The crystal structure of a Sec13–Sec16 complex reveals its functions in the early steps of COPII coat complex assembly.
Abstract
Ancestral coatomer element 1 (ACE1) proteins assemble latticework coats for COPII vesicles and the nuclear pore complex. The ACE1 protein Sec31 and Sec13 make a 2:2 tetramer that forms the edge element of the COPII outer coat. In this study, we report that the COPII accessory protein Sec16 also contains an ACE1. The 165-kD crystal structure of the central domain of Sec16 in complex with Sec13 was solved at 2.7-Å resolution. Sec16 and Sec13 also make a 2:2 tetramer, another edge element for the COPII system. Domain swapping at the ACE1–ACE1 interface is observed both in the prior structure of Sec13–Sec31 and in Sec13–Sec16. A Sec31 mutant in which domain swapping is prevented adopts an unprecedented laminated structure, solved at 2.8-Å resolution. Our in vivo data suggest that the ACE1 element of Sec31 can functionally replace the ACE1 element of Sec16. Our data support Sec16 as a scaffold for the COPII system and a template for the Sec13–Sec31 coat.
Introduction
The COPII coat complex mediates formation of transport vesicles that bud from the ER and traffic secretory proteins to other organelles (Antonny and Schekman, 2001; Bonifacino and Glick, 2004; Tang et al., 2005; Hughes and Stephens, 2008). COPII consists of an inner coat composed of the Sec23–Sec24 dimer and the small GTPase Sar1 and an outer coat composed of Sec31 and Sec13 (Stagg et al., 2007). Sec13 is a β-propeller protein and has a dual role, as it also is present in the nuclear pore complex (NPC; Siniossoglou et al., 1996). The ER and nuclear envelope (NE) form a contiguous lipid bilayer. The NPC coats the NE at nuclear pores, establishes the selective permeability barrier of the NE, and serves as the sole conduit for transport across the NE (Brohawn et al., 2009). It is composed of ∼30 proteins, termed nucleoporins, each present in 8 × n copies, which are organized into subcomplexes symmetrically arranged about a central axis. A subset of architectural nucleoporins comprises the core structural scaffold of the NPC. A crystallographic study has demonstrated that the central α-helical unit of Sec31 is structurally similar to four large architectural nucleoporins, one of which binds Sec13. This α-helical unit, common to COPII and the NPC, is therefore termed the ancestral coatomer element 1 (ACE1; Brohawn et al., 2008).
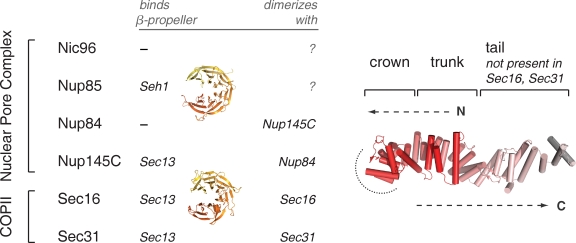
The ACE1 has a unique, irregular α-helical structure. It folds back on itself to form a J shape, divided into three modules (Fig. 1). The N-terminal and middle subdomains of the ACE1 fold together to constitute the trunk. The U turn between these subdomains is the crown. The ACE1 of nucleoporins has an additional module at the C terminus, the tail. Sequence similarity among the five known ACE1 proteins is weak: they have tolerated considerable mutation without compromising the overall structure. Their common ancestry was thus noted only after crystal structures were solved (Brohawn et al., 2008). Sec31 and Nup145C each bind Sec13 using the same mechanism, insertion of a single β-blade to close the open, six-bladed β-propeller of Sec13 in trans. The common ancestry of COPII and nucleoporin ACE1s provides strong evidence for the protocoatomer hypothesis, that various coat and coat-like protein complexes evolved from a small set of more versatile complexes (Devos et al., 2004). ACE1 is evidence that the NPC and the COPII coat derive from a common membrane-coating protein complex, already present in a primitive eukaryotic progenitor (Brohawn et al., 2008).
Figure 1.
Summary of ancestral coatomer element (ACE1) proteins. ACE1 was originally identified based on structural homology between Nup85, Nic96, Nup84, Nup145C, and Sec31. Sec16 is shown to contain an ACE1 in this study. Three ACE1 proteins bind Sec13, mutually exclusively; Nup85 binds its homologue Seh1. Nup84 and Nup145C form a heterodimer, and Sec16 and Sec31 form homodimers. The structure of Nic96 is shown to illustrate the three modules that compose the ACE1: crown, trunk, and tail. The COPII ACE1 domains might lack the tail module. The structure is colored red to white from the N to C terminus, as labeled. Dashed arrows show how the ACE1 forms a J shape. A dotted arc encircles the surface by which ACE1 dimerization occurs.
The COPII system is among the best-studied intracellular transport systems (Bonifacino and Glick, 2004; Fromme and Schekman, 2005; Mancias and Goldberg, 2005; Hughes and Stephens, 2008). Its components were identified genetically (Kaiser and Schekman, 1990) and can be reconstituted into a functional system in vitro (Salama et al., 1993; Barlowe et al., 1994; Shaywitz et al., 1997; Matsuoka et al., 1998; Sato and Nakano, 2004). The COPII outer coat, Sec31 and Sec13, can assemble into polyhedral cages without membrane or other proteins (Antonny et al., 2003; Stagg et al., 2006). The structure of this cage has been determined by cryo-EM (Stagg et al., 2006, 2008) and by x-ray crystallography (Fath et al., 2007). Sec31 homodimerizes via a crown to crown interface of its ACE1. This homodimer forms the central rod of a Sec13–Sec312–Sec13 tetramer, which self-assembles in multiple copies to form the complete cage. In addition to Sec13, Sec31, and three proteins of the inner coat, COPII transport also uses the essential accessory protein Sec16 (Kaiser and Schekman, 1990). However, Sec16 is much more poorly understood.
Sec16 interacts genetically and physically with components of COPII (Kaiser and Schekman, 1990; Espenshade et al., 1995; Gimeno et al., 1995, 1996; Shaywitz et al., 1997). In particular, temperature-sensitive alleles of Sec13 and Sec16 are synthetic lethal. Sec13 and Sec16 interact weakly by yeast two-hybrid (Shaywitz, 1997). A sequential mechanism for assembly of the COPII coat complex has been delineated, and Sec16 is implicated in early steps of this assembly process, but absence of structural data on Sec16 has impeded a full understanding of its function. At 242 kD in Saccharomyces cerevisiae, Sec16 is notoriously difficult to work with in vitro.
In this study, we show that Sec16 contains a central ∼50-kD domain of the ACE1 type. We report the 165-kD heterotetrameric crystal structure of Sec16984–1421 in complex with Sec13 and compare the related Sec13–Sec31 edge element to this novel Sec13–Sec16 edge element. Intriguingly, both Sec16 and Sec31 are shown to homodimerize by domain swapping. Furthermore, we report the crystal structure of a Sec13–Sec31 mutant with a compromised domain swap, showing it still forms an edge, however with a drastically different and unexpected topology. Mutants of Sec16 or Sec31 that compromise the domain swap, that exchange the ACE1 units of these proteins, or affect other aspects of their structures were created and tested in vivo. Together, structural and in vivo results suggest that Sec16 templates assembly of the COPII outer coat.
Results
Structure of the Sec13–Sec16 tetramer
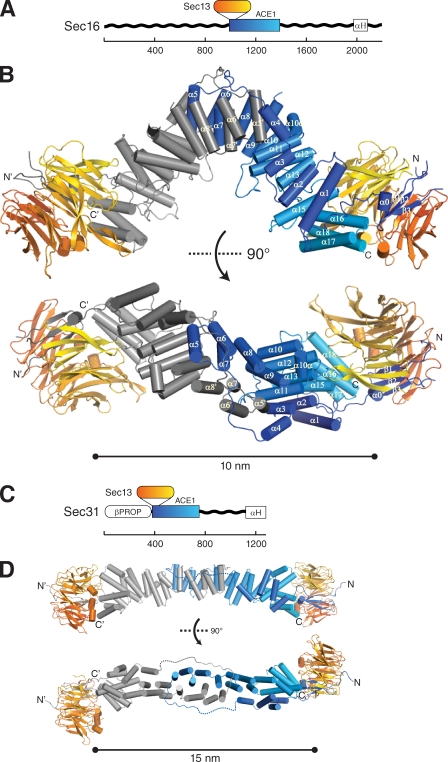
Sec16 is proposed to scaffold COPII assembly, but its precise function is ill defined. It has never been purified in high yield as a full-length protein. A central conserved domain (CCD), ∼400 aa, is well conserved among Sec16 homologues in various organisms (Connerly et al., 2005). A sequence alignment for the CCD is shown (Fig. S1). Secondary structure prediction suggested that the CCD of Sec16 is largely α-helical and flanked by unstructured regions (Fig. 2 A). Noting that it shares sequence similarity with the ACE1 of Sec31 (16% identity and 34% similarity over 391 residues), we inferred that the CCD of Sec16 might comprise two structural elements: a single N-terminal β-blade, which would bind the open Sec13 β-propeller by insertion, followed by an α-helical ACE1. (While this manuscript was in preparation, it was reported that the CCD of human Sec16A interacts with Sec13 by yeast two-hybrid [Hughes et al., 2009].) The CCD of Sec16 (residues 984–1421) and full-length Sec13, from S. cerevisiae, were coexpressed in Escherichia coli from a bicistronic plasmid and purified to homogeneity. Soluble and highly expressed (∼40 mg of pure protein per liter of culture, final yield) Sec16984–1421 and Sec13 formed a stable, equimolar complex that eluted at 11.6 ml on a Superdex 200 10/300 column, indicating an apparent molecular mass of ∼375 kD, when compared with globular protein standards (Fig. S2 A). Gel filtration systematically overestimates the molecular weight of elongated macromolecules because an elongated molecule passes through the column in less volume than a globular protein of the same molecular weight (Siegel and Monty, 1966). To determine the oligomeric state of the complex more accurately, sedimentation velocity ultracentrifugation was performed and indicated this species was tetrameric and homogenous (s = 6.5; Mf = 177 kD; Fig. S2 B). Collectively, these experiments indicated that Sec16 and Sec13 form a 2:2 tetramer of 165 kD, whose shape is elongated, not globular.
Figure 2.
Structural comparison of Sec16 and Sec31. (A) Domain organization of Sec16. Unstructured regions (wavy black lines), CCD (blue–cyan box), and C-terminal α-helical domain (white box, labeled αH) are shown. Scale bar shows number of amino acids residues. The β-propeller protein Sec13 binds Sec16 at the position indicated. (B) Crystal structure of Sec16 in complex with Sec13. Sec13 is colored orange to yellow from the N to C terminus, as in A. One Sec16 monomer is colored blue to cyan, as in A, the other dark to light gray from the N to C terminus. N and C termini are labeled, as well as β-strands β1–β3 and helices α0–α18. Helices α5–α7 of the Sec16 monomer domain swap at the homotypic interface. The bottom view is rotated 90° toward the viewer as indicated. (C) Domain organization of Sec31. Unstructured region (wavy black line), β-propeller (white oval, labeled βPROP), central structured domain (blue–cyan box), and C-terminal α-helical domain (white box, labeled αH) are shown. Scale bar shows number of amino acids residues. The β-propeller protein Sec13 binds Sec31 at the position indicated. (D) Structure of the Sec13–Sec31 tetramer. Color scheme as in B, with Sec31 coloring matching that of Sec16. The bottom view is rotated 90° toward the viewer as indicated.
Crystals of Sec16 in complex with Sec13 grew in the orthorhombic space group P212121 or its monoclinic subgroup P21 and diffracted to 2.7-Å resolution. The structure was solved by single-wavelength anomalous dispersion, using selenomethionine-substituted protein (Se-SAD). The asymmetric unit in P212121 contains one Sec13–Sec162–Sec13 tetramer. For simplicity, we refer to this complex as the Sec13–Sec16 tetramer. Two Sec13 molecules were placed by phased molecular replacement. Two Sec16 molecules were built using the experimental map. The model was refined to Rwork/Rfree of 19.4%/25.0% (Table I). The Sec13–Sec16 tetramer is arranged such that Sec13 forms the two ends of the elongated structure, whereas the two Sec16 molecules homodimerize in the center. The tetramer has overall dimensions of 165 Å × 70 Å × 50 Å (Fig. 2 B). The N terminus of Sec16984–1421 is a β-blade that completes the open, six-bladed β-propeller of Sec13 in trans, creating a 2,820-Å2 interface. Sec16 provides three β-strands of this seventh β-blade, whereas residues 1–6 of Sec13 donate the outermost β-strand, forming a velcro closure, as seen in many β-propeller structures (Chaudhuri et al., 2008). The remainder of Sec16 forms an α-helical block, with dimensions ∼70 Å × 30 Å × 30 Å. Two Sec16 molecules form a homodimer centered on the twofold axis of the tetramer. The dimer interface is composed of identical residues from each Sec16 molecule, measuring 2,960 Å2 in size.
Table I.
Data collection and refinement statistics
| Data collection and refinement statistics | Sec13–Sec16 | Sec13–Sec16 | Sec13–Sec31ΔL |
| Data collection | |||
| Data set | Native | Selenomethionine | Native |
| Space group | P212121 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 56.7, 139.0, 205.4 | 56.0, 144.1, 204.3 | 156.2, 46.6, 192.0 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 93.5, 90 |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 |
| Resolution range (Å) | 30.0–2.65 (2.74–2.65) | 30.0–2.70 (2.80–2.70) | 30.0–2.80 (2.87–2.80) |
| Total reflections | 168736 | 430502 | 396307 |
| Unique reflections | 45333 | 88825 | 69366 |
| Completeness (%) | 99.8 (99.4) | 99.9 (100) | 99.6 (99.4) |
| Redundancy | 3.7 (3.5) | 3.6 (3.7) | 5.7 (5.5) |
| Rmerge (%) | 7.4 (65.3) | 8.1 (56.0) | 16.4 (47.7) |
| Rr.i.m. (%) | 8.6 (80.1) | 9.9 (78.7) | 18.1 (50.0) |
| Rp.i.m. (%) | 4.4 (42.0) | 4.5 (35.8) | 7.3 (20.8) |
| I/σ | 20.7 (1.9) | 25.5 (2.7) | 9.6 (3.3) |
| Wilson B factor (Å2) | 51.6 | 49.6 | 55.7 |
| Refinement | |||
| Resolution range (Å) | 30.0–2.69 | 30.0–2.80 | |
| Rwork | 19.8 | 26.7 | |
| Rfree | 24.3 | 30.0 | |
| Twin law | −h, −k, l | ||
| Twin fraction | 0.23 | ||
| Number of reflections | |||
| Total | 44,752 | 69,353 | |
| Rfree | 2,158 | 1,792 | |
| Number of atoms | |||
| Protein | 10,476 | 19,197 | |
| Water | 184 | 0 | |
| B factors (Å2) | |||
| Protein | 103.0 | 51.8 | |
| Sec13 (chain A) | 45.8 | ||
| Sec16 (chain B) | 74.3 | ||
| Sec16 (chain C) | 111.0 | ||
| Sec13 (chain D) | 191.0 | ||
| Water | 52.0 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.005 | 0.006 | |
| Bond angles (°) | 0.836 | 0.936 | |
| Ramachandran plot | |||
| Favored (%) | 95.5 | 94.4 | |
| Allowed (%) | 4.3 | 5.1 | |
| Outliers (%) | 0.2 | 0.5 |
The highest resolution shell is shown in parentheses. Rmerge is the merging R factor. Rr.i.m. is the redundancy independent merging R factor. Rp.i.m. is the precision-indicating merging R factor. For definitions, see Weiss (2001).
The β-blade connects through helix α0 to helices α1–α3, which form an antiparallel stack (Fig. 2 B). Helix α4 and a 14-residue loop extend out and around the other Sec16 molecule and connect to helix α5. Helices α5–α9 form a second antiparallel stack that returns toward the N terminus, forming a hydrophobic interface with helices α5′–α9′ in the other Sec16 molecule. The stack continues with helix α10. To be consistent with the ACE1 helix nomenclature (Brohawn et al., 2009), the next helix is labeled α10a. Helix α10a lies outside the stack, perpendicular to α10 and α11. Helices α11–α15 bind helices α1–α3, placing the C terminus near the N terminus. Helix α14 of a canonical ACE1 is absent. In place of helix α14, a structured but nonhelical stretch of 20 residues winds out, is exposed on the convex surface of the tetramer, traverses the stack, and connects back to the next helix, labeled α15. A final helical bundle, α16–α18, completes the stack and braces against Sec13. At the C terminus, residues 1391–1421 are not structured.
In crystallography, the quality of electron density and the temperature (B) factors for a portion of a model indicate how rigidly that portion of the model is held in place (Wlodawer et al., 2008). We observed much poorer electron density and twofold higher B factors for one copy of Sec13 than the other (Table I). The better-ordered copy of Sec13 is positioned near a crystallographic 21 axis, stabilized by 1,420 Å2 of crystal contacts. However, the poorly ordered Sec13 molecule is barely constrained by crystal packing interactions. Because the remainder of the model is rather well ordered, this observation suggests that Sec13 may pivot relative to Sec16 in vivo, when not constrained by crystal packing.
As anticipated from its sequence, the α-helical portion of the Sec16 CCD is an ACE1. The CCD as defined by Connerly et al. (2005) is residues 992–1420 in S. cerevisiae, nearly the same fragment as the crystallization construct. The CCD contains two structural elements: the β-blade that binds Sec13 and the α-helical ACE1. As in other ACE1s, helices α1–α3 and α13–α18 form the trunk, whereas helices α5–α11 form the crown. The crown creates a U turn that caps the α-helical block and binds the second Sec16 molecule. Each α-helix corresponds to an α-helix in the canonical ACE1, except α10a, which is added, and α14, which is replaced by nonhelical structure. There is very low sequence homology to ACE1 members other than Sec31. As is true of the entire ACE1 class, individual modules (i.e., crown, trunk, and tail) superimpose well, but their orientations vary. Specific structural differences such as insertion of α10a or unwinding of α14 dictate the placement of each module.
We mapped the amino acid substitutions that cause known temperature-sensitive alleles of Sec16 onto the structure (Fig. S3). The dot1 mutation from Pichia pastoris substitutes a proline with leucine in the connection between insertion blade and ACE1 (Connerly et al., 2005). Proline (as a cyclic secondary amino acid) is particularly suited to create a sharp turn in the polypeptide backbone. The proline replaced by the dot1 mutation is the first of two in the turn created by the motif FPGPL, which is strongly conserved among Sec16 orthologues. The five known temperature-sensitive alleles in S. cerevisiae are caused by four distinct point mutations (Espenshade et al., 1995). (sec16-2 and sec16-5 are the same.) Although these are dispersed in the sequence, they cluster together in the structure. All replace hydrophobic residues in the core of the interaction between the N- and C-terminal halves of the trunk, undoubtedly hindering the ACE1 from folding into its proper shape.
Structural comparison of ACE1 edge elements
The Sec13–Sec16 tetramer is similar to the previously reported structure of the Sec13–Sec31 tetramer (Fig. 2, C and D; Fath et al., 2007). Unlike Sec16, Sec31 has a structured N-terminal domain, a β-propeller, immediately preceding the Sec13 interaction site (Fig. 2 C). In the in vitro assembled Sec13–Sec31 cage, the N-terminal β-propeller forms the major vertex interactions that propagate the cage. Sec13 and the ACE1 of Sec31 form the edges of this cage, which was termed the Sec13–Sec31 edge element (Fig. 2 D; Fath et al., 2007). The Nup84–Nup145C–Sec13 trimer is an analogous edge element for the NPC (Brohawn and Schwartz, 2009). The crystal structure reported in this study shows that the Sec13–Sec16 tetramer is another edge element for the COPII system. In all three edge elements, ACE1 units form a central dimer by interaction of their crowns. However, the crown to crown interactions in Sec16, Sec31, and Nup84–Nup145C have important differences detailed below.
Central angle of the edge element.
The dimerized ACE1 units in the edge elements of COPII and the NPC create varying angles: 90° at the Sec16–Sec16 interface, 120° at the Nup84–Nup145C interface, and 165° at the Sec31–Sec31 interface (Fath et al., 2007; Brohawn and Schwartz, 2009). In the assembled Sec13–Sec31 cage, the angle of Sec31 is more acute than in the crystal structure, 135° (Stagg et al., 2006, 2008). It is striking that in its crystal structure, the crown to crown interface of Sec16 is more bent than Sec31 in either the crystal structure (Fath et al., 2007) or the assembled cage (Stagg et al., 2006, 2008). We asked whether the Sec13–Sec16 tetramer is indeed more bent than the Sec13–Sec31 tetramer in solution or whether crystal packing alone accounts for the discrepancy between Sec16 and Sec31. The hydrodynamic radius of each edge element was calculated from its crystal structure, using HYDROPRO (García De La Torre et al., 2000). The calculated hydrodynamic radii are 5.3 nm for Sec13–Sec16 and 5.5 nm for Sec13–Sec31. The smaller radius for Sec13–Sec16 reflects its more compact structure, even though it is 10% greater in mass than Sec13–Sec31, 165 versus 150 kD. Sec13–Sec16 and Sec13–Sec31 edge elements were compared by size exclusion chromatography (Fig. S2 A). Consistent with its smaller calculated hydrodynamic radius, Sec13–Sec16 elutes after Sec13–Sec31. We conclude that the crystal structures reflect a true difference between Sec13–Sec16 and Sec13–Sec31. The central angle of the Sec13–Sec16 edge element is more bent than the Sec13–Sec31 edge element in solution.
Domain swapping of the edge element.
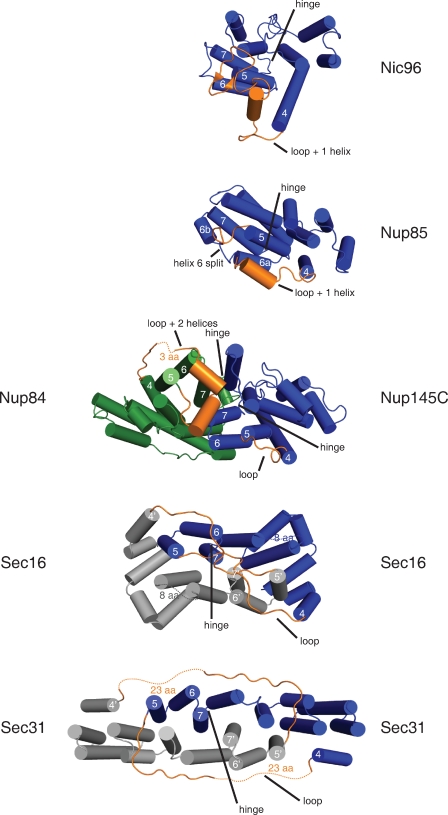
When the structure of Sec31 was first reported (Fath et al., 2007), no closely related structures were known. We compared Sec16 and Sec31 to the ACE1 structures solved in the interim. This showed that Sec16 and Sec31 are domain-swapped dimers, that is, they interact by exchange of identical subunits (Bennett et al., 1995; Liu and Eisenberg, 2002; Rousseau et al., 2003). Strictly speaking, domain swapping is said to occur only when a closed, monomeric form of the protein also exists (Gronenborn, 2009). Comparison to the closed form of a homologue can also be used. To define the closed form, we refer to the unswapped ACE1s, Nup85, Nic96, and Nup145C (Fig. 3). In the crowns of Nic96 and Nup85, helices α5–α7 pack against the trunk, forming a compact α-helical block (Jeudy and Schwartz, 2007; Brohawn et al., 2008; Debler et al., 2008; Schrader et al., 2008). Similarly, Nup145C adopts a closed form, whether or not bound to Nup84 (Hsia et al., 2007; Brohawn and Schwartz, 2009). In the Nup84–Nup145C–Sec13 trimer, Nup145C adopts a structure very similar to that in a Nup145C–Sec13 dimer (1.4-Å root mean square deviation over 420 Cα positions). In contrast, in Sec16 (this work) and Sec31 (Fath et al., 2007), helices α5–α7 exchange with the corresponding helices α5′–α7′ in the binding partner. We call helices α5–α7 the swap domain.
Figure 3.
Interactions formed by the ACE1 crown. The crown domain of each ACE1 is shown colored blue, except Nup84, which is green. Nic96 and Nup85 are not known to dimerize via crown–crown interaction. Nup84 and Nup145C form a heterodimer, and Sec16 and Sec31 form homodimers. The second copies of Sec16 and Sec31 are colored gray. Disordered loops are shown as dotted lines and labeled with the number of amino acids not observed. Sec16 and Sec31 dimerize by domain swapping. Helices α5–α7 exchange positions with helices α5′–α7′ in the binding partner. The domain swap requires extension of the swap loop (labeled loop) that connects helix α4 to α5 and rotation of the swap hinge (labeled hinge) that connects helix α7 to α8. The positions of the corresponding loop and hinge are labeled in all ACE1s. Nic96, Nup85, and Nup84 have one or two α-helices inserted into this loop (labeled loop + n helices).
Among known examples of domain swapping, rarely is the swap domain a central element (Schwartz et al., 2006; Gronenborn, 2009). More commonly, the N or C terminus of the protein is exchanged. To swap a central element, typically at least one of the two linkers to the remainder of the protein needs to be flexible to allow the rearrangement. In Sec16 and Sec31, a swap loop connects helices α1–α4 to the swap domain. This loop must be long enough to reach out and around the other molecule. In closed monomers, no such loop is needed because helix α4 already lies near helix α5. Nevertheless, all ACE1s except Nup145C have some insertion between α4 and α5. Nic96, Nup84, and Nup85 each have inserted α-helical segments. These may be capable of unfolding and extending. At its opposite end, the swap domain is connected to helices α8–α18 by a swap hinge. This hinge rotates out from its position in the closed monomers, in which the swap domain is not swapped but instead is tucked against the rest of the α-helical stack. It is a tight turn in all ACE1s, except Nup84. Nup84 has three short α-helical segments inserted between α7 and α8 (these segments interact directly with Nup145C).
Domain swapping substantially increases interface areas. We computed interface areas in swapped or hypothetical unswapped conformations for Sec16 and Sec31. The swap loop and swap hinge were excluded from calculations because it is not possible to know their position in the unswapped conformation. In swapped Sec16, the interface is ∼2,500 Å2, compared with ∼800 Å2 in unswapped Sec16. Similarly, in swapped Sec31, the interface is ∼3,000 Å2, compared with ∼1,000 Å2 in unswapped Sec31. Domain swapping triples the interface area in the Sec16 and Sec31 crown to crown interfaces. Nup84–Nup145C, although unswapped, has an interface surface area of ∼2,000 Å2 because addition sites in conserved loops frame the primary interaction surface.
Interlocking of the edge element.
Though Sec16 and Sec31 both dimerize by domain swapping, the path taken by the swap loop causes a key difference. The Sec31 dimer is interlocked, but the Sec16 dimer is not. In Sec31, the swap loops extend around and over the binding partner to connect back to the swap domain (Fath et al., 2007). 23 residues of the swap loop are disordered and not visible in the crystal structure. The gap left by these unmodeled 23 residues is 25 Å. In comparison, an adjacent segment of the swap loop spans 25 Å in only nine residues. If we accept that the unstructured segment does indeed connect the two ends of the swap loop across this gap, the crystal structure shows that the Sec31 dimer is interlocked. In contrast, in Sec16, symmetric swap loops extend over the top of the crown, touch in the middle, and then turn back to connect to their respective swap domains (Fig. 3). This swap loop is entirely ordered and modeled in the final structure. To confirm the path of this loop, a simulated annealing omit map for the loop was calculated, unequivocally showing the complete amino acid trace (Fig. S4). The path of the swap loop leaves the two Sec16 copies not interlocked; i.e., if pulled by the ends, the Sec16 dimer would break apart. Because the Sec31 dimer is interlocked, if pulled by the ends, the Sec31 dimer would remain entangled with itself. Thus, the Sec31 dimer can only be formed or broken by disrupting the interaction between the N- and C-terminal halves of the trunk. This suggests that creation of the Sec31 dimer is coordinated with folding of the ACE1 unit.
Structure-based mutants of Sec31
The structures of the Sec13–Sec16 edge element, reported in this study, and that of the Sec13–Sec31 edge element (Fath et al., 2007) were used as a basis to design specific mutations to probe the functional and structural integrity of these related complexes.
Solution behavior of Sec31 mutants Sec31EE and Sec31ΔL.
The difference between the unswapped ACE1s found in the NPC and the domain-swapped dimers observed in Sec16 and Sec31 is intriguing. To investigate whether domain swapping is physiologically relevant, we designed a mutation to prevent domain swapping of Sec31 in the Sec13–Sec31 edge. The swap loop of Sec31 was deleted to form Sec31ΔL. This deletion prevents helix α5 from swinging out from helix α4, otherwise a strict requirement for domain swapping to occur. We also designed a mutation to prevent Sec31 dimerization. In previous work on the Nup84–Nup145C edge element, it was shown that charged residues introduced into helix α7 of either protein prevent dimerization, when these are chosen to replace conserved hydrophobic residues crucial to high-affinity binding (Brohawn et al., 2008; Brohawn and Schwartz, 2009). A similar mutant of Sec31 was designed. Two residues, methionine 540 and leucine 544 in helix α7, were mutated to a charged residue, glutamic acid (E), to generate Sec31EE.
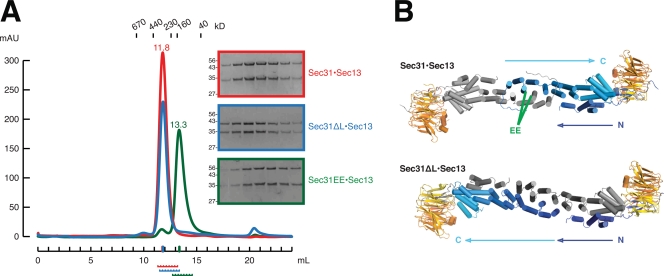
The Sec13–Sec31 edge element was expressed and purified in three forms: wild type, Sec31ΔL, or Sec31EE. The purified complexes were compared by size exclusion chromatography on a Superdex 200 10/300 column (Fig. 4 A). Sec31ΔL is 4 kD (9%) smaller than Sec31 and therefore migrates slightly faster on SDS-PAGE. Despite its reduced mass, Sec13–Sec31ΔL forms a tetramer with the same hydrodynamic radius as wild-type Sec13–Sec31. The interface mutant Sec13–Sec31EE elutes from the column later, indicating that, indeed, the double point mutation from Sec31 to Sec31EE is sufficient to disrupt tetramer formation. Sec13–Sec31EE instead forms a 1:1 heterodimer.
Figure 4.
Solution behavior and crystal structure of Sec13–Sec31 mutants. (A) Sec13–Sec31 (red) compared with Sec13–Sec31 mutants ΔL (blue) and EE (green) by size exclusion chromatography. Sec13–Sec31 and Sec13–Sec31ΔL are heterotetramers, whose structures are shown in B, with the same hydrodynamic radii. The EE mutation disrupts the crown interface. Sec13–Sec31EE is a heterodimer in solution. Elution volume in milliliters is plotted against absorbance at λ = 280 nm. Elution volumes for standard globular proteins are as indicated. SDS-PAGE analysis of each sample is shown, with fractions indicated below the x axis. (B) Crystal structure of Sec13–Sec31ΔL compared with Sec13–Sec31 (Protein Data Bank ID 2PM6). The tetramers form extended rods with the same 165° central angle. Because of deletion of the swap loop, Sec13–Sec31ΔL forms a laminated structure, rather than a U turn, as indicated by arrows labeled N and C. Both copies of Sec31 are colored blue to cyan from the N to C terminus. The green label EE indicates residues M540 and L544, in helices α7 and α7′, which were mutated to glutamic acid to generate Sec13–Sec31EE.
Crystal structure of Sec13–Sec31ΔL.
To determine definitively whether deletion of the swap loop prevents domain swapping, we solved the crystal structure of Sec13–Sec31ΔL, by molecular replacement, in space group P21 at 2.8-Å resolution (Table I). The structure of wild-type Sec13–Sec31 was used as a search model. To obtain an unbiased electron density map of the mutated swap loop (connecting helices α4/5) and the swap hinge (connecting helices α7/8), these were deleted from the search model. Furthermore, the model was split into two parts, the ACE1 of Sec31 and the completed β-propeller (six-bladed Sec13 and the insertion β-blade of Sec31). The asymmetric unit contains two copies of a Sec13–Sec31ΔL tetramer. The crystal lattice of Sec13–Sec31ΔL is different from that of wild-type Sec13–Sec31. Yet, compared with wild-type Sec13–Sec31 (Fath et al., 2007), the shape of the tetramer changes only slightly. The angle of the edge, 165°, is the same in both structures. The C-terminal three-helix bundle of the ACE1 and the Sec13 β-propeller are displaced ∼15 Å with respect to the central rod, in the plane parallel to a face of a Sec13–Sec31 cage.
The structure of Sec13–Sec31ΔL at the crown to crown interface revealed an unexpected result. In rebuilding the structure, strong difference density at the swap hinge was apparent (Fig. S5). Because deletion of the swap loop prevents domain swapping, we expected the swap hinge to rotate inward to accommodate an unswapped conformation. Surprisingly, we find the swap hinge in the same place as in wild-type Sec13–Sec31. In consequence, deletion of the swap loop does prevent domain swapping but in an unexpected manner. To have helix α5 adjacent to α4, as it is dictated by the truncated swap loop, and to retain the swap hinge still in the same conformation, Sec31ΔL laminates with its binding partner across its entire length (Fig. 4 B). Instead of folding back into a J shape as in wild-type Sec31, the two copies of Sec31ΔL extend completely and lie flat against one another. Because the entire transverse section of the dimer is an interface between two molecules, the interface surface area of Sec31ΔL is 6,260 Å2. The final model was refined to Rwork/Rfree = 26.7%/30.0% (Table I).
The structure of Sec13–Sec31ΔL suggests that there is a large energetic penalty for rotating the swap domain into the closed (not swapped) conformation, as is observed in monomeric ACE1s and the Nup84–Nup145C dimer. Shortening the swap loop should draw the swap domain back to the closed conformation; but despite shortening the swap loop, the swap domain stays extended, as in the domain-swapped conformation of wild-type Sec13–Sec31. The laminated structure of Sec13–Sec31ΔL is therefore indirect evidence that the domain-swapped conformation of Sec31 is favored over the closed conformation and thus that the crystal structure of Fath et al. (2007) is the physiological structure.
Complementation of sec16Δ or sec31Δ by structure-based mutants
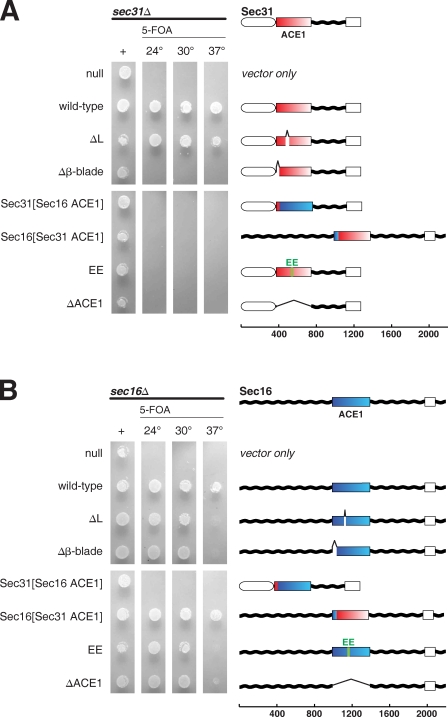
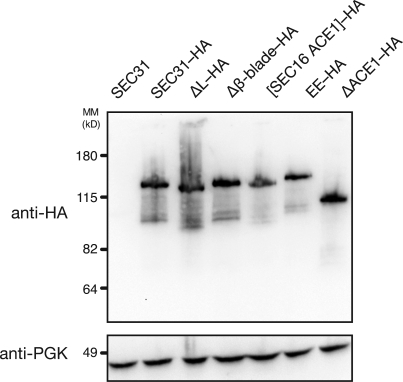
SEC16 and SEC31 are both essential genes. To assess the physiological relevance of structural characteristics of COPII edge elements, we tested in a plasmid shuffle assay whether several designed mutants complement null alleles of SEC16 or SEC31 (Fig. 5). Haploid strains sec16Δ × pRS316[SEC16] or sec31Δ × pRS316[SEC31] were isolated, transformed with plasmids containing designed mutants of SEC16 or SEC31, and tested for growth on media containing 5-fluoroorotic acid, which selects against pRS316, causing the strain to lose the wild-type gene. This experiment shows that the ACE1 is required for the essential function of SEC31 (Fig. 5 A). Furthermore, failure of Sec31EE to rescue sec31Δ proves that Sec31 must form a proper edge element. Deletion of the insertion blade that binds Sec13 also disables the gene. (A Western blot to epitope-tagged versions of these mutants of SEC31 shows that although they do not rescue growth, they are indeed expressed [Fig. 6].) In contrast, domain swapping is not required for the essential function of SEC31. Sec31ΔL complements the null at all temperatures tested. Evidently, the radical rearrangement observed in laminated Sec31ΔL does not prevent Sec31 from assembling a functional edge element for the COPII cage.
Figure 5.
Complementation of Sec16 or Sec31 by structure-based mutations. (A) Structure-based mutants of Sec31 assayed by plasmid shuffle. A sec31Δ plasmid shuffle strain was prepared using endogenous SEC31 cloned into a URA3 plasmid. Mutations were introduced into SEC31 on a LEU2 plasmid, transformed into the shuffle strain, spotted onto media lacking leucine (first lane) or lacking leucine and supplemented with 5-fluoroorotic acid (5-FOA; subsequent lanes), and grown for 36 h at 24, 30, or 37°C. Sec31 domain architecture is diagramed: N-terminal β-propeller (white oval), central insertion β-blade and ACE1 (red–white box), unstructured region (wavy black lines), and C-terminal α-helical domain (white box). Fragments of Sec16 used in chimeric genes are shown in blue–cyan. (B) Structure-based mutants of Sec16 assayed by plasmid shuffle. Mutations were introduced into SEC16 on a LEU2 plasmid and tested as in A. Genes are diagramed: unstructured regions (wavy black lines), CCD (blue–cyan box), and C-terminal α-helical domain (white box). Fragments of Sec31 used in chimeric genes are shown in red–white.
Figure 6.
Western blot of structure-based mutants of Sec31. Structure-based mutants of Sec31 were tagged with the HA epitope and transformed into the sec31Δ plasmid shuffle strain (as in Fig. 5 A). Untagged Sec31 is shown as a control for specificity of the anti-HA antibody. The blot was reprobed with an antibody to 3-phosphoglycerate kinase (PGK) as a control for equal sample loading. MM, molecular mass.
The corresponding mutations were also generated for SEC16 (Fig. 5 B). Expectedly, purified Sec13–Sec16ΔL behaves as a tetramer in solution, and Sec13–Sec16EE forms a dimer, as determined by size exclusion chromatography (unpublished data). We find that SEC16 is more robust to mutation than SEC31. A strain in which the ACE1 of Sec16 is deleted is able to grow normally at 30°C but grows very slowly at 37°C. Deletion of the swap loop to prevent domain swapping, deletion of the β-blade to prevent binding of Sec13, or the EE mutation to prevent dimerization is each likewise tolerated at 30°C but not at 37°C.
Because Sec16 and Sec31 form similar edge elements, we asked whether the ACE1 units of these two proteins are interchangeable. To test this, chimeric genes were designed that place the ACE1 of each gene into the middle of the other. Sec31[Sec16 ACE1] and Sec16[Sec31 ACE1] were tested in the plasmid shuffle assay (Fig. 5). sec31Δ is not complemented by Sec31[Sec16 ACE1]. We found, though, that sec16Δ is complemented at all temperatures tested by Sec16[Sec31 ACE1]. Unlike the other Sec16 mutants, Sec16[Sec31 ACE1] exhibits no temperature sensitivity. Remarkably, despite only 34% sequence similarity, the ACE1 of Sec31 can substitute for the ACE1 of Sec16.
Discussion
Our study of ACE1 has revealed that the CCD of Sec16 comprises a β-blade and 19 α-helices that closely resemble the core of the COPII vesicle coat protein Sec31. Sec16 is considered a scaffold for the COPII system because it localizes to the cytoplasmic surface of the ER, binds the major components of the COPII system, and promotes vesicle coating (Espenshade et al., 1995; Gimeno et al., 1996; Shaywitz et al., 1997; Supek et al., 2002) These studies have shown that Sec16 binds the cargo adaptor Sec24, the GTPase-activating protein Sec23, and the GTPase Sar1 and additionally that it binds Sec31 and possibly Sec13. Additional studies on Sec16 in yeast, fly, and mammalian systems have substantiated its role in COPII transport (Connerly et al., 2005; Watson et al., 2006; Bhattacharyya and Glick, 2007; Ivan et al., 2008; Hughes et al., 2009).
We show that Sec16 and Sec13 form a stable, tetrameric complex. The crystal structure of Sec16984–1421 in complex with Sec13 reveals that Sec16 forms a Sec13–Sec162–Sec13 tetramer. This tetramer is similar to the Sec13–Sec312–Sec13 edge element, which assembles into the COPII outer coat (Stagg et al., 2006, 2008; Fath et al., 2007). It is also analogous to the Nup84–Nup145C–Sec13 edge element of the NPC (Brohawn and Schwartz, 2009). Because Sec16 and Sec31 (as well as Nup145C) occupy the same binding interface on Sec13, their interactions with Sec13 are mutually exclusive. It is likely that these proteins bind distinct pools of Sec13. This explains the observation that Sec16 is present at the transitional ER in lower abundance than Sec13 (Connerly et al., 2005).
The discovery that Sec16 and Sec31 are related proteins provides novel insight into the function of Sec16 in the COPII system. Sec16 and Sec31 form with Sec13 analogous heterotetrameric edge elements. Unlike Sec31, secondary structure prediction shows that Sec16 does not have an N-terminal β-propeller domain. In the COPII cage, the Sec13–Sec31 tetramers self-assemble through contacts primarily mediated by the N-terminal β-propeller domains of adjacent Sec31 molecules (Stagg et al., 2006, 2008; Fath et al., 2007). It is important to note that without such a β-propeller, Sec16 lacks a vertex element and thus is unlikely to form a cage in a similar manner.
One unresolved question about the Sec13–Sec31 edge element has been the nature of the central angle of the Sec31 homodimer (Stagg et al., 2006, 2008; Fath et al., 2007). This angle is more acute in the cryo-EM structure of the assembled Sec13–Sec31 cage than in the crystal structure of the edge element. Several reasons to explain this difference have been proposed (Fath et al., 2007; Stagg et al., 2008): (a) flexing this angle allows the Sec13–Sec31 cage to accommodate cargoes of different size; (b) a conformational change occurs upon cage assembly making the angle more acute; (c) the edge element in the crystal structure is distorted from its native conformation by crystal packing; (d) the yeast protein in the crystal structure differs from the human protein used for cage assembly; (e) the C-terminal half of Sec31, which is not included in the crystal structure, influences the angle. Stagg et al. (2008) show that the angle of Sec31 stays the same whether the cage formed is a cuboctahedron 60 nm in diameter or an icosidodecahedron 100 nm in diameter. In other words, the adjustments that change the shape of the cage occur primarily at the interfaces between vertex elements rather than within the ACE1 blocks, arguing against the first explanation that variation in this angle is used to form cages of different sizes.
In the structure of Sec13–Sec162–Sec13, the Sec16 homodimer forms the analogous central angle. We noted with interest that this angle is 90°, much more bent than in Sec13–Sec31. Because one end of the Sec13–Sec16 edge element is not held by significant crystal contacts, we believe that the conformation of Sec13–Sec16 in the crystal structure is near to the native conformation in solution. Our data show that, consistent with the crystal structures, in solution, the Sec13–Sec16 tetramer is more bent than the Sec13–Sec31 edge element. Additionally, our crystal structure of Sec13–Sec31ΔL reveals that this mutant has the same angle at the Sec31–Sec31 interface, 165°, as wild-type Sec13–Sec31.
A striking feature of the COPII edge elements is domain swapping, which is rarely observed in the middle of a polypeptide chain. In reporting the crystal structure of Sec13–Sec31, Fath et al. (2007) noted that the two molecules of Sec31 interlock. It becomes apparent that domain swapping occurs in Sec31, when this structure is compared with those of ACE1 nucleoporins, which were solved later and which form compact J-shaped helical units. Before solving the structure of Sec16, it remained unclear whether domain swapping in Sec31 is caused by crystallization alone or is physiologically relevant. The crystal structure of Sec13–Sec16 shows that like Sec31, Sec16 homodimerizes by domain swapping. This is strong evidence that domain swapping in Sec31 is physiologically relevant. It is unlikely to be a coincidence that domain swapping occurs in two different ACE1 proteins in unrelated crystal lattices.
Domain swapping triples the interface surface area of this crown to crown interaction to ∼3,000 Å2, suggesting that the domain-swapped interaction is very strong. Indeed, each of the COPII edges is observed to remain stably associated in solution. Sec13–Sec16 and Sec13–Sec31 edge elements are likely similarly stable in vivo. Their structures support the notion that cage assembly and disassembly is mediated at the vertices of the cage, not by making and breaking the edge itself.
In attempting to prevent domain swapping in Sec16 and Sec31, we generated and solved the crystal structure of a variant of the Sec13–Sec31 edge element that laminates across its entire length, rather than forming two J-shaped molecules. This laminated variant provided a means to test the physiological importance of domain swapping. We find that the laminated Sec31 variant, Sec31ΔL, is able to complement deletion of Sec31, but a mutant designed to prevent dimerization, Sec31EE, fails to complement this null. We further show with the chimeric gene Sec16[Sec31 ACE1] that the ACE1 of Sec31 can functionally replace the ACE1 of Sec16. This result suggests that its architectural role, forming an edge element, is the most important function of the ACE1 of Sec16.
Sec16 as a template for the COPII coat
Sec16 has been called a scaffold for the COPII coat. The term scaffold refers to a protein that binds several factors to bring them together in the cell. It is clear that by binding many components of the COPII system, Sec16 performs this function. In other contexts, the term scaffold also refers to proteins that organize a system into a predetermined structure. For example, in viral assembly, scaffolding proteins recruit elements of the naive viral capsid, organize them, and establish the desired size and shape of the capsid. Such viral scaffolding proteins also act as chaperones for capsid proteins and exclude host proteins from the capsid. Scaffolding proteins are then left out of the mature viral capsid (Thuman-Commike et al., 1998; Fane and Prevelige, 2003). Sec16 is similarly thought to assist in organizing and shaping the assembly of the COPII coat and to be excluded from the final coat because it is present substoichiometrically and is not required for assembly of the coat in vitro (Matsuoka et al., 1998; Connerly et al., 2005).
The structure of Sec16 in complex with Sec13 now allows us to make this model more specific. Sec13–Sec16 is shown to form an alternative edge element for the COPII system. It dimerizes, allowing it to concentrate twice the number of COPII components in its vicinity. Furthermore, it mimics the Sec13–Sec31 edge element, which should allow it to position precisely COPII components with respect to the Sec13–Sec31 outer coat. For this reason, it could be termed a template for the COPII coat. Because the similarity between Sec16 and Sec31 was not previously known, this is a new interpretation of Sec16’s role in the COPII system, but one that is consistent with prior experiments. It suggests a few further directions for studying Sec16. Because Sec16 lacks a vertex element like the β-propeller domain of Sec31, it still is not clear whether or not Sec16 can form cage-like structures. It may form a precursor coat for the COPII system over which the Sec13–Sec31 cage is laid. Or, it may coassemble with Sec13–Sec31 at an early stage of COPII assembly, before being replaced. We hypothesize that Sec13 bound to Sec16 may form weak homotypic interactions with other copies of Sec13 or with the β-propeller of Sec31, organizing Sec13–Sec16 tetramers with respect to one another and with respect to Sec13–Sec31.
It has been noted that COPII coat assembly is delicately balanced between forces that drive assembly in the presence of Sar1–GTP and forces that promote disassembly once Sar1–GTP converts to Sar1–GDP (Supek et al., 2002). Because Sec31 promotes hydrolysis of GTP by Sar1, in vitro assembly of Sec13–Sec31 (counterintuitively) rapidly triggers its own disassembly (Antonny et al., 2001). Thus, it remains to be explained how Sec13–Sec31 is able to assemble a full coat without prematurely falling apart. By establishing an organized template, Sec16 may help tip the balance toward assembly in the early moments after Sec13–Sec31 recruitment.
Model for the function of Sec16 in the COPII coat
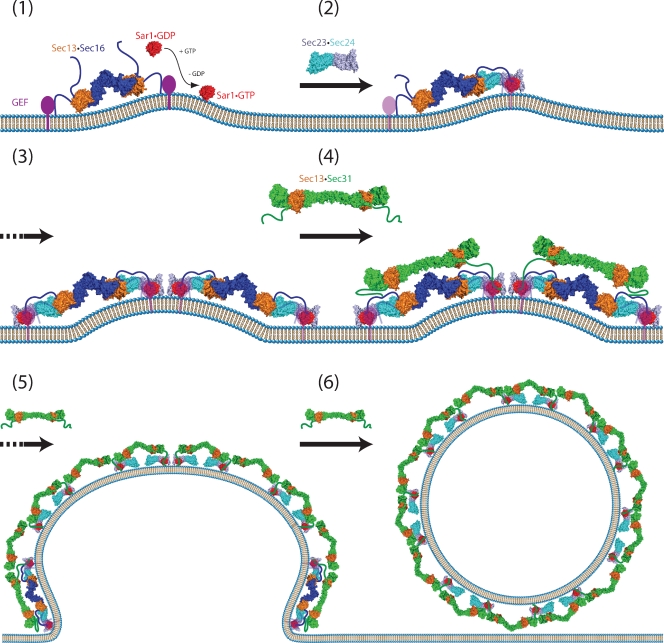
We propose the following model for COPII assembly (Fig. 7), based largely on experimental evidence and models proposed previously (Bonifacino and Glick, 2004; Fromme and Schekman, 2005; Mancias and Goldberg, 2005; Gürkan et al., 2006). The Sec13–Sec16 tetramer binds to the ER membrane through charged segments of the N-terminal unstructured region and by interaction with Sed4 (or Sec12) through its C-terminal helical domain. Sar1 is recruited when it is converted from the GDP- to GTP-bound state. Insertion of the N-terminal amphipathic α-helix of Sar1 into the membrane induces curvature of the ER membrane, assisted by Sec16-mediated clustering. The Sec23–Sec24 dimer binds both Sec16 and Sar1, joining them together. Because the Sec23–Sec24 dimer forms two independent interactions with Sec16, the Sec23–Sec24 dimer may help cross-link adjacent Sec13–Sec16 tetramers into higher-order oligomers. Cargo is concurrently recruited via interactions with Sec24. Once this precoat is formed, Sec13–Sec31 begins to assemble. Interactions of Sec31 with Sec23–Sec24 and Sar1 facilitate this assembly, as do direct interactions between Sec31 and Sec16 and, perhaps, homotypic interactions between Sec13 molecules on different edges. Sec13–Sec16 is similar in size and shape to Sec13–Sec31 and places all Sec31’s partners close at hand. Sec16 acts against Sec31-promoted hydrolysis of GTP by Sar1, either directly or by holding Sed4 (or Sec12) nearby to recycle Sar1–GDP to Sar1–GTP. Sec13–Sec16 is gradually displaced. It is included substoichiometrically, or not at all, in the final COPII coat, which then severs from the ER.
Figure 7.
Model for assembly of the COPII coat complex. The common model for assembly of the COPII coat complex is modified to include the role of Sec16. For simplicity, cargo molecules are omitted. (1) The Sec13–Sec16 tetramer is stably associated with the ER membrane and binds the integral membrane protein Sed4 or its homologue Sec12. Sar1 becomes associated with the membrane, when it is converted from the GDP- to GTP-bound state. Concentration of membrane-associated proteins begins to bend membrane. (2) Sec13–Sec16 and Sar1 collaborate to recruit the cargo adaptor Sec23–Sec24 dimer. (3) A precoat self-associates into higher-order oligomers. (4) Sec13–Sec16 and Sec23–Sec24–Sar1 form independent interactions with Sec13–Sec31, causing it to assemble near and/or in place of Sec16. (5) The forming coat contains progressively more Sec13–Sec31 and less Sec13–Sec16. Hand-off of Sec23–Sec24–Sar1 from Sec16 to Sec31 sets the stage for GTP hydrolysis by Sar1. (6) A final COPII coat is formed, and vesicle budding is complete. Sec13–Sec16 remains mostly associated with the ER.
The crystal structure of the Sec13–Sec16 tetramer, the structure of a laminated Sec13–Sec31 mutant, and supporting functional data support a new interpretation of Sec16’s role in assembly of the COPII coat. This hitherto mysterious protein contains an ACE1, structurally related to both nucleoporins and the COPII coat protein Sec31. The central domain of Sec16 allows the Sec13–Sec16 tetramer to act as an alternative edge element for COPII. The structure of the Sec13–Sec16 edge element may allow it to precisely template assembly of the COPII vesicle coat.
Materials and methods
Sequence analysis
PSI-BLAST was performed using the National Center for Biotechnology Information server (http://blast.ncbi.nlm.nih.gov). Secondary structure prediction was performed with the PredictProtein server (http://www.predictprotein.org/). Alignments were generated using the MAFFT algorithm in JalView (Waterhouse et al., 2009) and a figure prepared in ALINE (Bond and Schüttelkopf, 2009).
Protein expression constructs
Sec16 (residues 984–1421) and Sec13 were cloned into a bicistronic pET-Duet vector (EMD), engineered to encode a human rhinovirus 3C (HR3C)–cleavable His6 tag at the N terminus of Sec16. The construct was modified by PCR methods to create Sec16ΔL (deletion of residues 1115–1128) or Sec16EE (mutations A1159E and L1162E). Sec31 (residues 370–746) and Sec13 were cloned likewise, with Sec31 tagged at the N terminus, and modified to create Sec31ΔL (deletion of residues 474–507) or Sec31EE (mutations M540E and L544E).
Protein preparation and crystallization
The proteins were expressed in E. coli strain BL21 (DE3)-RIL (Agilent Technologies) in Luria-Bertani medium, induced with 200 µM isopropyl-β-d-1-thiogalactopyranoside at 18°C. Harvested cells were homogenized at 4°C in 50 mM potassium phosphate, pH 8.5, 400 mM NaCl, 40 mM imidizole, and 5 mM β-mercaptoethanol. The Sec13–Sec16 complex, Sec13–Sec31 complex, or each mutant complex was purified by Ni-affinity chromatography and then dialyzed against 10 mM Tris-HCl, pH 8.5, 250 mM NaCl, 0.5 mM EDTA, and 1 mM DTT. After removing the His6 tag, each complex was further purified on a Superdex 200 26/60 column (GE Healthcare), equilibrated in 5 mM Hepes, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, and 1mM DTT. Selenomethionine-substituted protein was expressed as described previously (Brohawn et al., 2008).
Sec13–Sec16 was concentrated to 70 mg ml−1. An initial crystallization condition was found by vapor diffusion using commercial screens. Crystals grew as bundles of rods each 50 µm × 100 µm × 200–400 µm, within days, at 16°C in 2-µl hanging drops, over 0.1 mM bis-Tris propane, pH 6.5, 0.2 M NaBr, and 12% polyethylene glycol (PEG) 3,350. Selenomethionine-substituted protein crystallized subsequent to streak seeding with microcrystals of the native protein. Crystals were cryoprotected by serial transfer through reservoir solution supplemented by increasing amounts of PEG 200, 16% final concentration, and then flash frozen in liquid nitrogen.
Sec13–Sec31ΔL was concentrated to 70 mg ml−1, and an initial crystallization condition was found likewise. Crystals grew as rods, 100 µm × 100 µm × 200–400 µm, within days, at 16°C in 2-µl hanging drops, over 0.1 mM bis-Tris propane, pH 6.9, 0.2 M Na citrate, and 16% PEG 3,350. Crystals were cryoprotected by serial transfer through reservoir solution supplemented by increasing amounts of PEG 200, 18% final concentration, and then flash frozen in liquid nitrogen.
Data collection and structure solution
For Sec13–Sec16, diffraction data to 2.7-Å resolution were collected at 100 K at beamline 24-IDC at the Advanced Photon Source (Argonne, IL) and processed with the HKL2000 package (Otwinowski and Minor, 1997). Merging factors Rr.i.m. and Rp.i.m. were calculated with RMERGE (Weiss, 2001). The majority of crystals were orthorhombic, belonging to space group P212121. The selenomethionine crystal that diffracted best belonged to the monoclinic subgroup P21. Phases were determined by Se-SAD. In the P21 crystals, the asymmetric unit contains two Sec13–Sec162–Sec13 heterotetramers. 50 of (4 × 13 =) 52 possible Se sites were identified by PHENIX AutoSol (Adams et al., 2002). Density-modified maps were used to build and assign the structure of the four Sec16 monomers. Phased molecular replacement, as implemented in MolRep (Vagin and Isupov, 2001), was used to position two of four Sec13 monomers (Protein Data Bank ID 2PM6). The other two Sec13 monomers are poorly ordered in the crystal. The symmetry relating the Sec16 monomers was used to infer the location of these Sec13 monomers, for which the observed electron density is weak, as explained in the Results. For refinement, the native dataset in space group P212121 was used. The two crystal forms are nearly the same: the noncrystallographic symmetry operator relating two heterotetramers in space group P21 becomes a crystallographic symmetry operator in P212121, and consequently, the asymmetric unit of the native crystal contains only one heterotetramer. In Sec16, the C terminus (residues 1391–1421) and one loop (residues 1185–1192) are not structured and were not modeled. The structure was refined against the native data with PHENIX (Adams et al., 2002), using noncrystallographic symmetry restraints and TLS refinement (Rwork/Rfree = 19.4%/25.0%).
For Sec13–Sec31ΔL, diffraction data to 2.8-Å resolution were collected and processed with XDS and XSCALE (Kabsch, 2010). The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) and refined in Refmac (Bailey, 1994; Murshudov et al., 1997), using keyword TWIN to account for partial merohedral twinning of the data (twin fraction = 0.23; Rwork/Rfree = 26.7%/30.0%). Data collection and refinement statistics are summarized in Table I. Figures were created in PyMOL (http://www.pymol.org/). Coordinates and native structure factors were submitted to the Protein Data Bank with accession codes 3MZK and 3MZL.
Analytical ultracentrifugation and size exclusion chromatography
Sedimentation velocity ultracentrifugation was performed on a Beckman Coulter XL-I, at 42 krpm, 20°C, using interference optics in 50 mM potassium phosphate, pH 7.0, 150 mM NaCl, 0.1 mM EDTA, and 1 mM DTT, using Sec13–Sec16 at 0.25 mg ml−1. Data were collected every 1.5 min and fit to a single species model by SEDFIT. No significant residuals were observed, confirming a single, pure species. Size exclusion chromatography was performed on a 10/300 Superdex 200 column in 10 mM Hepes, pH 7.0, 150 mM NaCl, 0.1 mM EDTA, and 1 mM DTT, using 100 µl of sample at 0.25 A280.
Complementation assay
SEC16 or SEC31, with flanking sequence −1500 bp and +500 bp, were cloned into pRS315 and pRS316 (Sikorski and Hieter, 1989). Diploid strains, SEC16/sec16Δ and SEC31/sec31Δ, from the EUROSCARF deletion collection were transformed using the lithium acetate/PEG method with pRS316[SEC16] or pRS316[SEC31]. Cells grown on media lacking uracil were sporulated for 1 wk in 0.02% raffinose and dissected for tetrads. Haploid null strains were selected by G418 resistance. Mutations were introduced to pRS315[SEC16] or pRS315[SEC31] by PCR methods, transformed into the respective null strains, and selected on media lacking leucine. Complementation was tested on media supplemented with 5-fluoroorotic acid, to select against pRS316[SEC16] or pRS316[SEC31]. To test expression of SEC31 variants, pRS316[SEC31] was modified to encode the HA epitope in the C-terminal extension (at aa 817). Protein from cells grown to log phase was extracted according to the method of Kushnirov (2000), blotted onto polyvinylidene fluoride membrane and probed with anti-HA antibody 3F10 (Roche) and an HRP-conjugated secondary antibody. An antibody to 3-phosphoglycerate kinase (Invitrogen) was used to control for equal sample loading.
Online supplemental material
Fig. S1 shows sequence alignment of the CCD of Sec16 homologues. Fig. S2 shows hydrodynamic characterization of Sec13–Sec16. Fig. S3 shows structural consequences of temperature-sensitive alleles of Sec16. Fig. S4 shows a simulated annealing omit map of the Sec16 swap loop. Fig. S5 shows electron density at the Sec31ΔL swap hinge. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003092/DC1.
Acknowledgments
We thank the staff at Beamlines 24-IDC/E at Argonne National Laboratory, especially K. Rajashankar for excellent assistance with data collection; C. Kaiser, E. Spear, and D. Morse for valuable advice; S. Brohawn for critically reading the manuscript; and members of the Schwartz laboratory for discussions. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and National Institutes of Health GM68762) is gratefully acknowledged.
This work was supported by a Pew Scholar award (to T.U. Schwartz). This work is based on research conducted at the Northeastern Collaborative Access Team Beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences under contract number DE-AC02-06CH11357.
Footnotes
Abbreviations used in this paper:
- CCD
- central conserved domain
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- PEG
- polyethylene glycol
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Antonny B., Schekman R. 2001. ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13:438–443 10.1016/S0955-0674(00)00234-9 [DOI] [PubMed] [Google Scholar]
- Antonny B., Madden D., Hamamoto S., Orci L., Schekman R. 2001. Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 3:531–537 10.1038/35078500 [DOI] [PubMed] [Google Scholar]
- Antonny B., Gounon P., Schekman R., Orci L. 2003. Self-assembly of minimal COPII cages. EMBO Rep. 4:419–424 10.1038/sj.embor.embor812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.; Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 10.1107/S0907444993011898 [DOI] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895–907 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Schlunegger M.P., Eisenberg D. 1995. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 4:2455–2468 10.1002/pro.5560041202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D., Glick B.S. 2007. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol. Biol. Cell. 18:839–849 10.1091/mbc.E06-08-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C.S., Schüttelkopf A.W. 2009. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 65:510–512 10.1107/S0907444909007835 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Glick B.S. 2004. The mechanisms of vesicle budding and fusion. Cell. 116:153–166 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Brohawn S.G., Schwartz T.U. 2009. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat. Struct. Mol. Biol. 16:1173–1177 10.1038/nsmb.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Leksa N.C., Spear E.D., Rajashankar K.R., Schwartz T.U. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 322:1369–1373 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Partridge J.R., Whittle J.R.R., Schwartz T.U. 2009. The nuclear pore complex has entered the atomic age. Structure. 17:1156–1168 10.1016/j.str.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri I., Söding J., Lupas A.N. 2008. Evolution of the beta-propeller fold. Proteins. 71:795–803 10.1002/prot.21764 [DOI] [PubMed] [Google Scholar]
- Connerly P.L., Esaki M., Montegna E.A., Strongin D.E., Levi S., Soderholm J., Glick B.S. 2005. Sec16 is a determinant of transitional ER organization. Curr. Biol. 15:1439–1447 10.1016/j.cub.2005.06.065 [DOI] [PubMed] [Google Scholar]
- Debler E.W., Ma Y., Seo H.S., Hsia K.C., Noriega T.R., Blobel G., Hoelz A. 2008. A fence-like coat for the nuclear pore membrane. Mol. Cell. 32:815–826 10.1016/j.molcel.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2:e380 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade P., Gimeno R.E., Holzmacher E., Teung P., Kaiser C.A. 1995. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J. Cell Biol. 131:311–324 10.1083/jcb.131.2.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane B.A., Prevelige P.E., Jr 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 64:259–299 10.1016/S0065-3233(03)01007-6 [DOI] [PubMed] [Google Scholar]
- Fath S., Mancias J.D., Bi X., Goldberg J. 2007. Structure and organization of coat proteins in the COPII cage. Cell. 129:1325–1336 10.1016/j.cell.2007.05.036 [DOI] [PubMed] [Google Scholar]
- Fromme J.C., Schekman R. 2005. COPII-coated vesicles: flexible enough for large cargo? Curr. Opin. Cell Biol. 17:345–352 10.1016/j.ceb.2005.06.004 [DOI] [PubMed] [Google Scholar]
- García De La Torre J., Huertas M.L., Carrasco B. 2000. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78:719–730 10.1016/S0006-3495(00)76630-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R.E., Espenshade P., Kaiser C.A. 1995. SED4 encodes a yeast endoplasmic reticulum protein that binds Sec16p and participates in vesicle formation. J. Cell Biol. 131:325–338 10.1083/jcb.131.2.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R.E., Espenshade P., Kaiser C.A. 1996. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol. Biol. Cell. 7:1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn A.M. 2009. Protein acrobatics in pairs—dimerization via domain swapping. Curr. Opin. Struct. Biol. 19:39–49 10.1016/j.sbi.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürkan C., Stagg S.M., Lapointe P., Balch W.E. 2006. The COPII cage: unifying principles of vesicle coat assembly. Nat. Rev. Mol. Cell Biol. 7:727–738 10.1038/nrm2025 [DOI] [PubMed] [Google Scholar]
- Hsia K.-C., Stavropoulos P., Blobel G., Hoelz A. 2007. Architecture of a coat for the nuclear pore membrane. Cell. 131:1313–1326 10.1016/j.cell.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Stephens D.J. 2008. Assembly, organization, and function of the COPII coat. Histochem. Cell Biol. 129:129–151 10.1007/s00418-007-0363-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Budnik A., Schmidt K., Palmer K.J., Mantell J., Noakes C., Johnson A., Carter D.A., Verkade P., Watson P., Stephens D.J. 2009. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J. Cell Sci. 122:2924–2934 10.1242/jcs.044032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V., de Voer G., Xanthakis D., Spoorendonk K.M., Kondylis V., Rabouille C. 2008. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell. 19:4352–4365 10.1091/mbc.E08-03-0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S., Schwartz T.U. 2007. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J. Biol. Chem. 282:34904–34912 10.1074/jbc.M705479200 [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., Schekman R. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 61:723–733 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. 2000. Rapid and reliable protein extraction from yeast. Yeast. 16:857–860 [DOI] [PubMed] [Google Scholar]
- Liu Y., Eisenberg D. 2002. 3D domain swapping: as domains continue to swap. Protein Sci. 11:1285–1299 10.1110/ps.0201402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias J.D., Goldberg J. 2005. Exiting the endoplasmic reticulum. Traffic. 6:278–285 10.1111/j.1600-0854.2005.00279.x [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. 1998. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 93:263–275 10.1016/S0092-8674(00)81577-9 [DOI] [PubMed] [Google Scholar]
- McCoy A., Grosse-Kunstleve R., Adams P., Winn M., Storoni L., Read R. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. In Macromolecular Crystallography, part A. Methods in Enzymology, Vol. 276 Carter C.W., Jr., Sweet R.M., Academic Press, New York: 307–326 [DOI] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz J.W.H., Itzhaki L.S. 2003. The unfolding story of three-dimensional domain swapping. Structure. 11:243–251 10.1016/S0969-2126(03)00029-7 [DOI] [PubMed] [Google Scholar]
- Salama N.R., Yeung T., Schekman R.W. 1993. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 12:4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nakano A. 2004. Reconstitution of coat protein complex II (COPII) vesicle formation from cargo-reconstituted proteoliposomes reveals the potential role of GTP hydrolysis by Sar1p in protein sorting. J. Biol. Chem. 279:1330–1335 10.1074/jbc.C300457200 [DOI] [PubMed] [Google Scholar]
- Schrader N., Stelter P., Flemming D., Kunze R., Hurt E., Vetter I.R. 2008. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell. 29:46–55 10.1016/j.molcel.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Schwartz T.U., Schmidt D., Brohawn S.G., Blobel G. 2006. Homodimerization of the G protein SRbeta in the nucleotide-free state involves proline cis/trans isomerization in the switch II region. Proc. Natl. Acad. Sci. USA. 103:6823–6828 10.1073/pnas.0602083103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz D.A. 1997. Towards an understanding of the role of COPII subunits in vesicle coat assembly. PhD thesis. Massachusetts Institute of Technology, Cambridge, MA: 171 pp [Google Scholar]
- Shaywitz D.A., Espenshade P.J., Gimeno R.E., Kaiser C.A. 1997. COPII subunit interactions in the assembly of the vesicle coat. J. Biol. Chem. 272:25413–25416 10.1074/jbc.272.41.25413 [DOI] [PubMed] [Google Scholar]
- Siegel L.M., Monty K.J. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362 10.1016/0926-6585(66)90333-5 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E.C. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84:265–275 10.1016/S0092-8674(00)80981-2 [DOI] [PubMed] [Google Scholar]
- Stagg S.M., Gürkan C., Fowler D.M., LaPointe P., Foss T.R., Potter C.S., Carragher B., Balch W.E. 2006. Structure of the Sec13/31 COPII coat cage. Nature. 439:234–238 10.1038/nature04339 [DOI] [PubMed] [Google Scholar]
- Stagg S.M., LaPointe P., Balch W.E. 2007. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 17:221–228 10.1016/j.sbi.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Stagg S.M., LaPointe P., Razvi A., Gürkan C., Potter C.S., Carragher B., Balch W.E. 2008. Structural basis for cargo regulation of COPII coat assembly. Cell. 134:474–484 10.1016/j.cell.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Madden D.T., Hamamoto S., Orci L., Schekman R. 2002. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J. Cell Biol. 158:1029–1038 10.1083/jcb.200207053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.L., Wang Y., Ong Y.S., Hong W. 2005. COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta. 1744:293–303 10.1016/j.bbamcr.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Thuman-Commike P.A., Greene B., Malinski J.A., King J., Chiu W. 1998. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys. J. 74:559–568 10.1016/S0006-3495(98)77814-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A.A., Isupov M.N. 2001. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 57:1451–1456 10.1107/S0907444901012409 [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Townley A.K., Koka P., Palmer K.J., Stephens D.J. 2006. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 7:1678–1687 10.1111/j.1600-0854.2006.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.S. 2001. Global indicators of X-ray data quality. J. Appl. Cryst. 34:130–135 10.1107/S0021889800018227 [DOI] [Google Scholar]
- Wlodawer A., Minor W., Dauter Z., Jaskolski M. 2008. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 275:1–21 10.1111/j.1742-4658.2008.06444.x [DOI] [PMC free article] [PubMed] [Google Scholar]