Abstract
This paper describes the application of a molecular construct of a photosensitizer and an aptamer for photo-therapeutically targeting tumor cells. The key step in increasing selectivity in chemotherapeutic drugs is to create effective molecular platforms that could target cancer cells but not normal cells. Recently, we have developed a strategy via cell-SELEX (Systematic Evolution of Ligands by Exponential Enrichment) to obtain cell specific aptamers using intact viable cells as targets to select aptamers that can recognize cell membrane proteins with high selectivity and excellent affinity. We have identified an aptamer TD05 that only recognizes Ramos cells, a Burkitt’s lymphoma cell line. Here, the high specificity of aptamers in target cell binding and an efficient phototherapy reagent, Ce6, are molecularly engineered to construct a highly selective Aptamer-photosensitizer conjugates (APS) to effectively destroy target cancer cells. Introduction of the APS conjugates followed by irradiation of light selectively destroyed target Ramos cells but not acute lymphoblastic leukemia and myeloid leukemia cell lines. This study demonstrates that the use of cancer specific aptamers conjugated to a photosensitizer will enhance the selectivity of photodynamic therapy. Coupled with the advantages of the cell-SELEX in generating multiple effective aptamers for diseased cell recognition, we will be able to develop highly efficient photosensitizer based therapeutical reagents for clinical applications.
Keywords: anti-tumour probes, aptamers, photodynamic therapy, photosensitizers, targeted therapy
We have molecularly engineered an aptamer photosensitizer using cell specific aptamers and a photosensitizer reagent, Ce6, for highly selective phototherapy targeting tumor cells. Photodynamic therapy (PDT) cleverly exploits the ability of a photosensitizer (PS) to generate reactive oxygen species upon irradiation with an appropriate wavelength of light.[1] Unlike typical drugs, the toxicity of a PS results from the interaction of excited photosensitizer with neighboring oxygen that produces cytotoxic reactive oxygen species (ROS).[2] Recently, PDT based therapeutic approaches have garnered much attention; however, one of the biggest challenges in using PS is the effective localization of the PS at the diseased site. By improving the co-localization, it is feasible to increase efficiency of PDT. Also, the hydrophobic nature of most photosensitizers makes it difficult to dissolve them in typically aqueous, biological fluids. The key in addressing these issues is the generation of water soluble, tumor selective probes coupled with PS prior to treatment. To address this problem, several strategies such as immuno-conjugates, peptide or protein conjugates, as well as photo-sensitizer encapsulated nano-carriers have been used.[3–8] However, these methods have drawbacks as conjugation with antibodies or proteins is tedious and the shelf life of protein conjugates in general is short. Also, cross-reactivity of the cargo can significantly lower the selectivity.
One way of generating effective molecular probes to differentiate target cells is to use aptamers, by which molecular level differences between a healthy cell and a diseased cell can be explored.[9] This strategy will generate a panel of molecular probes capable of differentiating various types of cells and make PDT specifically targeted. Owing to their many significant advantages (including small size, easy chemical synthesis with high reproducibility, easy chemical manipulation, and low immunogenity), aptamers can be applicable in cancer therapy. Aptamers, which are designer DNA/RNA probes,[10–11] are considered to be one of the most effective molecular probes in selective identification of one specific type of cancer cells. Furthermore several reports have shown that the selectivity of aptamers can be effectively used as a way to therapeutically target cancer cells.[12–14] These methods mainly focus on inducing cell toxicity by internalized aptamer-toxin conjugates which needed to be co-localized in a specific cellular compartment.
Recently, we have developed an effective method to generate aptamer based molecular probes for the specific recognition and targeting of cancer cells.[9] Using this new strategy, referred to as cell-SELEX (cell based Systematic Evolution of Ligands by EXponential enrichment), we have selected a panel of aptamers that can uniquely identify a given set of cancer cells in complex biological mixtures and even in patient samples.[15–16] The selectivity of these probes originates from the aptamer selection process where positive and negative selection processes are coupled to produce the most specific aptamers for targeting tumor cells. These probes can be optimized to have better binding properties.[17] They can also be used to identify different expression patterns of the membrane proteins in a variety of cell types. These unique recognition patterns will assist in developing effective therapeutic and diagnostic tools.
Using cell-SELEX, we have identified an aptamer TD05.[15] Through this work, we demonstrate that highly specific aptamers can be selected for targeting one particular cell line with high affinity. Interestingly, among a significant number of selected aptamers, TD05 was unique in that it specifically recognizes target Ramos cells, a Burkitt’s lymphoma cell line (Figure 1).[15]
Figure 1. Selectivity and affinity comparison of TD05 before and after Ce6 conjugation.

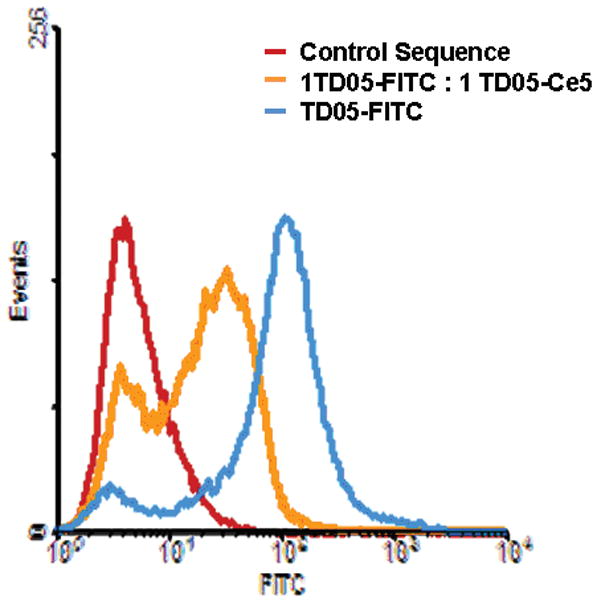
(Top) Flow cytometry analysis of FITC labeled TD05 with myeloid leukemia cell lines K-562, NB-4 and HL 60. TD05 only binds with target Ramos. (Bottom): Competitive binding of TD05 with Ramos cells for TD05 before and after Ce-6 conjugation. Shift to lower fluorescence after competitive binding indicates that both probes bind to the cells with similar affinities.
Compared to Ramos cells, TD05 binding with control cells such as K-562 cells, NB-4 cells, HL60 cells and CEM cell is low.[15] We hypothesized that this excellent specificity of TD05 could be used to selectively target Ramos cells by effectively localizing a photosensitizer on the cell membrane prior to light illumination. Advantages of the conjugation of a photosensitizer with a negatively charged aptamer are two-fold: (1) Effective co-localization of the photosensitizer will increase the efficiency and specificity of PDT; (2) Coupling of negatively charged DNA increases the aqueous solubility of a photosensitizer. Thus, we have combined the high selectivity of TD05 with easy chemical manipulation of DNA to develop a highly selective aptamer photosensitizer (APS) which can effectively destroy aptamer bound cancer cells.
The chemical conjugation of TD05 with chlorin e6 (Ce6), a porphyrin based photosensitizer, was achieved using an amine modified TD05 (Supplementary Figure 1). The confirmation of conjugation of TD05 with Ce6 was done by observing the absorbance at 260 nm for DNA, and 404 nm for Ce6 (Supplementary Figure 2). The effect of conjugation of TD05 aptamer with Ce6 on generation of reactive oxygen species (ROS) was evaluated according to the manufacture’s protocol using a commercially available singlet oxygen sensor green reagent. Compared with free ce6, TD05-ce6 conjugate did not show any loss in the generation of ROS (Supp. Figure 3).
Next, we evaluated whether this conjugation had affected the selective binding of TD05 aptamer with target Ramos cells. We performed an in vitro binding study using flow cytometry (FACS) with FITC labeled TD05 and Ce6 labeled TD05 (Figure 1B). By observing the fluorescence shift resulting from competition between the original TD05-FITC probes in the presence of TD05-Ce6 conjugate, it was found that the binding of TD05-FITC does shift about 50% lower in fluorescence, suggesting that the TD05-Ce6 still recognize Ramos cells with selectivity similar to that of FITC-TD05. In addition, binding of TD05-Ce6 conjugates were analyzed with control cells using FACS counting 10,000 events at 404 nm excitation (supp. Figure 4). The observed binding pattern of TD05-ce6 was similar to that of TD05-FITC confirming that the specificity of TD05 retained. After confirming the binding affinity of TD05-Ce6 conjugate, we next evaluated whether TD05-Ce6 can selectively target Ramos cells upon illumination of light. We used CEM, a T-cell acute lymphoblastic cell line and three myeloid leukemia cell lines K562, HL-60, NB-4 as the control. The cells were incubated with 250 nM of TD05-Ce6 conjugate in the dark for 30 min. The Kd for TD05 was evaluated to be 75 nM;[15] we thus used a TD05-Ce6 in excess of its Kd to ensure that the cells are properly stained with TD05-Ce6 conjugate. Then the unbound and non-specifically attached probes were washed away prior to illumination of light. Due to the long absorbance range of Ce6, we illuminated the samples using a white fluorescent light (fluence rate=2.88 J cm2) for 4 h. Cell viability was determined 36 h after illumination by measuring propidium iodide incorporation using flow cytometry (FACS, 10,000 cell events) to gauge the cytotoxicity of the treatments. Separate control experiments were carried out using unmodified TD05 and free Ce6. The free aptamer without Ce6, free Ce6 without aptamer and cells exposed to light did not show any significant toxicity.
Based on the initial selectivity studies with TD05 probe with control cell lines (Figure 1A), we expected to observe similar toxicity patterns with the TD05-Ce6 conjugate. We observed TD05-Ce6 toxicity had induced cell death in Ramos cells (71.3% ± 6.9) which strongly correlated with our initial binding patterns. The toxicity observed in control CEM cells (35.8% ± 7.4), K562 (30% ± 3. 37), NB4 (36.4±7.09) and HL60 (<1%) cell lines are over 50% less than the targeted cell lines. This suggests that the specific interaction of TD05-Ce6 on the cell membrane could induce selective cell death by phototoxicity. The calculated P value for the TD05-Ce6 induced cell toxicity is 0.02 suggesting the statistical significance of the TD05-ce6 induced cell death. The slight toxicity of aptamer PS conjugate toward the CEM, K-562, and NB-4 could be due to non-specific interactions with the cell membrane. While un-conjugated Ce6 is insoluble in aqueous media and is washed away; the conjugation of the DNA aptamer with Ce6 dramatically increases its aqueous solubility, resulting in an increased interaction of the hydrophobic portion of Ce6 with cell membrane.
We next investigated whether the demonstrated toxicity induced from APS conjugate by interaction of aptamer-with its target protein or due to toxicity induced by light. The un-irradiated samples did not show any significant toxicity towards targeting Ramos cells, suggesting that the APS conjugate is not toxic in the absence of light (Supplementary Figure 5). Also, we further confirmed specific toxicity using a random DNA sequence attached to the PS followed by treatment of respective cell lines (Supplementary Figure 6). Random DNA labeled with Ce6 did not show any toxicity towards Ramos cells; however, 26% ± 2.15% decrease in cell viability was observed for CEM cells. This could be due to non-specific interaction of DNA attached to Ce6 with the CEM cell membrane. Taken all these together, our observed results suggest that TD05-Ce6 conjugates could selectively induce cell death towards targeting Ramos cells. Also, the incubation of the aptamers at 4°C and washing away the free probe prevent endocytosis of the aptamer or free drug conjugates. The observed poor therapeutic efficacy can be addressed by conjugating multiple ce6 molecules using a multiple amino linker linked to the DNA aptamer. Despite the poor therapeutic efficacy, this study has demonstrated the feasibility of this approach in targeted cell treatment in a therapeutical setting. Photo-active therapeutic approaches have great potential for cancer therapy. However, due to limited therapeutic windows, the long-term administration of photo-active drugs is usually not possible. For this reason, conjugation of a photosenstizer with a targeting molecular probe that can be localized at the tumor site will enhance the therapeutic effectiveness. Here, we have exploited the specific recognition capability of DNA aptamers evolved from live cells to effectively target one type of cancer cells. Since the photosensitizer is covalently attached, it also can provide stability in the cellular environment. The observed selectivity of evolved aptamers can be further utilized to selectively kill target cells.
In conclusion, we have shown that aptamers selected using cell-SELEX strategy can selectively target one specific cell line. The binding affinity and selectivity of the aptamer after conjugation with a PS did not alter significantly. This suggests that the versatility of the aptamer TD05 for detection and selective targeting. Pronounced toxicity of TD05-Ce6 conjugate with target cells strongly correlates with observed initial binding patterns of TD05. Thus, by harnessing the advantages of cell-SELEX in generating multiple effective aptamers for diseased cell recognition, we will be able to develop highly efficient photosensitizer based therapeutic reagents for clinical applications. In the next phase of our studies, we will be using a few liver and/or lung cancer specific APS conjugates to target solid tumors.
Experimental Section
Synthesis of Conjugate
Amine modified TD05 was synthesized using standard phosphoimidite chemistry and probes were purified using reversed phase HPLC. The Conjugation of photodynamic ligand Chlorin e6 (Ce6) (Frontier Scientific, Logan, UT) with amine modified TD05 was done using N-hydroxysuccinimide ester (NHS) of Ce6 and using dicyclohexyl carbodiimide (DCC) as a coupling agent. Briefly, an equimolar of NHS and Ce6 along with equimolar of DCC was dissolved in anhydrous DMF in the dark for 30 min. Activated Ce6 was then added to amine modified TD05 in NaHCO3 at pH 7 by vigorously stirring overnight in the dark. The un-conjugated Ce6 was removed by ethanol precipitation of DNA for 5 times. Resulting crude mixture was separated by HPLC. Quantification of the conjugated DNA and Ce6 was done by measuring the absorbance at 260 nm and 404, 643 nm respectively.
Characterization of Conjugate
CCRF-CEM (CCL-119, T cell line, human Acute Lymphoblastic Leukemia), Ramos (CRL-1596, B-cell line, human Burkitt’s lymphoma), CA46 (CRL 1648, B-Cell line, Human Burkitt’s lymphoma), Jurkat (,TIB-152, human acute T cell leukemia) Toledo (CRL-2631, B-cell line, human diffuse large-cell lymphoma), K562 (CCL-243, chronic myelogenous leukemia (CML), and NB-4, and HL-60 (CCL-240, acute promyelocytic leukemia), were obtained from American Type Culture Collection. All of the cells were cultured in RPMI medium 1640 (American Type Culture Collection) supplemented with 10% FBS (heat-inactivated; GIBCO) and 100 units mL1 penicillin-streptomycin (Cellgro). Cells were washed before and after incubation with wash buffer (4.5 gL1 glucose and 5 mM MgCl2 in Dulbecco’s PBS with calcium chloride and magnesium chloride; Sigma). Binding buffer used for selection was prepared by adding yeast tRNA (0.1 mg mL1; Sigma) and 1 mg mL1 sheared salmon sperm DNA into wash buffer to reduce non-specific binding.
In Vitro Photolysis
Ramos cells and control cell lines were washed with the cold binding buffer, and incubated with 250 nM of TD05-ce6 probe at 4 °C for 20 min, Cells were then washed with wash buffer, exposed to white fluorescent light (fluence rate=2.88 J cm2) for 4 h, and then re-cultured in RPMI-1640 for 36 h. The cell viability was determined by popidium iodide incorporation (Invitrogen). Experiments were repeated three times and each time with triplicates. Highly concentrated free Ce6 solution was made by dissolving in DMF, stock solution was subsequently diluted appropriated experiments. The optimal exposure times of cells to TD05-Ce6 conjugate and fluorescent light were determined preliminary time-course experiments (data not shown).
Figure 2.
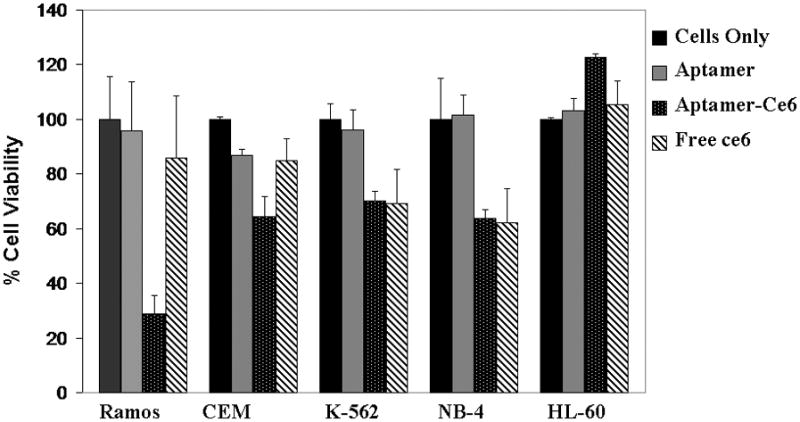
Cell toxicity assay results for Ramos cells (P <0.05) after 30 min incubation followed by irradiation of light for 4 h, and subsequent growth for 36 h. Target cell is Ramos, and control cell lines are CEM, K-562, NB-4 and HL-60.
Acknowledgments
We thank Dr. Ying Li and Dr. Zehui Charles Cao for interesting discussions during the process of this research. This work was supported by the NIH R01 GM079359 and NIH U54 NS 058185 grants and the State of Florida Bankhead-Coley Cancer Research Program, 07BB-10.
Footnotes
Supporting information for this article is available on the WWW under http://www.chemmedchem.org or from the author.
References
- 1.Dennis EJ. Nat Rev Cancer. 2003;3:380–387. doi: 10.1038/nrc1071. [DOI] [PubMed] [Google Scholar]
- 2.Castano AP. Nat Rev Cancer. 2006;6:535–545. doi: 10.1038/nrc1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oseroff AR. Proc Natl Acad Sci USA. 1986;83:8744–8748. doi: 10.1073/pnas.83.22.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogura SI. J Controlled Release. 2005;103:1–6. doi: 10.1016/j.jconrel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Laptev R. Br J Cancer. 2006;95:189–196. doi: 10.1038/sj.bjc.6603241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi Y. Cancer Res. 2006;66:7225–7229. doi: 10.1158/0008-5472.CAN-06-0448. [DOI] [PubMed] [Google Scholar]
- 7.Governatore MD. Br J Cancer. 2006;82:56–64. [Google Scholar]
- 8.Kim S. J Am Chem Soc. 2007;129:2669–2675. doi: 10.1021/ja0680257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shangguan D. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborn SE. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 11.Klussmann S. The Aptamer Handbook: Functional Oligonucleotides and Their Applications. Wiley-VCH; Weinheim: 2006. p. 150. [Google Scholar]
- 12.Chu TC. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 13.Chu TC. Nucleic Acids Res. 2000;28(10):e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagalkot V. Angew Chem. 2006;118:8329–8332. [Google Scholar]; Angew Chem Int Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 15.Tang Z. Anal Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 16.Shangguan D. Clin Chem. 2007;53:1153–1156. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 17.Shangguan D, et al. Chem Bio Chem. 2007;8:603. [Google Scholar]


