muscle progenitors in G0 either commit to terminal differentiation and fuse or replenish the undifferentiated progenitor population (Olguin and Olwin, 2004; Zammit et al., 2004). These cell fate decisions are altered in the absence of Syndecan-3. At 48 h and 72 h after serum withdrawal, sdc3−/− cultures showed larger myotubes (Fig. 1 H; Cornelison et al., 2004) and reduced Pax7+ reserve cells (Fig. 1, H and I) compared with wild-type cultures, which indicates that sdc3−/− SCs exiting the cell cycle prefer to undergo terminal myogenic differentiation as opposed to self-renewal. Our results suggest that a combination of cell cycle lengthening, increased cell death, and impaired balance between differentiation and self-renewal may be responsible for the reduced expansion of sdc3−/− cells in culture.
Syndecan-3 loss affects SC proliferation, differentiation, and self-renewal in vivo
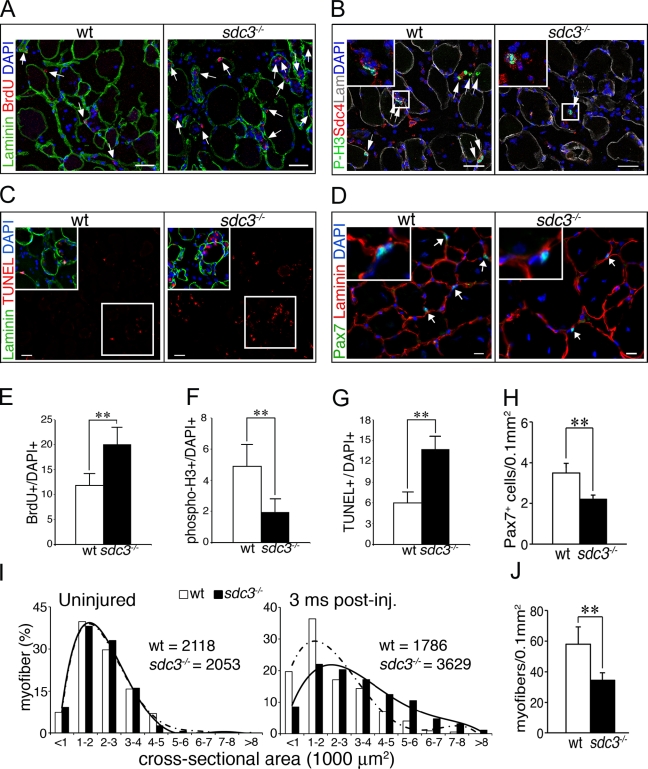
The effects of Syndecan-3 loss on SC proliferation and differentiation observed in culture are unlikely to be a culture artifact, as we observed similar phenotypes in vivo during injury-induced muscle regeneration. Injured sdc3−/− tibialis anterior (TA) muscles have more BrdU+ cells (Fig. 2, A and E), fewer P-H3+ cells (Fig. 2, B and F), and increased cell death (TUNEL+ cells; Fig. 2 C, G) compared to wild-type controls 36 h after injury. Moreover, similar to our in vitro data, we observe dysregulated cell fate choices in vivo (Fig. 2, D and H–J). Injured sdc3−/− TA muscles show fewer Pax7+ SCs when analyzed at 2 wk, 6 wk (Fig. S2), or 3 mo (Fig. 2, D and H) after injury, and an increase in myofiber size at 3 mo after injury when compared with wild-type controls (Fig. 2 I). This is accompanied by a decrease in the myofiber number (Fig. 2 J). Although the total number of nuclei per tissue area is unchanged in sdc3−/− regenerated TA muscles compared with wild-type controls (not depicted), more of the sdc3−/− nuclei are found in hypertrophic myofibers, whereas fewer cells are retained as undifferentiated Pax7+ progenitors (Fig. 2, D and H). Our in vitro and in vivo results indicate that loss of Syndecan-3 impairs SC progression through the cell cycle and alters cell fate choices, leading to myofiber hypertrophy and depletion of the undifferentiated (sublaminar, Pax7+) population upon muscle injury.
Figure 2.
Cell cycle progression and cell fate decisions are dysregulated in regenerating sdc3−/− muscles in vivo. (A–G) Proliferation is impaired in sdc3−/− SCs. Sections from wild-type TA muscles show fewer BrdU+ cells (arrows in A indicate BrdU+ cells; E shows a quantification of A), more P-H3+ cells (arrows in B indicate P-H3+ cells; F shows a quantification of B), and fewer TUNEL+ cells (C) than those from sdc3−/− TA muscles (G shows a quantification of C). (D and J) Cell fate decisions are altered in sdc3−/− SCs. The number of Pax7+ sublaminar cells (SCs) per tissue area is reduced (arrows in D indicate Pax7+ cells; H shows a quantification of D), the myofiber cross-sectional area is increased (I), and the total number of myofibers per tissue area is reduced (J) in sdc3−/− TA muscles 3 mo after injury compared with wild-type muscles. Medians of the myofiber cross-sectional area distribution for each genotype at 3 mo after injury are shown in I. Curves in I represent polynomial functions that best fit the plotted myofiber size distributions; broken line, wild type; solid line, sdc3−/−. A minimum of 1,000 myofibers and four sections per each biological replica were scored. Inset panels in B–D show enlarged views of the boxed regions. Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. **, P < 0.01. Bars, 30 µm.
Syndecan-3 is required for Notch signaling
Genetic ablation of Syndecan-3 leads to alterations in several signaling pathways involved in the regulation of adult myogenesis, but the molecular mechanisms involved are unknown (Cornelison et al., 2004). To identify the mechanisms involved, we used a global gene expression analysis approach. SCs were FACS-isolated from wild-type, sdc3−/− uninjured, and injured TA muscles 12 h after injury. Gene expression was then analyzed using Spotfire software after hybridization to Affymetrix gene chips, where we investigated which genes were up-regulated by ≥1.5-fold in wild-type SCs 12 h after injury but were not up-regulated in sdc3−/− SCs 12 h after injury (Table S1). Relative expression of Notch target genes appeared impaired in sdc3−/− versus wild-type SCs isolated 12 h after an induced muscle injury (Fig. 3 A). The Notch pathway is a key regulator of adult myogenesis, where Notch inhibition leads to reduced muscle progenitor cell proliferation (Conboy and Rando, 2002; Carlson et al., 2008), precocious terminal differentiation and fusion (Conboy and Rando, 2002; Holterman et al., 2007), and reduced progenitor self renewal (Kitzmann et al., 2006; Kuang et al., 2007). Remarkably, these are all phenotypes we observe in sdc3−/− SCs in vitro (Fig. 1) and in vivo (Fig. 2).
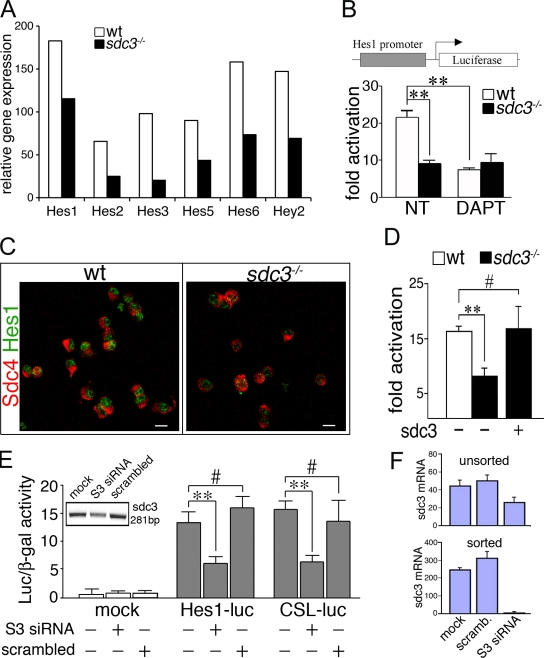
Figure 3.
Notch signaling is impaired in the absence of Syndecan-3 and rescued by ectopic Syndecan-3 expression. (A) Wild-type and sdc3−/− SCs were FACS-isolated from uninjured TA muscles and TA muscles 12 h after injury, then total mRNA was extracted and pooled from a minimum of three animals, and gene expression was assayed on three replicate Affymetrix gene chips. Relative gene expression (false discovery rate ≤ 0.01; P ≤ 0.01) and expression levels of several Notch target genes were plotted. (B) Notch reporter activity is increased in wild-type compared with sdc3−/− cultured SCs and reduced by DAPT treatment in wild-type but not sdc3−/− SCs (n = 8). (C) The number of Hes1+ cells is reduced in proliferating sdc3−/− SC cultures compared with wild-type cultures. Syndecan-4 (Sdc4) marks all SCs (n = 3). Bars, 10 µm. (D) Ectopic expression of Syndecan-3 (sdc3) in sdc3−/− SCs restores Notch reporter activity to wild-type levels. (E) Syndecan-3 knockdown reduces Notch reporter activity in MM14 cells. (E, inset) Syndecan-3 siRNAs (S3 siRNA) reduce Syndecan-3 mRNA. (F) The extent of Syndecan-3 knockdown reflects the efficiency of transfection and is proportional to the extent of Notch reporter activity reduction. GFP+ MM14 cells transfected with EGFP and Syndecan-3 siRNAs or scrambled siRNAs as indicated were harvested 24 h after transfection. Total mRNA was extracted from unsorted or GFP+ FACS–sorted cells, quantitative PCR (q-PCR) for Syndecan-3 and 18S was performed, and values for Syndecan-3 mRNA, normalized to 18S ribosomal mRNA, were plotted (n = 2). Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. **, P < 0.01; #, P > 0.05.
12 h after injury, expression of Notch target genes is reduced in sdc3−/− SCs compared with wild-type SCs (Fig. 3 A). Similarly, cultured sdc3−/− SCs fail to activate the Notch reporter Hes1-Luc (Fig. 3 B). Furthermore, the basal reporter activity observed in sdc3−/− cells is not affected by treatment with the Notch inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester (DAPT; Fig. 3 B). In agreement with these data, sdc3−/− SCs have reduced levels of Hes1 protein compared with their wild-type counterpart (Fig. 3 C). We obtained similar results when we used a different Notch reporter, CSL-Luc, which suggests that Notch signaling is inhibited in sdc3−/− SCs upstream of Notch target transcription (Fig. S3 A).
Inhibition of Notch signaling due to Syndecan-3 loss is not caused by reduced receptor expression, as Notch1 and Notch2 mRNA levels in cultured sdc3−/− SCs are comparable to levels in wild-type cells (Fig. S3 B). Reduction of Notch signaling in sdc3−/− SCs is directly due to Syndecan-3 loss because ectopic expression of Syndecan-3 in sdc3−/− SCs rescues activation of the Notch reporter CSL-Luc (Fig. 3 D). Similarly, knockdown of Syndecan-3 in the SC line MM14 (Lim and Hauschka, 1984) by transfection with a mix of three different Syndecan-3 siRNAs reduces activation of both Hes1-Luc and CSL-Luc Notch reporters (Fig. 3, E and F).
Molecular events that link Syndecan-3 to Notch
Expression levels of Notch targets are reduced in sdc3−/− SCs activated by induced muscle injury compared with wild-type controls (Fig. 3 A). To identify components of the Notch pathway that may account for the lack of Notch signaling in sdc3−/− SCs, we compared global gene expression in wild-type and sdc3−/− SCs isolated from uninjured TA muscles with SCs isolated from injured TA muscles 12 h after injury using IPA. IPA provided two appealing candidates: (1) Notch ligands (Delta/Jagged), induced in wild-type but not in sdc3−/− SCs upon injury, and (2) the Notch inhibitor Numb, which was induced in sdc3−/− but not in wild-type cells upon injury (Fig. 4 A).
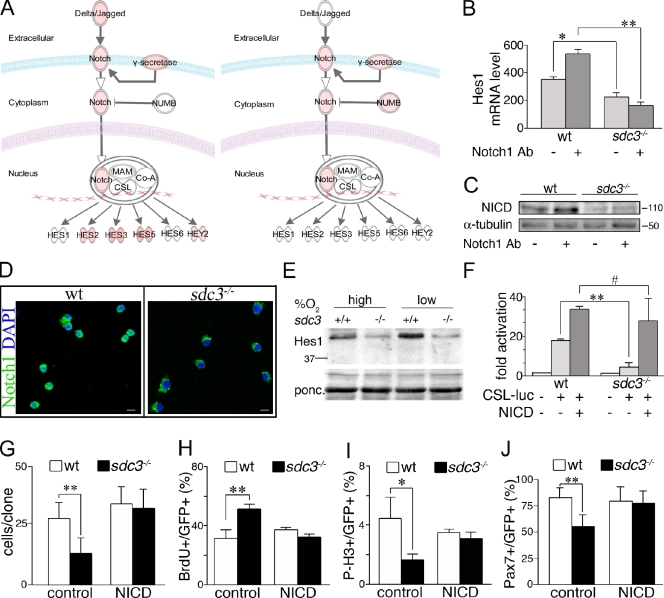
Figure 4.
Syndecan-3 is required for NICD generation. (A) Results from IPA analysis of genes differentially expressed in SCs from 12-h injured TA muscles versus uninjured TA muscles are shown for both wild-type (left) and sdc3−/− (right) mice: the intensity of the red color depicted in the figure is proportional to the relative increase in gene expression in SCs at 12 h after injury versus uninjured. Sdc3−/− SCs induce Numb at a higher extent than wild-type SCs and fail to induce Notch ligands and several Notch target genes in response to injury. (B) Sdc3−/− SCs fail to induce Hes1 mRNA when stimulated with an anti-Notch1 antibody. SCs cultured for 48 h on plates coated with a stimulating anti-Notch1 antibody were assessed by q-PCR for Hes1 mRNA (18S ribosomal RNA was used as a loading control; n = 3). (C) Western blot analysis for NICD of wild-type and sdc3−/− SCs cultured as in B shows that NICD is reduced in sdc3−/− SC extracts compared with wild-type cell extracts, and does not increase in response to Notch1 stimulation. Anti-tubulin is shown as a loading control. (D) Subcellular localization of Notch1 probed with an antibody specific to the intracellular domain of Notch1 reveals nuclear and extranuclear localization in wild-type SCs, but only extranuclear localization in sdc3−/− SCs. Bars, 10 µm. (E) Western blot analysis of Hes1 shows that Hes1 protein levels are reduced in sdc3−/− SC extracts compared with wild-type extracts and are not rescued by culturing the cells under low oxygen atmosphere. Molecular mass standards are indicated next to gel blots (kD). (F) CSL-Luc reporter activity is enhanced in wild-type and rescued in sdc3−/− SCs expressing the NICD (n = 3). (G) Ectopic expression of NICD rescues sdc3−/−-reduced expansion in culture. Wild-type and sdc3−/− SCs transfected with eGFP and either an empty vector (control) or NICD (NICD) were scored for the total number of GFP+ cells/clone 48 h after transfection (20 clones/plate scored, n = 3). (H–J) Ectopic expression of NICD rescues sdc3−/− cell cycle and cell fate phenotypes. Wild-type and sdc3−/− cultured SCs were transfected as in G. 48 h after transfection, they were scored for the number of GFP+BrdU+ cells (H), GFP+P-H3+ cells (I), or GFP+Pax7+ cells (J). Values normalized to the total number of GFP+ cells are plotted (n = 3). Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. **, P < 0.01; *, P < 0.05; #, P > 0.05.
To evaluate SC responsiveness to Notch receptor stimulation, we cultured wild-type and sdc3−/− SCs on tissue culture plates coated with an activating anti-Notch1 antibody that mimics ligand activation of Notch1 (anti-Notch1, clone 8G10; Conboy et al., 2003). Culture of sdc3−/− cells with the activating anti-Notch1 antibody fails to increase expression of either Hes1 (Fig. 4 B) or Hey2 (Fig. S4 A). Moreover, the expression of these two Notch targets is reduced in untreated sdc3−/− SCs compared with untreated wild-type cells, which confirms the Notch reporter activities (Figs. 4 B and S4 A). The reduction in Notch target gene induction was accompanied by impaired NICD accumulation (Fig. 4 C) and nuclear localization (Fig. 4 D) in sdc3−/− versus wild-type cultured SCs. Similar results where obtained when TA muscle sections were analyzed for Notch1 intracellular domain localization in SCs 24 h after injury (Fig. S4 B). Thus, all of our in vitro and in vivo data suggest that reduced NICD is responsible for reduced Notch signaling in sdc3−/− SCs.
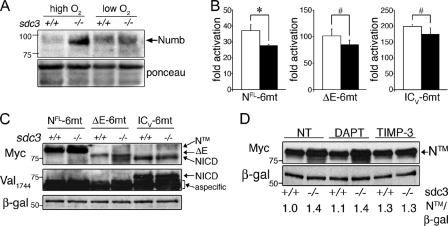
Decreased levels of NICD in sdc3−/− cells could be caused by impaired signal transduction or by a reduction in NICD stability. Nuclear NICD is a very short-lived protein (Chiba, 2006). Cofactors of the Mastermind family (MAML) and the hypoxia responding factor 1α (HIF1-α) directly interact with the CSL-NICD transcription activation complex and regulate NICD turnover (Keith and Simon, 2007; McElhinny et al., 2008). Culturing cells in a low oxygen atmosphere activates HIF1-α and stabilizes nuclear NICD, enhancing Notch signaling in primary SCs and myoblast cell lines (Gustafsson et al., 2005). Enhanced Notch signaling prevents differentiation and promotes expansion of undifferentiated myoblasts (Gustafsson et al., 2005; Keith and Simon, 2007). If the impaired NICD accumulation observed in sdc3−/− SCs is caused by reduced NICD stability, we predict that culturing under low oxygen will rescue Notch signaling in sdc3−/− cells to wild-type levels. Although capable of increasing Hes1 protein levels and Notch reporter activation in wild-type cells, culturing sdc3−/− cells in low oxygen failed to rescue Hes1 protein levels (Fig. 4 E) and Notch reporter activation (Fig. S4 C). Thus, we conclude that impaired NICD generation rather than reduced NICD stability is likely the primary cause of impaired Notch signaling in sdc3−/− SCs. Indeed, ectopic expression of a wild-type NICD containing the PEST domain that is responsible for its short half-life completely rescues Notch reporter activation in sdc3−/− SCs (Fig. 4 F). Furthermore, NICD ectopic expression in sdc3−/− SCs also rescued reduced myoblast expansion (Fig. 4 G), impaired cell cycle progression (Fig. 4, H and I), and rescued SC self-renewal (Fig. 4 J). Culture in low oxygen atmosphere only partially rescued sdc3−/− myoblast expansion (Fig. S4 D) and did not rescue the impaired cell cycle progression or self-renewal (not depicted). These results demonstrate that reduced NICD stability is not responsible for the reduced Notch signaling observed in sdc3−/− SCs but instead suggest a role for Syndecan-3 in NICD generation.
Syndecan-3 regulates Notch signal transduction
A defect in NICD generation may be caused by defective Notch receptor activation and processing or may be caused by inhibition of Notch signal transduction (Kopan et al., 1996; Logeat et al., 1998; Schroeter et al., 1998; Brou et al., 2000; Mumm et al., 2000). An analysis of gene expression data by IPA suggests that increased Numb expression is a possible candidate for inhibition of Notch signal transduction (Fig. 4 A). Cultured sdc3−/− SCs show dramatically increased levels of Numb compared with wild-type cells, which confirms our microarray data (Fig. 5 A). However, high levels of Numb appear to affect sdc3−/− cells in a Notch-independent manner, as increased Numb levels but not impaired Notch signaling are reversed by low oxygen (compare Fig. 5 A with Fig. 4 E). Thus, increased Numb levels cannot account for the loss of Notch target gene induction in sdc3−/− cells, and this finding supports a hypothesis involving Syndecan-3 in regulating Notch receptor processing after ligand binding.
Figure 5.
Syndecan-3 is involved in Notch processing by ADAM17/TACE. (A) Numb protein levels are increased in sdc3−/− SCs analyzed by Western blotting and rescued to wild-type levels by exposure to low oxygen. Ponceau staining is shown as a loading control. (B) Explanted wild-type and sdc3−/− SCs were transfected with NFL-6mt, ΔE-6mt, or ICV-6mt in addition to either empty vector or CSL-Luc and CMV-LacZ as a transfection efficiency control. Reporter activity analyzed 24 h after transfection revealed that ΔE-6mt and ICV-6mt, but not NFL-6mt, ectopic expression rescues Notch reporter activity in sdc3−/− SCs to wild-type levels. (C) Wild-type and sdc3−/− SCs were isolated, cultured, and transfected as in B. 24 h later, they were analyzed by Western blotting to detect the myc-tag epitope. Ectopic Notch1 accumulates in sdc3−/− cells transfected with NFL-6mt (NTM), but not in sdc3−/− cells transfected with either ΔE-6mt or ICV-6mt (n = 3). The same blot was stripped and reprobed to detect NICD generated by γ-secretase cleavage. Although NICD was detected in samples transfected with ICV-6mt, no NICD was detected upon NFL-6mt or ΔE-6mt ectopic expression (n = 2). β-galactosidase is shown as a loading control. (D) Explanted wild-type and sdc3−/− SCs transfected with NFL-6mt were treated as indicated 24 h after transfection. Western blots of cell lysates were probed for the myc-tag epitope and β-galactosidase. NTM band intensities were quantified and values were normalized to the corresponding β-galactosidase band intensities plotted, with the normalized value corresponding to wild-type nontreated cells set at 1. Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. *, P < 0.05; #, P > 0.05. Molecular mass standards are indicated next to gel blots (kD).
Upon ligand binding, Notch signaling is initiated by two proteolytic cleavages of Notch (Fig. S5 A). The first cleavage at the S2 site occurs in the extracellular domain and is mediated by the matrix metalloproteinase ADAM17/TNF-converting enzyme (TACE). The second cleavage at the S3 site occurs within the plasma membrane and is mediated by the γ-secretase complex (Fig. S5 A). To understand which Notch processing event is affected by loss of Syndecan-3, we used three different deletion mutants of Notch: (1) NFL-6mt, a mutant that signals in a ligand-independent, TACE- and γ-secretase–dependent manner; (2) ΔE-6mt, a mutant that requires only γ-secretase–mediated cleavage to signal; and (3) ICV-6mt, a mutant that is constitutively active (Fig. S5 B). In each of these three mutants, the PEST domain has been replaced by a 6-myc epitope tag, stabilizing the protein and providing detection of the exogenous Notch mutants. When cotransfected with a Notch reporter, ICV-6mt and ΔE-6mt, but not NFL-6mt, rescue Notch signaling in sdc3−/− SCs (Fig. 5 B), which suggests a role for Syndecan-3 in regulating TACE-mediated cleavage of Notch at the S2 site (Fig. S5 A). When wild-type and sdc3−/− SCs transfected with NFL-6mt were analyzed by Western blotting, the transmembrane Notch fragment before TACE cleavage, NTM, accumulated more in sdc3−/− than in wild-type cells, which supports a role for Syndecan-3 in promoting NTM processing by TACE (Fig. 5 C, lanes 1 and 2). In contrast, we found equal accumulation of ΔE and NICD upon ectopic expression of either ΔE-6mt or ICV-6mt (Fig. 5 C, lanes 3–6), which indicates that ΔE-6mt processing by γ-secretase and ICV-6mt stability are not affected by Syndecan-3 loss. The additional band running at a lower molecular weight in the sdc3−/− extracts transfected with ΔE-6mt (Fig. 5 C, lane 4) does not correspond to NICD generated by cleavage of ΔE-6mt, as only ectopically expressed ICV-6mt was detected by the anti-NICD antibody (Val-1744) that specifically recognizes NICD upon γ-secretase–mediated cleavage (Fig. 5 C). These results strongly support our hypothesis involving Syndecan-3 in regulating TACE-mediated cleavage of Notch at the S2 site.
To further investigate the involvement of Syndecan-3 in Notch processing at the S2 site, we transfected wild-type and sdc3−/− SCs with NFL-6mt, and quantified NTM accumulation upon treatment with either a γ-secretase inhibitor (DAPT) or a TACE inhibitor (TIMP-3). TIMP-3 treatment rescued the difference in NTM accumulation between wild-type and sdc3−/− cell extracts (Fig. 5 D, lanes 5 and 6 vs. lanes 1 and 2), whereas DAPT treatment was ineffective at altering the levels of either wild-type or sdc3−/− NTM (Fig. 5 D, lanes 3 and 4 vs. lanes 1 and 2). Furthermore, we found that ΔE-6mt ectopic expression yielded equivalent ΔE band intensities in both nontreated and DAPT-treated wild-type and sdc3−/− cells (Fig. S5 C). These data further support our hypothesis for the involvement of Syndecan-3 in Notch processing by ADAM17/TACE and rule out a role for Syndecan-3 in γ-secretase–mediated cleavage.
Syndecan-3 and Notch are present in a complex on the SC plasma membrane
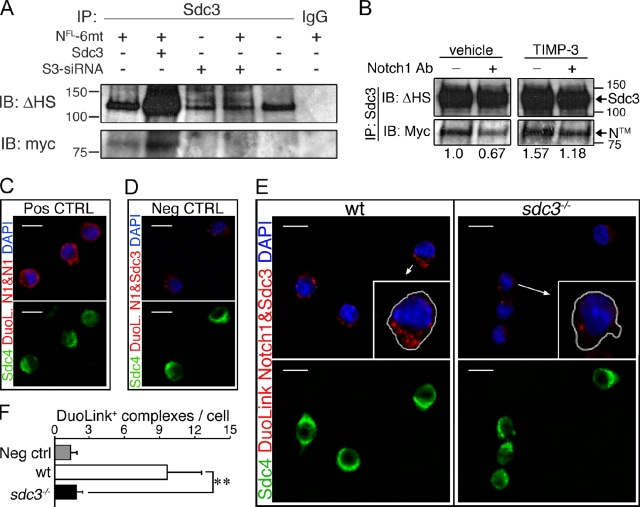
A requirement for Syndecan-3 in Notch processing suggests that Syndecan-3 and Notch may interact. Notch1 (NFL-6mt) ectopically expressed in MM14 cells coimmunoprecipitates with Syndecan-3 (Fig. 6 A, lane 1), which suggests that Syndecan-3 and Notch may physically interact. The disperse Syndecan-3 protein band (Fig. 6 A) is indicative of the incomplete GAG chain removal by heparitinase digestion required before gel electrophoresis (see Materials and methods). Increasing the stringency of washes after Syndecan-3 immunoprecipitation did not affect the ratio of Notch/Syndecan-3 coimmunoprecipitated (unpublished data), which indicates that the interaction between Syndecan-3 and Notch is specific. The amount of coimmunoprecipitated Notch1 increases when Syndecan-3 is overexpressed (Fig. 6 A, lane 2), and is undetectable when cells are cotransfected with Syndecan-3 siRNA (Fig. 6 A, lane 4). Our coimmunoprecipitation results strongly support a hypothesis for a specific Syndecan-3/Notch interaction. If Syndecan-3 and Notch coexist in the same molecular complex where Syndecan-3 facilitates Notch processing by TACE, we expect to find an increased abundance of Notch in immunocomplexes precipitated by an anti–Syndecan-3 antibody when Notch processing is inhibited. We also expect reduced Notch abundance in immunoprecipitated complexes when Notch processing is promoted. As expected, the abundance of Notch in anti–Syndecan-3 immunoprecipitates decreases when Notch processing is stimulated by an anti-Notch1–activating Ab, and increases when Notch processing is inhibited by TIMP-3 (Fig. 6 B). We were unsuccessful at attempts to coimmunoprecipitate endogenous Notch1 and Syndecan-3 in SCs using combinations of five different antibodies (unpublished data). However, using the DuoLink technology, an in situ proximity ligation assay (PLA) that detects two proteins only when they are in close proximity, we were able to demonstrate that Notch1 and Syndecan-3 are present in a complex on the SC plasma membrane. A positive control using two anti-Notch1 antibodies that recognize distinct epitopes was visualized as DuoLink complexes (red) in Syndecan-4+ cells (Fig. 6 C), whereas elimination of primary anti-Notch and anti–Syndecan-3 antibodies revealed few or no complexes (Fig. 6 D). If wild-type cells are processed for DuoLink using an extracellular anti-Notch1 antibody and an intracellular anti–Syndecan-3 antibody, abundant complexes are present (Fig. 6 E, left) that are absent in sdc3−/− cells (Fig. 6 E, right). The number of DuoLink reaction products was quantified, demonstrating the presence of abundant Notch1–Syndecan-3 complexes only in wild-type cells (Fig. 6 F). These data support the hypothesis that Syndecan-3 and Notch interact in a molecular complex involved in Notch processing.
Figure 6.
Syndecan-3 and Notch are in a complex on the SC membrane. (A) Syndecan-3 and Notch1 (NFL-6mt) were coimmunoprecipitated from MM14 cell extracts with an anti–Syndecan-3 antibody 24 h after transfection, and Western blots were probed for the myc-tag epitope and for Syndecan-3 using a terminal heparan sulfate attachment antibody (Δ-HS). (B) Extracts from MM14 cells cultured in the presence or absence of an anti-Notch1 antibody, transfected as in E, and treated with or without 200 nM TIMP-3 were immunoprecipitated with an anti–Syndecan-3 antibody. Blots probed for the myc-tag epitope, then stripped and reprobed for Δ-HS. The relative levels of NTM were quantified and plotted with the level of NTM in the untreated sample set to 1. Molecular mass standards are indicated next to gel blots (kD). (C–F) Endogenous Notch and Syndecan-3 are present in a protein complex on the SC surface, as revealed using DuoLink with an extracellular anti-Notch1 antibody and an intracellular Syndecan-3 antibody. Two distinct Notch1 antibodies from different species show as positive complexes in Syndecan-4 (sdc4)+ cells (green) after DuoLink labeling (C, red), whereas elimination of the primary anti-Notch1 and anti–Syndecan-3 antibodies yielded no positive complexes (D). Endogenous Notch1 and Syndecan-3 appear as complexes (red dots) in wild-type Syndecan-4+ cells (E) but are absent or at background levels in sdc3−/− cells (E). Arrows indicate which cells are magnified in the inset panels. (F) Quantification of C–E, where the mean number of DuoLink+ complexes/cell is plotted. n = 3. Open bars, wild type; shaded bars, sdc3−/−. Error bars indicate SEM. **, P < 0.01; #, P > 0.05. Bars, 10 µm.
Discussion
HSPGs play critical roles in regulating growth factor signaling pathways via a variety of mechanisms, including coreceptor functions, ligand sequestration, morphogenetic boundary regulation, and stem cell fate determination (Rapraeger et al., 1991; Lander, 1998; Muñoz et al., 2006; Dombrowski et al., 2009). Syndecan-3, a transmembrane HSPG expressed in adult SCs, has been previously described as playing a role in adult myogenesis (Fuentealba et al., 1999; Cornelison et al., 2001; Cornelison et al., 2004), but the mechanisms involved remain poorly understood. An in-depth characterization of sdc3−/− phenotypes in vitro and in vivo, combined with an unbiased analysis of gene expression and signaling, have allowed us to further explore the mechanisms involved in Syndecan-3–mediated regulation of adult myogenesis.
To identify signaling pathways contributing to the sdc3−/− phenotype, we performed a global gene expression analysis comparing wild-type and sdc3−/− SCs in uninjured muscle and 12 h after injury. IPA analysis of the gene expression data revealed a failure to induce Notch targets in sdc3−/− cells upon muscle injury and an increase in Numb expression. The increase in Numb expression could potentially account for the loss of Notch signaling, as Numb targets Notch for degradation by the proteasome (McGill and McGlade, 2003). In cultures under low oxygen atmosphere, we observed reduced Numb protein in sdc3−/− SCs but no rescue of Notch signaling, which suggests that increased Numb levels are unlikely to be responsible for impaired Notch signaling in sdc3−/− SCs. Instead Numb may function in SCs in a Notch-independent manner, similar to the role recently reported for Numb in embryonic myogenesis (Jory et al., 2009).
Proteoglycans are involved in binding and signaling for at least 200 secreted factors, often interacting with the proteoglycan GAG chains (Bass et al., 2009). Despite a prior publication postulating a role for a heparan sulfate 3-O-sulfotransferase in Drosophila Notch signaling (Kamimura et al., 2004), a requirement for proteoglycans in Notch signaling has not been investigated. We demonstrated an interaction between Syndecan-3 and Notch1 with a PLA in SCs, and by dose-dependent coimmunoprecipitation of both proteins that is further supported by the observation that the amount of Notch coimmunoprecipitated with Syndecan-3 varies when Notch processing is inhibited or promoted. Syndecan-3 appears to facilitate TACE-mediated cleavage of Notch, a key step in Notch signaling transduction upon ligand binding. Because the molecular mechanisms responsible for heparan sulfate–mediated regulation of signaling pathways often involves direct interaction of the HSPG GAG chains with either growth factors or their cognate receptors (Bass et al., 2009), we asked if the glycosaminoglycan chains, the core protein of Syndecan-3, or both were involved in regulating Notch signaling. Elimination of sulfation by chlorate treatment reduces Notch reporter activity, which supports the involvement of HSPG GAG chains; however, Notch reporter activity in sdc3−/− cells cannot be rescued by exogenous heparin (unpublished data). Our data demonstrate an important role for Syndecan-3 in regulating SC fate that is in part mediated via Notch signaling, where both the GAG chains and the core protein appear to play key roles.
Syndecan-3 and Notch are components of the SC niche. The finding that Syndecan-3 promotes Notch processing and signaling represents a further step toward understanding the mechanisms that regulate SC niche function. In this scenario, the Syndecan-3–Notch interaction seems to play important roles in the regulation of SC proliferation, commitment to terminal differentiation, and self-renewal. We tested this possibility and found that Syndecan-3 regulates the balance between SC proliferation, differentiation, and self-renewal. In the absence of Syndecan-3, SCs progress more slowly through S phase, leading to reduced progenitor cell expansion, delayed onset of differentiation, and increased cell death in addition to reduced self-renewal. Because constitutive activation of Notch signaling in sdc3−/− SCs rescues the altered cell cycle progression and impaired self-renewal, we postulate that Syndecan-3 and Notch cooperate in regulating cell cycle progression and cell fate determination of SCs.
Little is known to date regarding the possible mechanisms whereby Notch directly regulates SC proliferation (Gustafsson et al., 2005; Carlson et al., 2008). Notch signaling is known to inhibit myogenic differentiation, and, thus has been proposed to indirectly promote expansion of undifferentiated progenitors (Luo et al., 2005). However, recent work describes a role for Notch signaling in antagonizing TGF-β–dependent activation of the cyclin-dependent kinase inhibitors including p15, p16, p21, and p27 (Carlson et al., 2008). Because p21 and p15 play a key role in regulating intra–S phase checkpoints in a TGF-β–dependent manner in other progenitor cells (Zhu et al., 2004; Jirmanova et al., 2005; Kan et al., 2007; Petrov et al., 2008), it is possible that p15 and p21 regulate intra–S phase checkpoints in SCs as well. We believe that inhibition of Notch signaling in SCs, either because of an aged environment (Conboy et al., 2003; Carlson et al., 2008) or Syndecan-3 loss, results in increased expression of intra–S phase checkpoint inhibitors (including p15 and p21), leading to delayed progression through S-phase, increased cell doubling times, reduced cell survival, and ultimately reduced SC proliferation (Fig. 7; Carlson et al., 2008). Our results expand on previous studies, providing additional mechanistic insights into roles for Notch signaling in regulating SC proliferation (Fig. 7). Furthermore, our data provide evidence that regulation of the cell cycle directly impacts regulation of SC fate decisions, including commitment to terminal differentiation and cell death. Both the increased cell death and the delayed differentiation onset we observed are likely to be a direct consequence of aberrant progression through intra–S phase checkpoints, leading to cell cycle arrest and death, or delaying cell cycle exit and therefore the onset of terminal differentiation (Fig. 7; Carlson et al., 2008).
Figure 7.
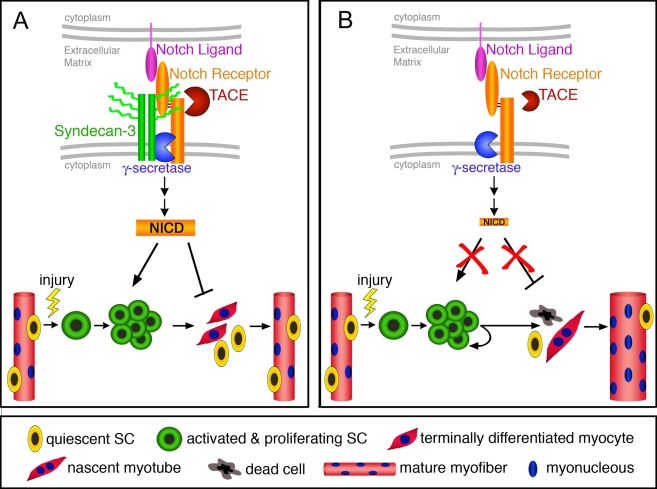
A model predicting SC behavior in sdc3−/− muscles. (A) In wild-type SCs, Syndecan-3 and Notch reside in a complex where Syndecan-3 facilitates Notch processing and NICD generation. Quiescent SCs (yellow cells) are activated and commit to myogenesis (green cells), where cooperation between Syndecan-3 and Notch promotes cell cycle progression and self-renewal, and inhibits terminal differentiation (red cells) and fusion to existing myofibers. Once muscle repair is completed, SCs cease proliferation and undifferentiated cells return to a quiescent state. Repaired myofibers appear indistinguishable from uninjured myofibers. (B) In sdc3−/− SCs, Notch processing is impaired and NICD generation is dramatically reduced. Once sdc3−/− SCs are activated and commit to myogenesis, the lack of Syndecan-3 and Notch signaling impairs cell cycle progression, delays onset of terminal differentiation, and reduces SC self-renewal. Activated sdc3−/− SCs fail to return to quiescence but remain in an activated state that leads to myofiber hypertrophy. An increase in cell death and the lengthening of cell cycle progression contribute in maintaining constant the total number of nuclei per tissue area. However, the balance between terminal differentiation and self-renewal is disrupted in sdc3−/− regenerating muscles, whereby more nuclei are found in hypertrophic myofibers and less in mononucleated sublaminar Pax7+ cells (SCs). Notch-independent Syndecan-3-dependent signaling pathways are not depicted in this diagram.
A reduction in Notch signaling in aged mice leads to aberrant SC function and severely affects muscle regeneration 5 d after muscle injury, but the effects of a reduction or loss of Notch signaling on later stages of muscle regeneration have not been tested (Conboy et al., 2003; Brack et al., 2008; Smythe et al., 2008; Carlson et al., 2009). Therefore we investigated muscle regeneration in sdc3−/− mice at 2 wk, 6 wk, and 3 mo after injury. Despite the impaired SC function we observed at early stages of muscle regeneration, which was similar to that observed in old injured muscles (Carlson et al., 2008), muscle regeneration in sdc3−/− mice was not impaired 3 mo after injury. Instead, as predicted from the cell cycle and cell fate phenotypes of cultured sdc3−/− SCs, regenerated sdc3−/− muscles display phenotypes consistent with a severe imbalance in cell fate decisions. Sdc3−/− cells prefer to differentiate as opposed to self-renew, creating an imbalance that gives rise to abnormally large myofibers accompanied by a reduction in the numbers of Pax7+ SCs. Although SC death is increased and self-renewal reduced in sdc3−/− injured muscles compared with wild-type controls, the total number of nuclei per tissue area and the total muscle weight are indistinguishable in sdc3−/− and wild-type injured TA muscles (unpublished data). These data are consistent with the data for cultured sdc3−/− SCs, where we predict that sdc3−/− SCs activated by an injury in vivo would maintain their proliferation state longer than wild-type cells due to their impaired progression through the cell cycle. In further support of this idea is the observation that myofiber hypertrophy in sdc3−/− regenerating muscles increases gradually over time when comparing sdc3−/− TA muscles to wild-type muscles. These sdc3−/− phenotypes appear to be largely Notch-dependent because ectopic expression of the constitutively active NICD rescues both sdc3−/− SC progression through the cell cycle and self-renewal in culture. However we cannot completely rule out the possibility of Syndecan-3 contributions to other signaling pathways.
Genetic ablation of Syndecan-3, a component of the cell glycocalix, was expected to affect several signaling pathways because the glycocalix is complex and integrates environmental signals such that changes or loss of one component affects several other components, magnifying downstream effects (Carey, 1996; Rapraeger, 2000; Tkachenko et al., 2005). Indeed, we previously demonstrated that FGF and HGF signaling in sdc3−/− cells was enhanced compared with wild-type SCs (Cornelison et al., 2004). Ongoing work will further address the mechanisms involved in Syndecan-3 interactions with Notch and will clarify to what extent other signaling pathways are affected when this important component of the SC niche is lost.
Materials and methods
Mice
Mice were housed in a pathogen-free facility, and all procedures and protocols were approved by the Institutional Animal Care and Use Committee at the University of Colorado. Control mice were littermate wild-type mice when possible or age- and sex-matched B6D2F1 (The Jackson Laboratory). Sdc3−/− mice were donated by H. Rauvala (University of Helsinki, Helsinki, Finland).
Cell cultures
Primary SCs or myofibers were obtained as described previously from 3–6-mo-old female mice (Cornelison et al., 2004). For clonal cultures, ∼500 cells/cm2 were plated in growth medium (GM; F12-C + 15% horse serum + 20 ng/ml FGF-2). MM14 myoblasts were plated at 100–500 cells/cm2 and cultured in GM at 37°C and 5% CO2. When required, 5 µM BrdU (Sigma Aldrich) was added for 30 min or for 4 h before fixation. For double label experiments, either CldU (United States Biological) or IdU (Sigma-Aldrich) were added at 6.5 µM 30 min before washing for further chase or fixation. Myogenic differentiation was induced by switching the growth medium to differentiation medium (F12-C + 2% horse serum).
Affymetrix arrays and analysis
Cells were isolated from mouse hindlimb muscle and sorted for Syndecan-4 expression as described previously (Tanaka et al., 2009). A minimum of at least three independent age-matched animals were analyzed per time point. Total RNA was isolated using a PicoPure RNA Isolation kit (Arcturus) followed by two rounds of linear T7-based amplification (RiboAmp HA kit; Arcturus) for an RNA equivalent of 5,000 dual-positive cells (based on the number of cells collected) per Affymetrix chip. Biotin-labeled antisense RNA was generated with an IVT Labeling kit (Affymetrix), and labeled cRNA was quantified and analyzed for size representation using a BioAnalyzer (Agilent Technologies). 5 mg of labeled cRNA was fragmented and hybridized to mouse microarrays (430 v. 2; Affymetrix) at the University of Colorado Core facilities; chips were scanned on a GeneChip Scanner 3000 (Affymetrix), and intensity data were recovered in GeneChip Operating Software (GCOS; Affymetrix). CEL files from three biological replicates on three gene chips were imported directly into Spotfire (Invitrogen) and normalized by GeneChip robust multiarray averaging. Analysis of variance (ANOVA) analysis set at a false discovery rate of 0.05% was used to identify differentially expressed probe sets. Probe sets with ≥1.5-fold changes between uninjured and 12-h injured in wild-ype and sdc3−/− datasets were further analyzed using IPA. Signaling pathways were analyzed first for pathways that exhibited significant changes between wild-type SCs from uninjured and 12-h injured muscles. These differences were then compared with signaling pathways in the uninjured versus 12-h injured dataset from sdc3−/− SCs. These data were plotted for the Notch pathway with the intensity of red signifying the fold change in gene expression.
Reagents
DAPT was obtained from EMD. Cell culture media was obtained from Sigma-Aldrich. Horse serum was obtained from HyClone. Syndecan-3 siRNA mix was obtained from Santa Cruz Biotechnologies, Inc., and RNA-induced silencing complex (RISC)-free control siRNA was obtained from Thermo Fisher Scientific. Recombinant tissue inhibitor of metalloproteinases-3 (TIMP-3) purified from transfected BHK cells was obtained from Abcam.
Cell treatments
For DAPT treatment, cells were incubated for 6–8 h with 10 µM DAPT or DMSO (untreated control) before harvesting. For TIMP-3 treatment, cells were detached using collagenase, resuspended in either GM alone or GM + TIMP-3 at 200 nM, and incubated for 40 min at 37°C before harvest.
Plasmids, transfections, and reporter assays
Hes1-Luc and CSL-Luc constructs were provided by N. Ahn (University of Colorado at Boulder, Boulder, CO) and used at 0.5 µg/ml. NICD, NFL-6mt, ΔE-6mt, and ICV-6mt constructs were provided by R. Kopan (Washington University, St. Louis, MO) and used at 50 ng/ml, except for NFL-6mt, which was used at 100 ng/ml. The Syndecan-3 (sdc3) expression vector was provided by A. Rapraeger (University of Wisconsin, Madison, WI) and used at 300 ng/ml. For reporter assays, primary SCs and MM14 myoblasts were cultured on gelatin-coated 12-well plates (Corning) and transfected using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Cells were lysed 24 h after transfection. A total of 1 µg/ml of DNA was transfected using an empty pGL2basic plasmid as a carrier when required. The CMV-LacZ expression vector (0.025 µg/ml) was cotransfected to normalize for transfection efficiency. For Syndecan-3 knockdown experiments, a mix of three different Syndecan-3 siRNAs (Santa Cruz Biotechnologies, Inc.) was transfected at 200 nM in MM14 cells using Lipofectamine 2000 transfection reagent (Invitrogen). RISC-free siRNA were used as controls (Thermo Fisher Scientific). Luciferase and β-galactosidase activities were determined as described previously (Olguin et al., 2007). For cell cycle and self-renewal rescue, primary SCs were transfected using Lipofectamine 2000 with a pEGFP expression vector (0.025 µg/ml) ± NICD; 48 h after transfection, coverslips were fixed and immunostained.
Immunofluorescence
Tissue samples, primary SCs, and MM14 cells were prepared as described previously (Cornelison et al., 2004; Olguin et al., 2007). For detection of labeled DNA, cells were fixed in methanol for 10 min at −20°C, washed in PBS, permeabilized in PBS + 0.2% Triton-X-100 + 1% BSA for 10 min, and denatured in 3M HCl for 20 min before incubation with primary antibodies overnight at 4°C in PBS + 10% horse serum. Primary antibodies used for immunofluorescence were: chicken polyclonal anti–Syndecan-4 at 1:1,000, rabbit polyclonal anti-Hes1 (Millipore) at 1:100, rabbit polyclonal anti-Notch1 cytoplasmic domain (Millipore) at 1:200, mouse monoclonal anti-Pax7 (Developmental Studies Hybridoma Bank at Iowa University) at 1:5, rabbit polyclonal anti-Pax7 (GeneTex, Inc.) at 1:250, rat polyclonal anti-BrdU (AbD Serotec) at 1:100, rabbit polyclonal anti-MyoD (Santa Cruz Biotechnology, Inc.) at 1:30, rabbit polyclonal anti-myogenin (Santa Cruz Biotechnology, Inc.) at 1:30, rabbit polyclonal anti–P-H3 (Millipore) at 1:100, mouse clone B44 recognizing IdU (and also BrdU, BD) at 1:400, and rat antibody clone BU1/75 (ICR1) recognizing CldU (and also BrdU; Novus Biologicals) at 1:200. Secondary antibodies conjugated with Alexa Fluor 594, Alexa Fluor 488, or Alexa Fluor 647 (Invitrogen) were used at 1:500 dilution. Vectashield with DAPI (Vector Laboratories) was used the mounting medium.
Primers
For Hes1 detection: forward, 5′-GACAAACCAAAGACGGCCTCT-3′; and backward, 5′-CCATGATAGGCTTTGATGACTTTCT-3′. For Hey2 detection: forward, 5′-GGACGAGACCATCGACGTG-3′; and backward, 5′-TTGTAGCGTGCCCAGGGTA-3′. For Notch1 detection: forward, 5′-CTGCCTCTTTGATGGCTTCGA-3′; and backward, 5′-ATCCCACTCACATTCGGCACT-3′. For Notch2 detection: forward, 5′-TTGCAGTGTCGAGGTGGTCAA-3′; and backward, 5′-ACCACCATTCTGACAGCGGTT-3′.
Immunoprecipitation
MM14 cells were lysed on plates with lysis buffer containing 1% Triton X-100 + 0.1% SDS + 5 mM EDTA supplemented with a protease inhibitor cocktail (Complete; Roche) for 15 min on ice. They were then collected, incubated 15 min on ice, and centrifuged for 10 min at 13,000 rpm, and supernatants were collected. For immunoprecipitation, samples were first cleared by incubating for 10 min at room temperature with protein G slurry (Thermo Fisher Scientific). Supernatants were collected and incubated overnight at 4°C with anti–Syndecan-3 antibody (Cornelison et al., 2004), 100 µl of protein G slurry was added and incubated for 2 h at room temperature, and pellets were processed for Western blot analysis.
Western blotting
Whole cell extracts were obtained by incubating cells in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium-deoxycholate, 150 mM NaCl, and 1 mM EDTA) supplemented with protease inhibitor cocktail (Complete; Roche) for 30 min on ice, then cleared by centrifugation at 13,000 rpm for 10 min at 4°C. Total protein (10–30 µg) was separated by 8% SDS-PAGE gels and transferred onto nitrocellulose membranes (Hybond; GE Healthcare) for Western blotting. For Western blot detection of Syndecan-3, cell extracts were treated with 20 CU/ml Heparitinase I (Seikagaku) for 8 h at 37°C followed by boiling with 1:1 volume loading buffer (Bio-Rad Laboratories). Primary antibodies for Western blotting were: rabbit polyclonal anti-Hes1 (Millipore) at 1:1,000, mouse monoclonal anti-myc (Cell Signaling Technology) at 1:1,000, chicken anti–β-gal (Abcam) at 1:3,000, rabbit polyclonal anti-Notch1 cytoplasmic domain (Millipore) at 1:2,000, and rabbit anti-NICD Val-1744 (Cell Signaling Technology) at 1:1,000. Syndecan-3 detection in immunocomplexes was performed using a mouse anti–Δ-HS antibody (Seikagaku) at 1:1,000 directed to the heparan sulfate GAG chain attachment sites, which are exposed upon enzymatic digestion of the heparan sulfate GAG chains. Anti–mouse, anti–rabbit, or anti–chicken HRP-conjugated secondary antibodies (Promega) were used at 1:5,000, and HRP activity was visualized using the ECL plus system (GE Healthcare).
Morphometric analysis
Myofiber cross-sectional area and numberin uninjured and injured TA muscle were quantified via manual masking using SlideBook (Intelligent Imaging Innovations, Inc.) based on laminin and DAPI staining in 20× fields. A minimum of 1,000 myofibers and four sections were analyzed per biological replica.
PLA
PLA (DuoLink) was performed to image protein–protein interactions using microscopy. In brief, oligonucleotide-conjugated secondary antibodies were directed against primary antibodies raised against cell surface receptors. Annealing of the oligonucleotides conjugated to secondary antibodies occurs only when the target proteins are in close proximity and initiates the amplification of a Texas red reporter signal. Cultured wild-type and sdc3−/− SCs were fixed with 4% PFA for 10 min at room temperature, blocked with PBS + 3% BSA for 1 h at room temperature, then incubated overnight at 4°C with an anti-Notch1 antibody (G20, goat polyclonal, see Western blotting section). The next day, samples were postfixed with 2% PFA for 5 min at room temperature, washed, permeabilized with 0.2% Triton X-100 for 10 min at room temperature followed by a blocking step with PBS + 3% BSA, then incubated with an anti–Syndecan-3 antibody to the intracellular domain of Syndecan-3 (rabbit polyclonal, see Western blotting section) for 2 h at room temperature. Secondary antibodies conjugated with oligonucleotides (anti-rabbit PLA probe PLUS and anti-goat PLA probe MINUS) were added to the reaction and incubated for 2 h at 37°C followed by incubation with a hybridization solution containing two oligonucleotides that hybridize to the two PLA probes only if they are in close proximity. After a ligation solution was added together with ligase to join the two hybridized oligonucleotides to a closed circle, samples were subjected to several cycles of rolling-circle amplification (RCA) using the ligated circle as a template and generating a concatemeric product extending from the oligonucleotide arm of the PLA probe. Lastly, a detection solution consisting of fluorescently labeled oligonucleotides was added, and the labeled oligonucleotides were hybridized to the RCA product. The signal was visible as a distinct fluorescent dot in the Texas red channel and analyzed by fluorescence microscopy. Hoechst is included in the detection solution. Negative controls consisted of wild-type samples treated as described except for primary antibodies. Positive controls consisted of wild-type samples where Notch1 was detected by two distinct primary antibodies produced in two different animal species. Anti-Notch1 G20 was produced in a goat then detected by anti–goat PLA probe MINUS and anti-Notch1 8G10 produced in a Syrian hamster, then detected by biotin-conjugated anti–Syrian hamster followed by anti-biotin PLA probe PLUS.
Microscopy, image processing, and figure preparation
Micrographs were taken with a confocal microscope (TCS SP2 AOBS; Leica) using dedicated Leica software, or with an epifluorescence microscope (Eclipse E800; Nikon) using Slidebook version 4.1 acquisition software (Intelligent Imaging Innovations Inc.) coupled to a digital camera (Sensicam; Cooke Optics). Lenses used with the confocal microscope were either HC Plan-Apochromat 20×/0.70 IMM CORR CS or HCX Plan-Apochromat 40×/1.25–0.75 NA. Lenses used with the epifluorescence microscope were either 40×/0.75 differential interference contrast M or 20×/0.50 Ph1 DLL Plan Fluor lenses (Nikon). All digital microscopic images were acquired at room temperature. The mounting medium for cells and sections used for imaging was Vectashield Mounting Medium (Vector Laboratories). The fluorochromes used were Alexa Fluor 488, Alexa Fluor 594, and Alexa Fluor 647 conjugated to secondary antibodies for immunofluorescence, as indicated in the Immunofluorescence section. Texas red was incorporated by polymerase reaction into DuoLink+ complexes, as indicated in the PLA section. For figure preparation, images were exported in Photoshop (Adobe). If necessary, the brightness and contrast were adjusted for the entire image, the image was cropped, and individual color channels were extracted (when required) without color correction or gamma adjustments.
Online supplemental material
Fig. S1 shows a schematic of the CldU-IdU pulse-chase experiment, impaired cell cycle progression, and delayed differentiation onset in sdc3−/− SCs compared with wild type SCs. Fig. S2 shows reduced Pax7+ SCs in sdc3−/− muscles compared with wild-type muscles 2 wk and 6 wk after injury. Fig. S3 shows reduced Notch signaling in sdc3−/− SCs compared with wild-type SCs despite similar expression levels of Notch receptors. Fig. S4 shows reduced Notch signaling in sdc3−/− injured muscles, that cultured sdc3−/− SCs do not respond to Notch stimulation, and that reduced Notch signaling and clonal expansion in sdc3−/− SCs are not rescued by low-oxygen culture conditions. Fig. S5 shows a schematic of Notch signaling transduction and a schematic of the Notch mutants used, and also shows that ΔE-6mt ectopic expression produces similar results in wild-type and sdc3−/− SCs. Table S1 shows differential gene expression values from Affymetrix chips for wild-type and sdc3−/− SCs isolated 12 h after muscle injury. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003081/DC1.
Acknowledgments
We thank Dr. Nathalie Ahn for the Notch reporter constructs, Dr. Raphael Kopan for the Notch mutant constructs, Dr. Alan Rapraeger for the anti–Syndecan-3 antibody and the Syndecan-3 expression vector, and Dr. Heikki Rauvala for the sdc3−/− mice. We thank Dr. Adam Cadwallader for optimizing protocols for the DuoLink Proximity Ligation Assay in SCs. We also thank Dr. Michelle Doyle and all members of the Olwin laboratory for critical reading of the manuscript and discussion.
This work was supported by National Institutes of Health grants AR39467 and AR049446.
Footnotes
Abbreviations used in this paper:
- CldU
- 5-chloro-2′-deoxyuridine
- CSL
- CBF1/suppressor of hairless/Lag-1
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester
- HSPG
- heparan sulfate proteoglycan
- IdU
- 5-iodo-2′-deoxyuridine
- IPA
- Ingenuity Pathway Analysis
- NICD
- Notch intracellular domain
- P-H3
- phospho–histone H3
- PLA
- proximity ligation assay
- q-PCR
- quantitative PCR
- SC
- satellite cell
- TA
- tibialis anterior
- TACE
- TNF-converting enzyme
References
- Bass M.D., Morgan M.R., Humphries M.J. 2009. Syndecans shed their reputation as inert molecules. Sci. Signal. 2:pe18 10.1126/scisignal.264pe18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U., Karlsson G., Karlsson S. 2008. Signaling pathways governing stem-cell fate. Blood. 111:492–503 10.1182/blood-2007-07-075168 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Horsley V., Fuchs E. 2007. Epithelial stem cells: turning over new leaves. Cell. 128:445–458 10.1016/j.cell.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen K.J., Post M.J. 2008. The muscle stem cell niche: regulation of satellite cells during regeneration. Tissue Eng. Part B Rev. 14:419–431 10.1089/ten.teb.2008.0045 [DOI] [PubMed] [Google Scholar]
- Brack A.S., Conboy I.M., Conboy M.J., Shen J., Rando T.A. 2008. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2:50–59 10.1016/j.stem.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J.R., Cumano A., Roux P., Black R.A., Israël A. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell. 5:207–216 10.1016/S1097-2765(00)80417-7 [DOI] [PubMed] [Google Scholar]
- Carey D.J. 1996. N-syndecan: structure and function of a transmembrane heparan sulfate proteoglycan. Perspect. Dev. Neurobiol. 3:331–346 [PubMed] [Google Scholar]
- Carlson M.E., Hsu M., Conboy I.M. 2008. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 454:528–532 10.1038/nature07034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M.E., Conboy M.J., Hsu M., Barchas L., Jeong J., Agrawal A., Mikels A.J., Agrawal S., Schaffer D.V., Conboy I.M. 2009. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 8:676–689 10.1111/j.1474-9726.2009.00517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S. 2006. Notch signaling in stem cell systems. Stem Cells. 24:2437–2447 10.1634/stemcells.2005-0661 [DOI] [PubMed] [Google Scholar]
- Collins C.A., Olsen I., Zammit P.S., Heslop L., Petrie A., Partridge T.A., Morgan J.E. 2005. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 122:289–301 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Rando T.A. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409 10.1016/S1534-5807(02)00254-X [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Conboy M.J., Smythe G.M., Rando T.A. 2003. Notch-mediated restoration of regenerative potential to aged muscle. Science. 302:1575–1577 10.1126/science.1087573 [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Filla M.S., Stanley H.M., Rapraeger A.C., Olwin B.B. 2001. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev. Biol. 239:79–94 10.1006/dbio.2001.0416 [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Wilcox-Adelman S.A., Goetinck P.F., Rauvala H., Rapraeger A.C., Olwin B.B. 2004. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 18:2231–2236 10.1101/gad.1214204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D.E., Mooney D.J., Zandstra P.W. 2009. Growth factors, matrices, and forces combine and control stem cells. Science. 324:1673–1677 10.1126/science.1171643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski C., Song S.J., Chuan P., Lim X., Susanto E., Sawyer A.A., Woodruff M.A., Hutmacher D.W., Nurcombe V., Cool S.M. 2009. Heparan sulfate mediates the proliferation and differentiation of rat mesenchymal stem cells. Stem Cells Dev. 18:661–670 10.1089/scd.2008.0157 [DOI] [PubMed] [Google Scholar]
- D’Souza B., Miyamoto A., Weinmaster G. 2008. The many facets of Notch ligands. Oncogene. 27:5148–5167 10.1038/onc.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiúza U.M., Arias A.M. 2007. Cell and molecular biology of Notch. J. Endocrinol. 194:459–474 10.1677/JOE-07-0242 [DOI] [PubMed] [Google Scholar]
- Fuentealba L., Carey D.J., Brandan E. 1999. Antisense inhibition of syndecan-3 expression during skeletal muscle differentiation accelerates myogenesis through a basic fibroblast growth factor-dependent mechanism. J. Biol. Chem. 274:37876–37884 10.1074/jbc.274.53.37876 [DOI] [PubMed] [Google Scholar]
- Gustafsson M.V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J.L., Poellinger L., Lendahl U., Bondesson M. 2005. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 9:617–628 10.1016/j.devcel.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Holterman C.E., Le Grand F., Kuang S., Seale P., Rudnicki M.A. 2007. Megf10 regulates the progression of the satellite cell myogenic program. J. Cell Biol. 179:911–922 10.1083/jcb.200709083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Kedes L., Hamamori Y. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255 10.1002/jcp.10208 [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E.H., Kopan R., Israel A. 1995. Signalling downstream of activated mammalian Notch. Nature. 377:355–358 10.1038/377355a0 [DOI] [PubMed] [Google Scholar]
- Jirmanova L., Bulavin D.V., Fornace A.J., Jr 2005. Inhibition of the ATR/Chk1 pathway induces a p38-dependent S-phase delay in mouse embryonic stem cells. Cell Cycle. 4:1428–1434 [DOI] [PubMed] [Google Scholar]
- Jory A., Le Roux I., Gayraud-Morel B., Rocheteau P., Cohen-Tannoudji M., Cumano A., Tajbakhsh S. 2009. Numb promotes an increase in skeletal muscle progenitor cells in the embryonic somite. Stem Cells. 27:2769–2780 10.1002/stem.220 [DOI] [PubMed] [Google Scholar]
- Kamimura K., Rhodes J.M., Ueda R., McNeely M., Shukla D., Kimata K., Spear P.G., Shworak N.W., Nakato H. 2004. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J. Cell Biol. 166:1069–1079 10.1083/jcb.200403077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Q., Jinno S., Yamamoto H., Okayama H. 2007. Chemical DNA damage activates p21 WAF1/CIP1-dependent intra-S checkpoint. FEBS Lett. 581:5879–5884 10.1016/j.febslet.2007.11.075 [DOI] [PubMed] [Google Scholar]
- Keith B., Simon M.C. 2007. Hypoxia-inducible factors, stem cells, and cancer. Cell. 129:465–472 10.1016/j.cell.2007.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M., Bonnieu A., Duret C., Vernus B., Barro M., Laoudj-Chenivesse D., Verdi J.M., Carnac G. 2006. Inhibition of Notch signaling induces myotube hypertrophy by recruiting a subpopulation of reserve cells. J. Cell. Physiol. 208:538–548 10.1002/jcp.20688 [DOI] [PubMed] [Google Scholar]
- Kopan R., Nye J.S., Weintraub H. 1994. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 120:2385–2396 [DOI] [PubMed] [Google Scholar]
- Kopan R., Schroeter E.H., Weintraub H., Nye J.S. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA. 93:1683–1688 10.1073/pnas.93.4.1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 129:999–1010 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Gillespie M.A., Rudnicki M.A. 2008. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2:22–31 10.1016/j.stem.2007.12.012 [DOI] [PubMed] [Google Scholar]
- Kuroda K., Tani S., Tamura K., Minoguchi S., Kurooka H., Honjo T. 1999. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274:7238–7244 10.1074/jbc.274.11.7238 [DOI] [PubMed] [Google Scholar]
- Lander A.D. 1998. Proteoglycans: master regulators of molecular encounter? Matrix Biol. 17:465–472 10.1016/S0945-053X(98)90093-2 [DOI] [PubMed] [Google Scholar]
- Lim R.W., Hauschka S.D. 1984. A rapid decrease in epidermal growth factor-binding capacity accompanies the terminal differentiation of mouse myoblasts in vitro. J. Cell Biol. 98:739–747 10.1083/jcb.98.2.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N.G., Israël A. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA. 95:8108–8112 10.1073/pnas.95.14.8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Renault V.M., Rando T.A. 2005. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin. Cell Dev. Biol. 16:612–622 10.1016/j.semcdb.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Maddika S., Ande S.R., Panigrahi S., Paranjothy T., Weglarczyk K., Zuse A., Eshraghi M., Manda K.D., Wiechec E., Los M. 2007. Cell survival, cell death and cell cycle pathways are interconnected: implications for cancer therapy. Drug Resist. Updat. 10:13–29 10.1016/j.drup.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Mauro A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny A.S., Li J.L., Wu L. 2008. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 27:5138–5147 10.1038/onc.2008.228 [DOI] [PubMed] [Google Scholar]
- McGill M.A., McGlade C.J. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196–23203 10.1074/jbc.M302827200 [DOI] [PubMed] [Google Scholar]
- Mumm J.S., Schroeter E.H., Saxena M.T., Griesemer A., Tian X., Pan D.J., Ray W.J., Kopan R. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell. 5:197–206 10.1016/S1097-2765(00)80416-5 [DOI] [PubMed] [Google Scholar]
- Muñoz R., Moreno M., Oliva C., Orbenes C., Larraín J. 2006. Syndecan-4 regulates non-canonical Wnt signalling and is essential for convergent and extension movements in Xenopus embryos. Nat. Cell Biol. 8:492–500 10.1038/ncb1399 [DOI] [PubMed] [Google Scholar]
- Nye J.S., Kopan R., Axel R. 1994. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 120:2421–2430 [DOI] [PubMed] [Google Scholar]
- Olguin H.C., Olwin B.B. 2004. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 275:375–388 10.1016/j.ydbio.2004.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin H.C., Yang Z., Tapscott S.J., Olwin B.B. 2007. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 177:769–779 10.1083/jcb.200608122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V.V., van Pelt J.F., Vermeesch J.R., Van Duppen V.J., Vekemans K., Fagard R.H., Lijnen P.J. 2008. TGF-beta1-induced cardiac myofibroblasts are nonproliferating functional cells carrying DNA damages. Exp. Cell Res. 314:1480–1494 10.1016/j.yexcr.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Rapraeger A.C. 2000. Syndecan-regulated receptor signaling. J. Cell Biol. 149:995–998 10.1083/jcb.149.5.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapraeger A.C., Krufka A., Olwin B.B. 1991. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 252:1705–1708 10.1126/science.1646484 [DOI] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger J.A., Kopan R. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 393:382–386 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- Schultz E., McCormick K.M. 1994. Skeletal muscle satellite cells. Rev. Physiol. Biochem. Pharmacol. 123:213–257 10.1007/BFb0030904 [DOI] [PubMed] [Google Scholar]
- Shimizu K., Chiba S., Saito T., Kumano K., Hamada Y., Hirai H. 2002. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem. Biophys. Res. Commun. 291:775–779 10.1006/bbrc.2002.6528 [DOI] [PubMed] [Google Scholar]
- Shinin V., Gayraud-Morel B., Gomès D., Tajbakhsh S. 2006. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell Biol. 8:677–687 10.1038/ncb1425 [DOI] [PubMed] [Google Scholar]
- Smythe G.M., Shavlakadze T., Roberts P., Davies M.J., McGeachie J.K., Grounds M.D. 2008. Age influences the early events of skeletal muscle regeneration: studies of whole muscle grafts transplanted between young (8 weeks) and old (13-21 months) mice. Exp. Gerontol. 43:550–562 10.1016/j.exger.2008.02.005 [DOI] [PubMed] [Google Scholar]
- Tanaka K.K., Hall J.K., Troy A.A., Cornelison D.D., Majka S.M., Olwin B.B. 2009. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell. 4:217–225 10.1016/j.stem.2009.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko E., Rhodes J.M., Simons M. 2005. Syndecans: new kids on the signaling block. Circ. Res. 96:488–500 10.1161/01.RES.0000159708.71142.c8 [DOI] [PubMed] [Google Scholar]
- Zammit P.S., Golding J.P., Nagata Y., Hudon V., Partridge T.A., Beauchamp J.R. 2004. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166:347–357 10.1083/jcb.200312007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Alvarez C., Doll R., Kurata H., Schebye X.M., Parry D., Lees E. 2004. Intra-S-phase checkpoint activation by direct CDK2 inhibition. Mol. Cell. Biol. 24:6268–6277 10.1128/MCB.24.14.6268-6277.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]