Abstract
Purpose
To develop a reliable preimplantation genetic diagnosis protocol for couples who both carry a mutant PKHD1 gene wishing to conceive children unaffected with autosomal recessive polycystic kidney disease (ARPKD).
Methods
Development of a unique protocol for preimplantation genetic testing using whole genome amplification of single blastomeres by multiple displacement amplification (MDA), and haplotype analysis with novel short tandem repeat (STR) markers from the PKHD1 gene and flanking sequences, and a case report of successful utilization of the protocol followed by successful IVF resulting in the birth of an infant unaffected with ARPKD.
Results
We have developed 20 polymorphic STR markers suitable for linkage analysis of ARPKD. These linked STR markers have enabled unambiguous identification of the PKHD1 haplotypes of embryos produced by at-risk couples.
Conclusions
We have developed a reliable protocol for preimplantation genetic diagnosis of ARPKD using single-cell MDA products for PKHD1 haplotyping.
Keywords: Preimplantation genetic diagnosis, Autosomal recessive polycystic kidney disease, Multiple displacement amplification, PKHD1 haplotype analysis, Linkage analysis with linked STR markers
Introduction
Polycystic Kidney Diseases (PKD) comprise a group of monogenic disorders that result in renal enlargement and functional impairment secondary to the development of cystic lesions and fibrosis. In addition, characteristic extrarenal manifestations occur with the two major genetic cystic diseases, namely Autosomal Dominant PKD (ADPKD) and Autosomal Recessive PKD (ARPKD) [1, 2]. Worldwide, it is estimated that over 12.5 million individuals have PKD, making it a major source of morbidity and mortality. Despite intense active research, there is currently no disease-specific therapy for PKD. Clinicians can improve symptoms and prolong life through treatment of hypertension, anticipation and monitoring of extrarenal complications, and utilization of end-stage renal disease therapy (i.e. dialysis and transplantation) [1].
Considering all age ranges, ADPKD makes up 95–98% of all cases of PKD [3, 4]. ADPKD generally causes symptoms in adulthood, but is increasingly recognized in children where it can be difficult to distinguish from ARPKD [2]. ARPKD makes up 2–5% of all cases of PKD, but more than 75% of all PKD cases that present clinically in the first month of life [4]. Indeed, ARPKD is an important cause of kidney failure and biliary plate abnormalities leading to congenital hepatic fibrosis in neonates and infants, and has an estimated mortality rate of 25% in the first 30 days of life despite optimal neonatal care [2, 4]. The prevalence of ARPKD is approximately 1 in 20,000 live births, with a carrier frequency of 1 in 70 individuals [4, 5].
ADPKD is genetically heterogeneous with two genes identified, namely, PKD1 and PKD2 [6–8]. The PKD1 gene is located in the chromosome 16p13.3 region and encodes polysystin-1 [8]. The PKD2 gene is located in the chromosome 4q21 region and encodes polysystin-2 [7]. The gene for ARPKD, known as Polycystic Kidney and Hepatic Disease 1 (PKHD1), is approximately 472 kb and consists of 67 exons [9, 10]. It is located in the chromosome 6p12.2 region, and encodes a unique 450 kDa protein, “fibrocystin”, which consists of 4,074 amino acid residues.
Preimplantation genetic diagnosis (PGD) is a procedure to analyze the genetic make-up of embryos formed through in vitro fertilization (IVF) for chromosome abnormalities, single gene disorders and HLA haplotypes [11, 12]. Conventionally, PGD for single gene disorders is performed by linkage analysis with linked short tandem repeat (STR) markers. This can be combined with direct detection of mutations when there are mutational hotspots and when the specific gene mutations are known [13].
Mutation testing is generally used when the genetic disorder is the result of a single mutation that occurs in most affected families such as spinal muscular atrophy [14] and sickle cell anemia [15]. This allows a single assay to be used repeatedly. In order to avoid misdiagnosis due to allele drop-out (ADO) and potential contamination, STR linkage analysis is indispensible even when the approach of direct mutation detection is employed.
Most single gene disorders, e.g. ARPKD [16], are the result of many different mutations so that most families harbor private mutations. PGD of these disorders is performed by linkage analysis exclusively [16, 17]. Because of the large number of mutations, the development of customized PGD assay protocols for specific mutations is impractical, labor intensive, expensive and time consuming [18, 19].
In order to carry out linkage analysis several polymorphic STR markers must be identified within and flanking the gene of interest. Family-specific informative linked STR markers are then identified and the haplotypes associated with the normal and mutant alleles determined. The most common method involves multiplex PCR of short tandem repeats (STR) in either one [20–22] or two rounds of PCR [23–25]. If only a single round of PCR is used in PGD, it is necessary to set up specific PCR protocols on single cells for every combination of PCR primers used, since optimal amounts of individual PCR primers and numbers of PCR cycle are critical in multiplex PCR [20, 21]. If nested PCR is used in PGD, one still need to test on single cells to determine whether the particular combination of PCR primers selected for a family will yield reliable results for each loci tested [25].
When nested and fluorescent PCR are used in PGD it is often difficult to choose a set of PCR primers for mutation-specific loci and linked genetic markers because of the interaction among primers in multiplex PCR. This interaction often results in a long and tedious optimization process before a robust mixture of primers can be found for each patient family [25]. An alternative approach that bypasses the need for multiplex PCR is whole genome amplification (WGA) of the DNA from a single biopsied blastomere. In this universal method, multiplex PCR is replaced by whole genome amplification of single blastomeres, e.g. using multiple displacement amplification (MDA) with phosphorothioate-modified random hexanucleotide primers [26–29]. MDA generates enough DNA from a single cell for the evaluation of many individual loci, each in separate PCR reactions. The amplified products can be used for STR genotyping and specific mutation detection. We have adapted this approach to PGD of the PKHD1 gene. The use of MDA products for STR genotyping results in a simple and generalized method for linkage analysis of PKHD1 for all families with ARPKD.
Using DNA amplified from single cells by MDA as templates, the average allele drop-out (ADO) rates of STR genotyping by PCR is reportedly 20.8–34% [26, 30–33]. Despite the fact that high ADO rates of PGD testing using MDA products derived from single cells could lead to misdiagnosis [34], the reliability of this approach has been demonstrated by many successful PGD protocols using single-cell MDA products provided that a sufficient number of STR markers are used [32, 35, 36].
The reportedly high ADO rates for single-cell genotyping using MDA products as template are the consequences of amplification bias among loci and incomplete genomic coverage by MDA. This is a known shortcoming of this technology [37]. Because single genome amplification is not a standardized application of MDA by the manufacturers, we report optimization of the conditions for cell lysis and every step of the MDA protocol for single cell application. Other critical factors in the development of an integrated PGD procedure that lowers the ADO rates allowing unambiguous PGD will be discussed.
Linkage analysis of the PKHD1 gene by STR markers has been used in prenatal diagnosis of ARPKD [16]. Our immediate goal was to identify additional highly polymorphic STRs within the PKHD1 gene and flanking regions for PGD linkage analysis. While whole genome amplification has been used in PGD of other genetic diseases [36, 38–40], this is the first time it has been used in PGD for ARPKD.
Methods and materials
Patient
The patient was 29 year old who presented to an academic infertility service for recommendations, pre-conceptual counseling and planning. The patient and her male partner were carriers for ARPKD. Her first pregnancy ended in a neonatal death secondary to complications associated with ARPKD. Autopsy, renal histology and DNA studies confirmed the diagnosis of ARPKD. A second pregnancy was terminated after prenatal testing confirmed an affected child. Both pregnancies were found to be homozygous for the c.107C>T (GenBank entry: NM_138694.3; p.T36M (GenBank entry: NP_619639.3)) missense mutation in the PKHD1 gene. The patient and her partner were counseled about and agreed to in vitro fertilization (IVF) with PGD for ARPKD.
The patient underwent an IVF protocol that consisted of a birth control pill preparatory cycle for 3 weeks, followed by pituitary down regulation with a gonadotropin hormone releasing agonist. This was then followed by continued use of the gonadotropin hormone releasing agonist with the addition of a combination of 75 IU of recombinant follicle stimulating hormone (FSH) and 75 U of a highly purified human menopausal gonadotropin (HMG) for a total of 10 days of stimulation. Transvaginal ultrasound monitoring and estradiol measurements were performed to assess follicular maturity with a peak estradiol level of 4327 pg/mL on the last day of stimulation. Human chorionic gonadotropin was administered when there were two lead follicles of 18 mm in average diameter and transvaginal ultrasound guided oocyte retrieval was performed 34 h later under monitored anesthetic care. Fourteen oocytes were recovered and 11 mature oocytes underwent intra-cytoplasmic sperm injection (ICSI). All 11 oocytes fertilized and 7 embryos were subsequently biopsied at the 8-cell stage.
Screening for polymorphic STRs from PKHD1 gene and flanking regions
STRs for PKHD1 gene and flanking sequences were identified from human genomic sequence using the online program Tandem Repeats Finder [41], and PCR primers were designed for selected STRs using Oligo™ software (Molecular Biology Insights, Cascade, CO) as previously described [17]. Unlabeled PCR primers used for STR genotyping and research studies were synthesized and desalted by Invitrogen (Carlsbad, CA). The 5′ end of these reverse PCR primers carries a pig-tail [42].
All specimens were collected following institutional review board approval and after informed consents were obtained. STR analysis of patient families and controls were performed using genomic DNA isolated from patient peripheral whole blood specimens or control lymphoblastoid cell lines by the salting-out method (Roche DNA Isolation Kit for Mammalian Blood, Indianapolis, IN). A 2X Multiplex PCR PreMix (Qiagen Multiplex PCR system, Valencia, CA) was used in all PCR reactions, which were carried out in GeneAmp Veriti or PCR System 9800 (Applied Biosystems, Foster City, CA). For STR genotyping of individuals, a 20-µL PCR reaction was carried out for each STR by combining 5 ng of genomic DNA sample with 10 µL of Qiagen Multiplex PCR PreMix and 20 pmol of each PCR primer. These STR primer pairs were optimized at annealing temperatures of 50°C 53°C, 55°C, 58°C and 61°C. For primers optimized at annealing temperature of 50°C, the initial denaturation lasted 14 min at 95°C followed by 30 PCR cycles. Cycles 1–10 consisted of 1 min at 95°C, 30 s at 50°C, and 30 s at 72°C. Cycles 11–30 consisted of 1 min at 94°C, 30 s at 50°C, and 30 s at 72°C. The cycles were followed by a final extension of 30 min at 60°C. For PCR primers optimized at other annealing temperatures, the same thermal profile was used except that the annealing temperature was changed.
For preliminary screening for polymorphic STRs, PCR products were separated on 8% or 10% (w/v) Novex 1X TBE polyacrylamide gels that were 8 cm long and 1 mm thick (Invitrogen, Carlsbad, CA) by electrophoresing at 200 V for 50 min to 1 h 30 min, as previously described [17]. The gels were post-stained with 1X SYBR Green I Nucleic Acid Gel Stain (Lonza, Rockland, ME). Gel images were captured by AlphaImager 3400 (Alpha Innotech, San Leandro, CA) using a camera equipped with SYBR Green I filter (Alpha Innotech) under UV 302 nm trans- and epi-illumination.
Our initial collection of 121 STRs consisted of 24 STRs within the PKHD1 gene, 52 STRs within 1000 kb of the 5′ end of the gene, and 45 STRs within 500 kb of the 3′ end of the gene. From this initial collection, 21 highly polymorphic STRs were selected based on their heterozygosity in the human population. These resulting STRs consisted of 6 intragenic STRs, 8 STRs in the 5′ flanking sequence, and 7 STRs in the 3′ flanking sequence.
PKHD1 haplotype analysis of control and patient families
Fluorescent PCR was used in the STR genotyping analysis of control and patient families, and in the subsequent PGD assays. The 5′ end of forward PCR primers were labeled with 6-FAM, and the 5′ end of reverse PCR primers carried a pig-tail. These PCR primers were synthesized, HPLC purified and characterized by mass spectrometry by Idaho Technology IT-BioChem (Salt Lake City, Utah).
The 21 polymorphic STRs identified were validated by examining the inheritance of alleles in ten anonymous local control families (each family consists of the parents and one child). A STR is considered to be informative for a family if both parents were heterozygous for that marker, but were different in allelic sizes of one or both STRs, so that it would be possible to distinguish paternal and/or maternal alleles of PKHD1 in the children.
STR genotyping of control and patient families by fluorescent PCR was similar to the initial screening for STRs with unlabeled PCR primers, except that 50 ng of genomic DNA was used as template for PCR. The thermal profiles were also similar to that for STR genotyping with unlabeled PCR primers, except that a total of 24 PCR cycles were used instead. The purification of PCR products by Montage* PCR µ96 Filter Plate (Millipore, Billerica, MA) and CE analysis on an 3730xl DNA Analyzer (Applied Biosystems) were performed as previously described [17].
One of the STRs located in the 5′ flanking region was uninformative in all 10 families and was not subject to further study. The remaining 20 informative STRs were further evaluated in single lymphoblastoid cell analysis. Alkaline lysis of single lymphoblastoid cells, whole genome amplification by MDA, and STR genotyping using MDA products were performed exactly as described for single blastomere cells in the following section of “Preimplantation Genetic Diagnosis”. These 20 STR markers, consisted of 6 intragenic markers, 7 markers within 1000 kb of the 5′ end of the gene, and 7 markers within 500 kb of the 3′ end of the gene, were found to be suitable for single cell analysis and thus used in subsequent PGD for PKHD1.
For each patient family, STR genotyping was performed as described for the control families by fluorescent PCR. From the collection of 20 polymorphic STRs, eight informative STR markers covering the intragenic PKHD1 region and flanking regions were selected for the subsequent PGD. We used 8 linked STR markers for our PGD testing, but smaller combinations of the described markers may be used. Using the selected linked STR markers PKHD1 haplotypes can be inferred for the affected child or fetus and parents provided that there is no meiotic recombination between the chosen STR markers in either parent. This information can then be used to reliably determine the PKHD1 haplotypes inherited from each parent to embryos. If the affected child, whose haplotypes are used to deduce the haplotypes of the parents, has a recombinant allele between the linked STR markers, the embryos will appear to be recombinants, and their haplotypes are inconsistent with the inferred parental haplotypes [43]. Identification of a recombination in the affected child or fetus can be aided by genotyping the grandparents. Care must be taken to avoid maternal cell contamination when using fetal samples for genotyping to identify the affected haplotype in a family.
Preimplantation genetic diagnosis
Cleavage-stage embryos were obtained using standard procedures of IVF and ICSI. Embryo biopsy was performed on day 3 post-injection [12]. One or two blastomeres were removed from embryos with 8 cells, and one blastomere was removed from embryos with less than 8 cells. Individual blastomeres from each embryo were washed twice in fresh droplets of HEPES-buffered, modified HTF medium, pH 7.4 (Irvine Scientific, Santa Ana, CA) and 5 μL of the last wash droplets were used as blank controls of the assay. The whole genomes of single blastomeres were amplified by MDA.
Each blastomere was transferred to 4 μL of alkaline lysis buffer that was prepared by mixing 1 volume of 1 mol/L KOH (Sigma, St. Louis, MO; Cat. # P4494-50 mL) / 150 mmol/L DTT (Pierce Chem/Thermo Fisher Scientific, Rockford, IL; Cat #20291) with 3 volumes of MDA Sample Buffer (a component of GE illustra GenomiPhi V2 DNA amplification kit, Cat. #25-6600-31). After incubating at 61°C for 10 min, the cell lysate was neutralized by adding 1 μL of 1 mol/L HCl and 0.9 M Tris-HCl, pH 7.4. An additional 15 μL of MDA Sample Buffer was then added to the cell lysate, which was further incubated at 4°C for at least 5 min. MDA reactions of 50 μL were initiated by adding additional 22.5 μL of Reaction Buffer, 5 μL water, and 2.5 μL of Enzyme Mixture to each cell lysate mixture. After incubating at 30°C for 4 h, MDA reactions were terminated by heating at 65°C for 10 min.
The 50-μL MDA reactions were diluted with 50 μL of water, and 3 μL aliquots of the 1:1 diluted products were then used as templates in separate STR genotyping reactions by fluorescent PCR. Forward PCR primers were labeled at the 5′end with 6-FAM, and reverse PCR primers carried a pig-tail at the 5′ end.
The following fluorescent PCR conditions were optimized for single-cell STR genotyping using MDA products. PCR reactions of 20 μL were prepared by adding 20 pmol of each PCR Primer, 10 μL of Qiagen Multiplex PCR PreMix, and 3 μL of the diluted MDA product. STR primer pairs were optimized at annealing temperature of 50°C, 53°C, 55°C, 58°C or 61°C.
For PCR primers optimized at 50°C, the initial denaturation was 95°C for 14 min followed by 25 PCR cycles (except for STR 3-263 k, for which the optimal no. of PCR cycle was 31). Cycles 1–20 consisted of 1 min at 94°C, 90 s at 50°C, and 1 min at 72°C. Cycles 21–25 consisted of 1 min at 94°C, 45 s at 50°C, and 45 s at 72°C. Then a final extension was performed for 30 min at 60°C. The above thermal profile was also used for PCR primers optimized at other annealing temperatures, except that the annealing temperature for all of the PCR cycles was changed accordingly. Purification of PCR products for CE analysis was performed as described in the previous paragraph.
The PCR primer sequences and annealing temperatures of the eight STRs used in our PGD case were the following:
- 5–326 k (58°C):
- [Forward] 5′- GAAAGCTCCAGCCATCTAGCC
- [Reverse] 5′ - GTTTCTTAAGGGTTTGGTTTGTGTGAGTTAAGTCT
- 5–85 k (61°C):
- [Forward] 5′ - AGTTGGGCATGGTGGCGTACAC
- [Reverse] 5′ - GTTTCTTGGTTGCAGCTTACTAATCATTGGATGTT
- 28 k (55°C):
- [Forward] 5′ - TAGGTTGGTTGTTCCTTCTCTGAA
- [Reverse] 5′ - GTTTCTTGTTTTGAACTGGTCTGCCTTTTTAACAT
- 329 k (61°C):
- [Forward] 5′ - CATTGTTAGGTGCTATGGGGTTTAG
- [Reverse] 5′ - GTTTCTTGGTGGGAGGATGGCTTAGGC
- 3–104 k (55°C):
- [Forward] 5′ - TTGGGACCTGCTGGGCATAGTG
- [Reverse] 5′ - GTTTCTTGCTCTCCTGGTGTGTTAGATTGGATA
- 3–85 k (55°C):
- [Forward] 5′ - GGCTTGATGGGGGAAGAGTATTG
- [Reverse] 5′ - GTTTCTTTTTTCTCATGTCTCTGCTGATGCTC
- 3–263 k (55°C):
- [Forward] 5′ - ACTTAGCCAAACCTTTAAATAGATAG
- [Reverse] 5′ - GTTTCTTTTCATGAGATTTGTACCTGGCTTTAAGTTATT
- 3–204.2 k (58°C):
- [Forward] 5′ - TCCTCAGAGTTCCACCTTGTGT
- [Reverse] 5′ - GTTTCTTGACATTATGAAAAGCAGACTTAGGCAAAT
Results
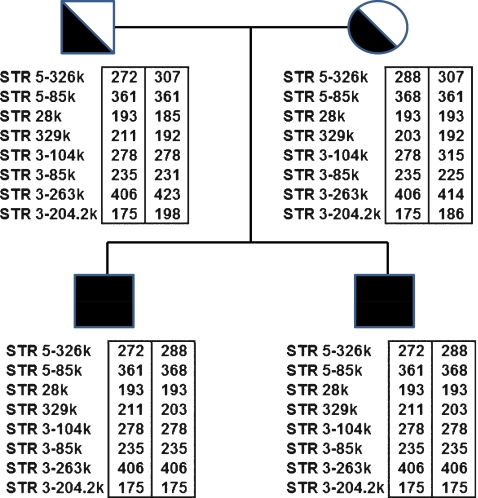
Utilization of this newly described protocol is demonstrated by application to a clinical case of ARPKD. Both parents were carriers of a common PKHD1 missense mutation, T36M [1, 16], and had two previously affected pregnancies both homozygous for the T36M mutation. In order to establish the PKHD1 haplotypes, STR genotyping analysis was carried out on the genomic DNA of the affected pregnancies and parents for our collection of 20 STR markers (Table 1). We selected eight linked STR markers that were informative for that family (Fig. 1). STR 28 k and STR 329 k were intragenic, STRs 5-326k and 5-85k were located in the 5′ flanking region of PKHD1 gene, while STRs 3–104 k, 3–85 k, 3–263 k, and 3–204.2 k were located in the 3′ flanking region.
Table 1.
Polymorphic STR markers for preimplantation PKHD1 haplotyping
| STR Name | STR Amplicon Location | Forward Primer (5′ --> 3′) | Reverse Primer (5′ --> 3′) |
|---|---|---|---|
| At the 5′ flanking region of PKHD1 gene: | |||
| 5–909 k | 52,969,730-52,970,172 | TGTGTCTGTGTAAATTAAAGTAGCACCA | GTTTCTTCTTTGTCTTCCTTCTCCGCTCT |
| 5–838 k | 52,899,197-52,899,465 | CTGGCCCAGTAACAATTTCGAG | GTTTCTTCTGTTAAAGCAAAGTTATCTTGAGGCAT |
| 5–570 k | 52,630,577-52,630,884 | TCCTCTTCTCTATTCAAAGGCAAC | GTTTCTTCACATTCTTGCCAGATTTGGTG |
| 5–564 k | 52,624,476-52,624,900 | TACAAAAAAAAATTAAAATTAGCTGGGCATCGT | GTTTCTTCACCATTTGGGAATAAGCCGAAG |
| 5–438 k | 52,498,725-52,499,021 | ATTTACAAGCCAAGAGAGACCTAGAACA | GTTTCTTCAATGTGGGGTTAAAATGGTAACTCTG |
| 5–326 k | 52,386,622-52,386,902 | GAAAGCTCCAGCCATCTAGCC | GTTTCTTAAGGGTTTGGTTTGTGTGAGTTAAGTCT |
| 5–85 k | 52,145,161-52,145,531 | AGTTGGGCATGGTGGCGTACAC | GTTTCTTGGTTGCAGCTTACTAATCATTGGATGTT |
| Within the PKHD1 gene: | |||
| 22 k | 52,037,793-52,038,039 | CCCAGCCCTTTGCCTTAGAATAG | GTTTCTTTATCACATGGAATGCCTAAAGCGAATTA |
| 28 k | 52,031,453-52,031,633 | TAGGTTGGTTGTTCCTTCTCTGAA | GTTTCTTGTTTTGAACTGGTCTGCCTTTTTAACAT |
| 72 k | 51,987,190-51,987,608 | AAATTATACCCTGTCTATTCGTCCG | GTTTCTTCACCAGTACCTGTCACAACACCT |
| 162 k | 51,898,142-51,898,312 | GCTCATCTACATTAAGGAAGGCAATCTT | GTTTCTTACTTGACTGGGCTATGGGGTAC |
| 329 k | 51,730,671-51,730,865 | CATTGTTAGGTGCTATGGGGTTTAG | GTTTCTTGGTGGGAGGATGGCTTAGGC |
| 387 k | 51,672,619-51,672,764 | ATACATCCTTAGAATGAAAAATTACTCAGGTA | GTTTCTTTATTATGCTGGGAATAGTATTAAATAGAACAA |
| At the 3′ flanking region of PKHD1 gene: | |||
| 3–104 k | 51,492,459-51,492,742 | TTGGGACCTGCTGGGCATAGTG | GTTTCTTGCTCTCCTGGTGTGTTAGATTGGATA |
| 3–101 k | 51,489,061-51,489,254 | TTCCTGGTTCTCCTTGTTTTTCTATCTT | GTTTCTTACCCTGGAGCAAGTAATCAATATCTGT |
| 3–85 k | 51,473,601-51,473,814 | GGCTTGATGGGGGAAGAGTATTG | GTTTCTTTTTTCTCATGTCTCTGCTGATGCTC |
| 3–32 k | 51,420,166-51,420,386 | GGAAAACATAATGAGTTAATCTCTGAATGC | GTTTCTTTCTCTCCCTGGATAATCTCTTTCATACT |
| 3–266 k | 51,354,413-51,354,612 | CCTATCTTCTTAGATTATTCTTGATTGTGCT | GTTTCTTCTGGATTATGCCTAGATAAAGGACTTTC |
| 3–263 k | 51,351,797-51,352,253 | ACTTAGCCAAACCTTTAAATAGATAG | GTTTCTTTTCATGAGATTTGTACCTGGCTTTAAGTTATT |
| 3–204.2 k | 51,292,282-51,292,464 | TCCTCAGAGTTCCACCTTGTGT | GTTTCTTGACATTATGAAAAGCAGACTTAGGCAAAT |
Fig. 1.
STR analysis of father, mother, and affected fetuses (black square) and resulting PKHD1 haplotypes. Informative STRs are ordered from the 5′ end (top) to the 3′ end (bottom). The numbers within the boxes next to each STR represent allele sizes in base-pairs (bp). Haplotypes are enclosed in boxes
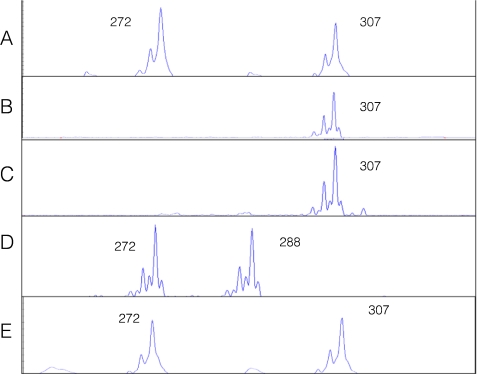
On day 3 post fertilization, 7 embryos were biopsied, and the 8 blastomeres from 7 biopsied embryos were genotyped for the 8 selected STR markers. Table 2 summarizes the genotyping results of all eight linked STR markers and the PKHD1 haplotypes deduced from these STR genotypes, revealing that three unaffected embryos (#2, #4 and #6) were suitable for transfer. There was no evidence of meiotic recombination within the PKHD1 gene and between all these linked STR markers. No contamination was detected in the last wash droplets of the blastomere cells. Embryos #1, #3 and #5, had PKHD1 haplotypes identical to those of the previously affected ARPKD pregnancies (with homozygous mutations). Embryos #4 and #6 were unaffected and not carriers of ARPKD. Embryo #2 was unaffected but a carrier of ARPKD. Embryo #7 gave inconclusive results and that was correlated with morphologically poor development (Table 2). For all seven embryos (8 blastomeres), the combined rates of ADO and amplification failure of eight STR markers were 7.8% (10/128 loci) and 6.2% (8/128 loci), respectively. Figure 2 illustrates the STR 5–326 k genotyping of embryos #2, #4, #6 and #3, as well the unaffected PGD infant.
Table 2.
Preimplantation PKHD1 haplotyping by PCR using STR markers and capillary electrophoretic detection
| STR Genotypes of Patient Family | ||||||||||
| STR: | 5–326 k | 5–85 k | 28 k | 329 k | 3–104 k | 3–85 k | 3–263 k | 3–204.2 k | ||
| Type: | Di | Tetra | Tetra | Di | Tetra | Di | Tetra | Tri | ||
| Father: | 272, 307 | 361, 361 | 193, 185 | 211, 192 | 278, 278 | 235, 231 | 406, 423 | 175, 198 | ||
| Fetuses: | 272, 288 | 361, 368 | 193, 193 | 211, 203 | 278, 278 | 235, 235 | 406, 406 | 175, 175 | ||
| Mother: | 288, 307 | 368, 361 | 193, 193 | 203, 192 | 278, 315 | 235, 225 | 406, 414 | 175, 186 | ||
| PGD infant: | 272, 307 | 361, 361 | 193, 193 | 211, 192 | 278, 315 | 235, 225 | 406, 414 | 175, 186 | ||
| STR Genotypes of Blastomeres | ||||||||||
| STR: | 5–326 k | 5–85 k | 28 k | 329 k | 3–104 k | 3–85 k | 3–263 k | 3–204.2 k | ||
| Type: | Di | Tetra | Tetra | Di | Tetra | Di | Tetra | Tri | ||
| Embryos | Cells | Predicted Haplotype | ||||||||
| E1 | −1 | ADO, 288 | 361, ADO | 193, 193 | 211, 203 | 278, 278 | 235, 235 | 406, 406 | 175, 175 | mut |
| E2 | −1 | 272, 307 | 361, ADO | 193, 193 | 211, 192 | 278, 315 | 238*, 225 | 406, 414 | [173]*, 186 | het |
| E3 | −1 | 272, 288 | 361, 368 | 193, 193 | 211, 203 | 278, 278 | 235, 235 | 406, 406 | FA | mut |
| E4 | −1 | Cell lysed after biopsy | ||||||||
| −2 | 307, 307 | 361, 361 | 185, 193 | 193, 193 | ADO, 315 | 231, 225 | 423, 414 | ADO, 186 | wt | |
| E5 | −1 | FA | 360, ADO | 193, 193 | 211, 203 | 278, 278 | 235, 235 | 406, 406 | 175, 175 | mut |
| E6 | −1 | 307, 307 | 361, 361 | 185, [193] | 192, 192 | 278, 315 | 231, 225 | [423], 414 | 198, 186 | wt |
| −2 | 307, 307 | 361, 361 | [185], 193 | 192, 192 | 278, 315 | 231, 225 | 423, [414] | [198], 186 | wt | |
| E7 | −1 | FA | 360, -- | 185, 193 | 198*, -- | 282*, -- | 233*, 225 | 415, -- | FA | Indeterminate; Morphologically poorly developed |
*: Anomalous mobility
[ ]: Weak signal due to preferential amplification
-- : The expected allele was not detected
E1–E7 Embryos #1–7, STR Short tandem repeat, ADO Allele drop-out, FA Amplification failure
Remark: Paternally derived markers in each cell are shown on the left and maternally derived markers on the right (not applicable to father and mother).
Test results are reported as follows:
wt Wildtype, mut Homozygous mutation, het Heterozygous mutation (or a carrier)
Fig. 2.
Capillary electropherograms of the PKHD1 STR 5–326 k for a blastomere biopsied from embryo #2 (a), a blastomere from embryo #4 (b), a blastomere biopsied from embryo #6 (c), a blastomere biopsied from embryo #3 (d), and the PGD infant (e). The numbers represent the allele sizes in base pairs (bp). The affected embryo #3 inherits the mutant alleles of 272 bp and 288 bp from the parents. The unaffected embryos #2, #4 and #6, and the PGD infant inherit the wildtype alleles of 307 bp from the parents
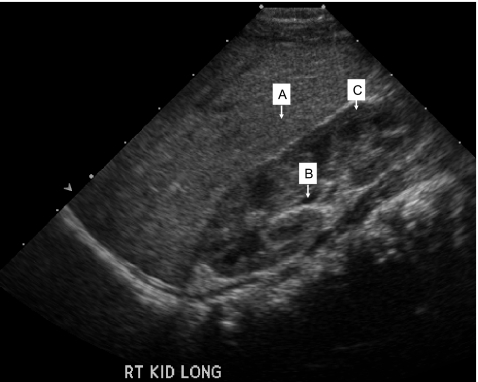
On day 5 post fertilization, embryo #2 was well developed into a blastocyst, while embryos #4 and #6 were poorly developed. The three embryos (#2, #4 and #6) diagnosed as not being affected were subsequently transferred using trans-abdominal ultrasound guidance. At 7 weeks of gestation a viable twin pregnancy was confirmed by transvaginal ultrasound. The development of one of the fetuses, however, arrested after 10 weeks. A female infant was born at 38 weeks of gestation. Follow-up ultrasound examination has confirmed that she is unaffected with ARPKD (Fig. 3). The STR genotype (Fig. 2) and haplotype analysis (Table 2) of the infant was identical to that of embryo #2 for the eight STR markers.
Fig. 3.
Renal sonogram (right kidney longitudinal view) of infant at 4 months of age born following PGD for ARPKD. Kidney measures 5.3 cm in length (normal for patient age is 5.28 cm ± 1.32 cm). Note striking corticomedullary differentiation and normal echogenicity when compared to the liver, which are characteristic of normal renal ultrasound for this age. (a) liver, (b) medullary pyramid, and (c) renal cortex
Discussion
Initial screening of 121 STR loci located in the intragenic and flanking regions of PKHD1 gene identified 41 polymorphic STR repeats for further study. Ten of these 41 STRs are intragenic, 11 of them are located at the 5′ 1000-kb flanking region, and 20 of them are located at the 3′ 500-kb flanking region. Of these 41 STRs, only four intragenic STRs and one STR in the 5′ flanking region were previously published [16, 25]. Out of these 41 STRs, 20 highly polymorphic STRs in the population were selected for further studies.
These 20 STRs were either fully or partially informative for at least one of ten arbitrarily selected anonymous families consisting of father, mother and child and were selected for further studies (Table 1). Six of these 20 STRs are intragenic, 7 of them are located at the 5′ 1000-kb flanking region, and 7 of them are located at the 3′ 500-kb flanking region. Only 3 of these 20 STRs were previously reported and used in linkage analysis [16, 25]. These three STRs were STR 28 k (also designated as D6S1344) [16], STR 387 k (also designated as D6S243) [16, 25], and STR 5–438 k (also designated as KIAA0057) [25].
A 4-h MDA reaction generated sufficient amounts of high quality amplified DNA to allow subsequent STR genotyping by PCR. By shortening the MDA reactions to 3 h, there was significant increase in the ADO rates for some STRs (Data not shown). It was critical to determine the optimal number of PCR cycles for each individual STR. By using suboptimal number of PCR cycles, the ADO rates were significantly higher. With the exception of the STR 3–263 k, 25 PCR cycles were appropriate for the remaining 19 STRs. For these 19 STRs, the combined ADO rates were determined to be 2.3% (3 in 128 loci), and the rate of amplification failure were also 6.2% (4 in 128 loci). STR 3–263 had the lowest ADO rate at 31 PCR cycles, but it was still significantly higher than the other 19 STRs.
A previous report utilizing PGD for ARPKD used six linked STR markers [25]: Two STR markers (D6S243 and D6S1714) were within the PKHD1 gene and thus less than 470 kb from the mutations. Two STR markers were on the centromeric side of the gene. One located in the 5′ flanking region of PKHD1, and one (D6S1662) located approximately 1.6 Mb from the 5′ end of PKHD1 gene. The remaining two STR markers were on the telomeric side of the gene, located between 500-1,000 kb from the 3′ end of PKHD1. In our analysis we have identified a total of 20 highly polymorphic STR markers for the PKHD1 gene: 6 intragenic STRs, 7 STRs within 1,000 kb from the 5′ end of the gene, and 7 STRs within 500 kb from the 3′ end of the gene. Thus, our STR markers in the flanking regions are located closer to the PKHD1 gene than previously described STRs [25]. As a result, the probability that a suitable embryo will be discarded due to recombination between STR markers has been reduced.
As the PKHD1 gene is approximately 470 kb in size, all of our intragenic STR markers are <470 kb from any mutation for ARPKD, and therefore the probability that a recombination event will occur between an intragenic STR marker and any mutation is <0.5%. The most distant STR marker that we identified in the 5′ flanking region is <1,940 kb from any given mutation in the ARPKD gene, and therefore the probability that a recombination event would occur between the farthest STR marker and any mutation is <2%. The most distant STR marker that we identified in the 3′ flanking region is <1440 kb from any mutation in the ARPKD gene, and therefore the probability that a recombination event would occur between the farthest STR marker and any mutation is <1.5%. The aforementioned rates of recombination are based on the assumption of 1% recombination per 1000 kb.
Preliminary screening for polymorphic STRs of the PKHD1 gene and flanking regions was carried out on polyacrylamide gels of 8 cm in length, as this gel system permits an inexpensive screening method for a large number of STR candidates using unlabeled PCR primers. Polymorphic STR alleles that were readily resolved by this gel system most likely differed by more than one repeat unit, and this helped with the selection of polymorphic STRs which had alleles differing by more than one repeat unit. It is difficult to distinguish shadow bands [17] from an allele which differed by only a single repeat unit.
These polymorphic STRs were then evaluated by studying the inheritance of alleles in ten arbitrarily chosen families by fluorescent PCR. Using our collection of 20 resulting STRs, the majority of families were informative for both parents for at least 5 linked markers. Linked STR markers were selected from both sides of the PKHD1 mutation(s) in order to detect possible meiotic recombination events. This also ensures successful assessment of embryos without misinterpretation due to potential amplification failure or ADO of a given STR.
Following our single cell MDA-STR genotyping protocol, the ADO rates (2.3% for lymphoblastoid cells, 7.8% for blastomeres) and amplification failure rates (7.8% for lymphoblastoid cells, 6.2% for blastomeres) were significantly lower than previously reported. Handyside et al. [26] reported ADO rate of 31% and amplification failure rate of 8%. Burlet et al. [35] found ADO rates between 7% and 34%, and failure in amplification between 5% and 15% for lymphocytes. In other studies of blastomeres, the ADO rates were 28% by Renwick et al. [30] and 34% by Ren et al. [32].
The conventional technique for PGD has been single-cell multiplex PCR, wherein the PCR primers for all the selected STR markers are present in the reaction. Interaction between PCR primers is not uncommon. This has been ameliorated by nested PCR, in which the first round amplification is single-cell multiplex PCR, while the second round uses primer pairs for each locus carried out as individual PCR reactions. The need for two sets of PCR primers, however, increases the cost of the assay. If the first round amplification is replaced by whole genome amplification using MDA, a single protocol can be used to make copies of the entire genome. Following our MDA procedure, we were able to obtain >80 ng of template-dependent amplified DNA from a single human cell in 4 h according to comparison of signal intensity with known DNA quantities (Data not shown). This amount of amplified DNA is generated from >12,000 fold amplification of a single genome, and is sufficient to analyze two dozen loci in an embryo according to our STR genotyping protocol. If one wished to examine more genetic loci, the MDA product can be re-amplified using the MDA kit (GE Healthcare, Piscataway, NJ) by following the manufacturer’s instructions for genomic DNA amplification. Alternatively, one could take advantage of a primase-based whole genome DNA amplification system to re-amplify the MDA product derived from a single cell [44]. After 1 h of incubation at 37°C, the Rapisome™ pWGA kit (Biohelix Corp., Beverly, MA) will generate several µg of amplified DNA from a 5-µL aliquot of 4-h single-cell MDA reaction. This amount of DNA would be sufficient for the analysis of a large number of genetic loci in an embryo, such as array-comparative genome hybridization (aCGH) in preimplantation genetic screening (PGS) for chromosomal aneuploidy [45], karyomapping [46], or other complex genetic mutation detection methods.
Our results agree with the findings of some PGD teams that approximately 4 h of MDA reaction generates sufficient amounts of high quality, amplified DNA from single cells for use as template for STR genotyping [28, 29, 38, 47]. With the exception of a report by Chow et al. [48] who used 1.5 h for MDA, other previous MDA protocols varied from 6 h [27, 33], 8 h [31, 32, 35] and 10 h [46] to 16 h [39].
With the goal of reducing ADO rates, we have streamlined the PGD procedure by optimizing every step from cell lysis to STR genotyping. We did not use the cell lysis solution and condition recommended by the kit manufacturer. Instead, single cells were lysed at 61°C in an alkaline cell lysis buffer free of EDTA as even a low concentration of EDTA has inhibitory consequence for single-cell PCR. In our protocol, we also scaled up the MDA reaction volume to 50 μL, altered the individual volumes of kit components added to the MDA reactions, and also limited the MDA reactions to 4 h to reduce the amounts of de novo synthesis of random template-independent products [26]. In order to minimize unbalanced amplification between alleles, we used a multiplex PCR kit which was found to be very useful for genotyping analysis of STRs. Finally, we have determined the optimal number of PCR cycles for each STRs used and carried out detection by capillary electrophoresis.
In order to carry out STR genotyping, aliquots equivalent to 1.5 μL of undiluted MDA products were transferred to each PCR reaction. These relatively large amounts of MDA amplified DNA allow a smaller number of PCR cycles for capillary electrophoretic detection. This has the effect of decreasing preferential amplification and ADO rates. In addition, the use of Qiagen Multiplex PCR System facilitates the co-amplification of STR alleles with differential lengths. For each individual STRs, we determined the smallest numbers of PCR cycles that will produce both the lowest ADO and amplification failure rates, and allow the fluorescent intensity to fall within reliable detection range of the capillary electrophoresis instrument where the ideal peak heights fell between 200 RFU and 25,000 RFU.
Conclusions
We have developed 20 informative STR markers for the PKHD1 gene and flanking sequences which are suitable for ARPKD linkage analysis and PGD by haplotyping. Most of the 20 STRs have not been previously reported. In addition to PGD, our newly discovered STRs can be used in prenatal diagnosis of ARPKD when linkage analysis is desired [16]. We have also developed a unique and reliable protocol for preimplantation ARPKD testing using DNA amplified by MDA for STR haplotyping, which was utilized to permit the birth of an unaffected infant to parents who previously had two pregnancies affected with ARPKD.
Acknowledgments
The excellent support of our colleagues Barbara Szlendakova, Amy Granlund, Dr. Peter vanTuinen, Brent Wells, Sarah Bick and Bridget Lawler are gratefully acknowledged.
Footnotes
Capsule Preimplantation Genetic Diagnosis of Autosomal Recessive Polycystic Kidney Disease using Multiple Displacement Amplification and Haplotype Analysis of PKHD1 Gene with Linked Short Tandem Repeat Markers
References
- 1.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–37. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dell KM, Sweeney WE, Avner ED. Polycystic kidney disease. In: Avner ED, Harmon WE, Niaudet P, Yoshikawa N, editors. Pediatric nephrology. 6. Heidelberg: Springer-Verlag; 2009. pp. 849–88. [Google Scholar]
- 3.Sweeney WE, Avner ED. Molecular and cellular pathophysiology of Autosomal recessive polycystic kidney disease. Cell Tissue Res. 2006;326:671–85. doi: 10.1007/s00441-006-0226-0. [DOI] [PubMed] [Google Scholar]
- 4.Dell KM, Avner ED (July 2008) Autosomal recessive polycystic kidney disease. In: GeneClinics: Online Clinical Genetic Information Resource; www.geneclinics.org
- 5.Sweeney WE, Avner ED. Renal cystic disease: new insights for the clinician. Pediatr Clin North Am. 2006;53:889–909. doi: 10.1016/j.pcl.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 6.European Polycystic Kidney Disease Consortium The polycystic kidney disease 1 gene encodes a 14kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77:881–94. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 7.Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–42. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 8.Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millán JL, et al. The polycystic kidney disease (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet. 1995;10:151–60. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 9.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet. 2002;30:259–69. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- 10.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, et al. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel ß-helix 1 repeats. Am J Hum Genet. 2002;70:1305–17. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bick DP, Lau EC. Preimplantation genetic diagnosis. Pediatr Clin North Am. 2006;53(4):559–77. doi: 10.1016/j.pcl.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Swanson A, Strawn E, Lau E, Bick D. Preimplantation genetic diagnosis: technology and clinical applications. WMJ. 2007;106(3):145–51. [PubMed] [Google Scholar]
- 13.Spits C, Sermon K. PGD for monogenic disorders: aspects of molecular biology. Prenat Diagn. 2008;29(1):50–6. doi: 10.1002/pd.2161. [DOI] [PubMed] [Google Scholar]
- 14.Fallon L, Harton GL, Sisson ME, Rodriguez E, Field LK, Fugger EF, et al. Preimplantation genetic diagnosis for spinal muscular atrophy type I. Neurology. 1999;53(5):1087–90. doi: 10.1212/wnl.53.5.1087. [DOI] [PubMed] [Google Scholar]
- 15.Xu K, Shi ZM, Veeck LL, Hughes MR, Rosenwaks Z. First unaffected pregnancy using preimplantation genetic diagnosis for sickle cell anemia. JAMA. 1999;281:1701–6. doi: 10.1001/jama.281.18.1701. [DOI] [PubMed] [Google Scholar]
- 16.Consugar MB, Anderson SA, Rossetti S, Pankratz S, Ward CJ, Torra R, et al. Haplotype analysis improves molecular diagnostics of autosomal recessive polycystic kidney disease. Am J Kidney Dis. 2005;45(1):77–87. doi: 10.1053/j.ajkd.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Bick SL, Bick BP, Wells BE, Roesler MR, Strawn EY, Lau EC. Preimplantation HLA haplotyping using tri-, tetra-, and pentanucleotide short tandem repeats for HLA matching. J Assist Reprod Genet. 2008;25:323–31. doi: 10.1007/s10815-008-9233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verlinsky Y, Rechitsky S, Verlinsky O, Chistokhina A, Sharapova T, Masciangelo C, et al. Preimplantation diagnosis for neurofibromatosis. Reprod Biomed Online. 2002;4(3):218–22. doi: 10.1016/S1472-6483(10)61809-3. [DOI] [PubMed] [Google Scholar]
- 19.Michaelides K, Tuddenham EGD, Turner C, Lavender B, Lavery SA. Live birth following the first mutation specific pre-implantation genetic diagnosis for haemophilia A. Thromb Haemost. 2006;95:373–9. doi: 10.1160/TH05-08-0574. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Garcia JF, Fallardo D, Ramirez L, Vidal F. Multiple fluorescent analysis of four short tandem repeats for rapid haemophilia A molecular diagnosis. Thromb Haemost. 2005;94(5):1099–103. doi: 10.1160/TH05-05-0360. [DOI] [PubMed] [Google Scholar]
- 21.Spits C, Rycke M, Verpoest W, Lissens W, Steirteghem A, Liebaers I, et al. Preimplantation genetic diagnosis for Marfan syndrome. Fertil Steril. 2006;86(2):310–20. doi: 10.1016/j.fertnstert.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 22.Christofidou C, Sofocleous C, Vrettou C, Destouni A, Traeger-Synodinos J, Kekou K, et al. PGD for X-linked and gender-dependent disorders using a robust, flexible single-tube PCR protocol. Reprod Biomed Online. 2009;19(3):418–25. doi: 10.1016/S1472-6483(10)60178-2. [DOI] [PubMed] [Google Scholar]
- 23.Spits C, Rycke M, Ranst N, Joris H, Verpoest W, Lissens W, et al. Preimplantation genetic diagnosis for neurofibromatosis type 1. Mol Hum Reprod. 2005;11(5):381–7. doi: 10.1093/molehr/gah170. [DOI] [PubMed] [Google Scholar]
- 24.Malcov M, Ben-Yosef D, Schwartz T, Mey-Raz N, Azem F, Lessing JB, et al. Preimplantation genetic diagnosis (PGD) for Duchenne muscular dystrophy (DMD) by triplex-nested PCR. Prenat Diagn. 2005;25:1200–5. doi: 10.1002/pd.1317. [DOI] [PubMed] [Google Scholar]
- 25.Gigarel N, Frydman N, Burlet P, Kerbrat V, Tachdjian G, Fanchin R, et al. Preimplantation genetic diagnosis for autosomal recessive polycystic kidney disease. Reprod Biomed Online. 2008;16(1):152–8. doi: 10.1016/S1472-6483(10)60569-X. [DOI] [PubMed] [Google Scholar]
- 26.Handyside AH, Robinson MD, Simpson RJ, Omar MB, Shaw M-A, Grudzinskas JG, et al. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. Mol Hum Reprod. 2004;10(10):676–72. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- 27.Hellani A, Coskun S, Benkhalifa M, Thakhi A, Sakati N, Al-Odaib A, et al. Multiple displacement amplification on single cell and possible PGD applications. Mol Hum Reprod. 2004;10(11):847–52. doi: 10.1093/molehr/gah114. [DOI] [PubMed] [Google Scholar]
- 28.Spits C, Caignec C, Rycke M, Haute L, Steirteghem A, Liebaers I, et al. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum Mutat. 2006;27(5):496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- 29.Spits C, Caignec C, Rycke M, Haute L, Steirteghem A, Liebaers I, et al. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1(4):1965–70. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 30.Renwick PJ, Lewis CM, Abbs S, Ogilvie CM. Determination of the genetic status of cleavage-stage human embryos by microsatellite marker analysis following multiple displacement amplification. Prenat Diagn. 2007;27:206–15. doi: 10.1002/pd.1638. [DOI] [PubMed] [Google Scholar]
- 31.Ren Z, Zhou C, Xu Y, Deng J, Zeng H, Zeng Y. Mutation and haplotype analysis for Duchenne muscular dystrophy by single cell multiple displacement amplification. Mol Hum Reprod. 2007;13(6):431–6. doi: 10.1093/molehr/gam020. [DOI] [PubMed] [Google Scholar]
- 32.Ren Z, Zeng H-T, Xu Y-W, Zhuang G-L, Deng J, Zhang C, et al. Preimplantation genetic diagnosis for Duchenne muscular dystrophy by multiple displacement amplification. Fertil Steril. 2009;91(2):359–64. doi: 10.1016/j.fertnstert.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 33.Glentis S, SenGupta S, Thornhill A, Wang R, Craft I, Harper JC. Molecular comparison of single cell MDA products derived from different cell types. Reprod BioMed Online. 2009;19:89–98. doi: 10.1016/S1472-6483(10)60051-X. [DOI] [PubMed] [Google Scholar]
- 34.Qubbaj W, Al-Aqeel AI, Al-Hassnan Z, Al-Duraihim A, Awartani K, Al-Rejjal R, et al. Preimplantation genetic diagnosis of Morquio disease. Prenat Diagn. 2008;28(10):900–3. doi: 10.1002/pd.2081. [DOI] [PubMed] [Google Scholar]
- 35.Burlet P, Frydman N, Gigarel N, Kerbrat V, Tachdjian G, Feyereisen E, et al. Multiple displacement amplification improves PGD for fragile X syndrome. Mol Hum Reprod. 2006;12(10):647–52. doi: 10.1093/molehr/gal069. [DOI] [PubMed] [Google Scholar]
- 36.Obradors A, Fernández E, Rius M, Oliver-Bonet M, Martínez-Fresno M, Benet J et al (2009) Outcome of twin babies free of Von Hippel-Lindau disease after a double-factor preimplantation genetic diagnosis: monogenetic mutation analysis and comprehensive aneuploidy screening. Fertil Steril 91(3):933.e1–7. [Epub 2009 Jan 10] [DOI] [PubMed]
- 37.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–6. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellani A, Sammour A, Johansson L, El-Sheikh A. Delivery of a normal baby after preimplantation genetic diagnosis for non-ketotic hyperglycinaemia. Reprod BioMed Online. 2008;16(6):893–7. doi: 10.1016/S1472-6483(10)60158-7. [DOI] [PubMed] [Google Scholar]
- 39.Lledo B, Ten J, Rodriguez-Arnedo D, Llacer J, Bernabeu R. Preimplantation genetic diagnosis of X-linked retinoschisis. Reprod BioMed Online. 2008;16(6):886–92. doi: 10.1016/S1472-6483(10)60157-5. [DOI] [PubMed] [Google Scholar]
- 40.Hellani A, Abu-Amero K, Azouri J, Al-Sharif H, Barblet H, El-Akoum S. Pregnancy after preimplantation genetic diagnosis for brachydactyly type B. Reprod Biomed Online. 2009;18(1):127–31. doi: 10.1016/S1472-6483(10)60434-8. [DOI] [PubMed] [Google Scholar]
- 41.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques. 1996;20:1004–10. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 43.Altarescu G, Geva TE, Brooks B, Margalioth E, Levy-Lahad E, Renbaum P. PGD on a recombinant allele: crossover between the TSC2 gene and ‘linked’ markers impairs accurate diagnosis. Prenat Diagn. 2008;28(10):929–33. doi: 10.1002/pd.2070. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Kim H-J, Zheng C, Chow WHA, Lim J, Keenan B, et al. Primase-based whole genome amplification. Nucleic Acids Res. 2008;36(13):e79. doi: 10.1093/nar/gkn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellani A, Abu-Amero K, Azouri J, El-Akoum S. Successful pregnancies after application of array-comparative genomic hybridization in PGS-aneuploidy screening. Reprod BioMed Online. 2008;17(6):841–7. doi: 10.1016/S1472-6483(10)60413-0. [DOI] [PubMed] [Google Scholar]
- 46.Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw M-A et al (2009 Oct. 25) Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. [Epub ahead of print] [DOI] [PubMed]
- 47.Kumar G, Garnova E, Reagin M, Vidali A. Improved multiple displacement amplification with phi29 DNA polymerase for genotyping of single human cells. BioTechniques. 2008;44(7):879–90. doi: 10.2144/000112755. [DOI] [PubMed] [Google Scholar]
- 48.Chow JF, Yeung WS, Lau EY, Lam ST, Tong T, Ng EH, Ho PC (2009) Singleton birth after preimplantation genetic diagnosis for Huntington disease using whole genome amplification. Fertil Steril 92(2):828.e7–10. [Epub 2009 Jun 9]. [DOI] [PubMed]