Abstract
We have recently reported the X-ray structure of the cyclic nucleotide regulated potassium channel, MlotiK1. Here we describe the application of both electron and X-ray crystallography to obtain high quality crystals. We suggest that the combined application of these techniques provides a useful strategy for membrane protein structure determination. We also present negative stain projection and cryo-data projection maps. These maps provide new insights about the properties of the MlotiK1 channel. In particular, a comparison of a 9 Å cryo-data projection with calculated model maps strongly suggests that there is a very weak interaction between the pore and the S1-S4 domains of this 6 TM tetrameric cation channel and that the S1-S4 domains can adopt multiple orientations relative to the pore.
Introduction
The most successful approach for determination of membrane protein structure involves the crystallization of protein in detergent and application of X-ray crystallography. However, the importance of phospholipids for the structure and function of a membrane protein is obvious and their absence from most three-dimensional (3D) crystals raises interesting questions about the relationship between the molecular structures determined in a detergent micelle and their physiological conformations in the lipid bilayer (Lee et al., 2005). Electron crystallography has the advantage of using two-dimensional protein crystals formed in lipid bilayers, a better mimic of the biological environment. Structures of membrane proteins, such as bacteriorhodopsin (Grigorieff et al., 1996), aquaporin (Gonen et al., 2005) and acetylcholine receptor (Unwin, 2005), have been successfully determined at medium to high resolution by this method. These structures resemble their X-ray counterparts and also provide structural and functional insights that result from the interaction between protein and phospholipid molecules.
We have recently reported the structure of the MlotiK1 potassium channel, determined by X-ray crystallography (Clayton et al., 2008) (PDB code: 3BEH). The prokaryotic MlotiK1 channel is a member of the family of tetrameric cation channels that are regulated by cyclic nucleotide monophosphate (usually cAMP or cGMP) (Nimigean et al., 2004). This family also includes the eukaryotic Hyperpolarisation activated Cyclic Nucleotide regulated (HCN) channels and the Cyclic Nucleotide Gated (CNG) channels (Craven and Zagotta, 2006). All of these are tetrameric channels where each subunit has 6 transmembrane helices (TMs) and a C-terminal cytoplasmic ligand-binding domain. The mechanism by which activity of cyclic nucleotide regulated channels is modified in response to cyclic-nucleotides is a matter of intense investigation for which structural information is desired. Here we describe how we combined electron crystallography and X-ray crystallography to obtain 2D and 3D crystals of MlotiK1. We also show how this combination has provided new insights about the structural properties of this potassium channel. In particular, we provide evidence of lipid bilayer-dependent structural effects in the channel.
Results
Growing MlotiK1 crystals
We approached the determination of the molecular structure of the MlotiK1 channel by attempting to grow three-dimensional crystals for X-ray crystallography. The protein was prepared according to a method previously described (Clayton et al., 2004; Nimigean et al., 2004). The crystals grew from a variety of conditions but were difficult to reproduce in a consistent manner. Diffraction patterns did not extend beyond 6-7Å, revealed multiple lattices and included streaky Bragg diffraction spots.
In an effort to improve crystal quality we used a range of standard approaches. We tested fourteen different detergents alone or in combinations; we attempted to co-crystallize the channel with heavy atoms, classical channel blockers (tetrabutyl-ammonium) or with a variety of single or mixed phospholipids (POPC, DOPC, DMPC, PC, E. coli polar and total lipids). None of these approaches improved the crystals. We also modified the protein by mutating lysines to histidines in potentially solvent exposed loops (Derewenda, 2004), or by introducing the epitope recognised by the KcsA channel antibody (Zhou et al., 2001). In general these protein modifications resulted in a reduction in protein expression levels or decreased protein stability, and did not improve crystal quality. Throughout this process we faced a common obstacle in membrane protein X-ray crystallography: due to weak intensity of diffraction patterns crystal screening was performed at synchrotron X-ray sources, resulting in a slow turn-around time in evaluation of crystal quality and optimization of crystal growth conditions.
Our lack of success in obtaining high quality three-dimensional crystals led us to attempt to solve the structure of MlotiK1 by electron-crystallography. Initial trials to form two-dimensional (2D) crystals were highly promising. To improve crystal quality we screened crystal growth conditions by testing different detergents, temperature, pH, salts, and lipids. Because protein aggregates were visualized by negative-stain electron microscopy of protein preparations we decided to alter our initial purification method. We introduced a second step of size-exclusion chromatography and reduced the overall purification time. Importantly, while determining the effect of solutes and solvents on the 2D crystals we found that water quality was reflected in the quality of the crystal morphology and diffraction limit. Switching from double distilled to deionized-distilled water for all buffers used during protein purification or crystal growth improved the quality of diffraction data. We do not know the reason for this effect but we presume that deionisation removes contaminants left by distillation. Crucially, evaluation of the 2D crystals was done in-house and under negative stain, allowing a fast turn-around time and resulting in relatively quick improvement in 2D crystal quality.
The best 2D crystals (Fig. 1) were obtained in the following conditions: protein at 1mg/ml in 50mM Tris pH7.5, 150mM KCl, 2mM DTT, 100 μM cAMP, 5 mM LDAO (lauryldimethylamine-N-oxide), 1mg/ml polar E. coli polar lipid extract (stock dissolved in 60 mM β-OG (β-octylglucoside)), dialysed against 50mM Tris pH 7.5, 150mM KCl, 200 μM cAMP. These conditions produced both vesicular and planar crystals with the largest measuring over 10μm in size. Fourier transforms of the best negatively stained crystals extended to the interpretable stain limit at ~12Å resolution, with low mosaicity.
Figure 1. Two-dimensional MlotiK1 crystals.
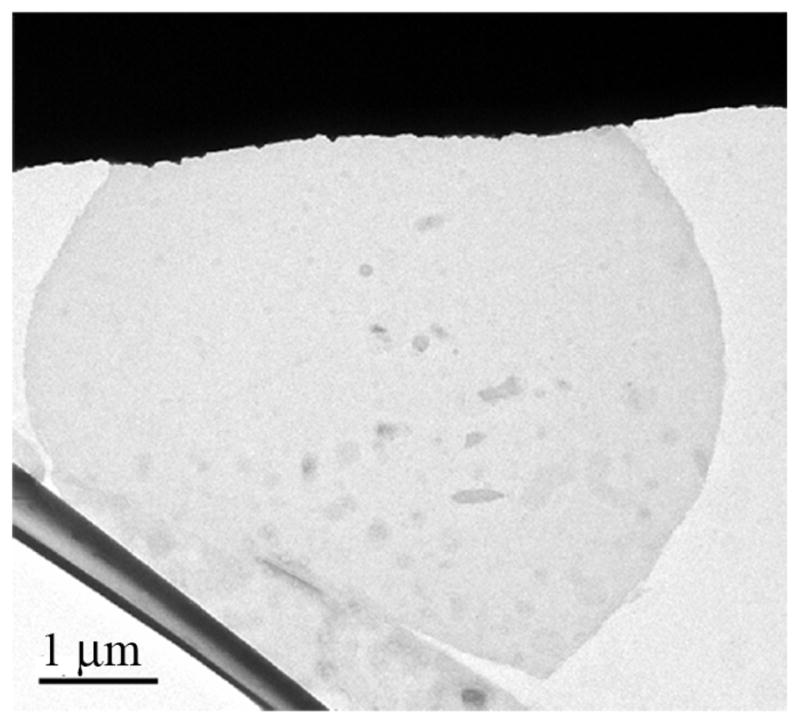
A wide-filed view of a two-dimensional crystal stained with 1% uranyl acetate.
Three-dimensional crystals
During the screening of crystal growth conditions for electron-microscopy we realized that the improvements achieved on 2D crystal quality might be applicable to the 3D crystals. We performed 3D crystal screens that included the optimal conditions defined during 2D crystallization. In particular, we prepared protein according to the new purification protocol and used deionized-distilled water in all solutions. We also focused our efforts with the best detergent used in 2D crystallization, LDAO. 3D crystals grew readily in different crystallization conditions but unlike the initial attempts the diffraction pattern showed a single lattice and was easily processed. At this stage resolution was still limited but improvements were achieved by performing a screen with a battery of additives from Hampton Research. These were tested at the manufacturer’s recommended concentrations with the conditions defined in the previous step that provided the best diffracting crystals. The most effective additives were in general organic compounds (phenol, 2,2,2-trifluoroethanol, 1,3-butanediol, dichloromethane) and with their inclusion we were able to extend resolution to 3.1Å.
The final crystal growth conditions were: 2 μl of MlotiK1 channel (10 mg/ml, 5 mM LDAO, 5 mM DTT, 150 mM KCl, 20 mM Tris pH 7.5) mixed with 2 μl reservoir (100 mM sodium citrate pH 5.6, 10% PEG 2000 MME, 20 % glycerol) plus 0.5 μl of additive. Several different crystal forms (in space groups P21, R3 or P4212) can be grown in these conditions, but the most common were crystals in the monoclinic or rhombohedral lattices. The crystals are in general large and not very sensitive to radiation exposure. As seen with other membrane proteins, diffraction quality varies between and within batches.
Comparing crystal lattices
The relatively simple translation of the 2D crystal improvements to the 3D crystal growth conditions made us suspect of a shared property between the two types of crystal. We have therefore analysed and compared their respective lattices.
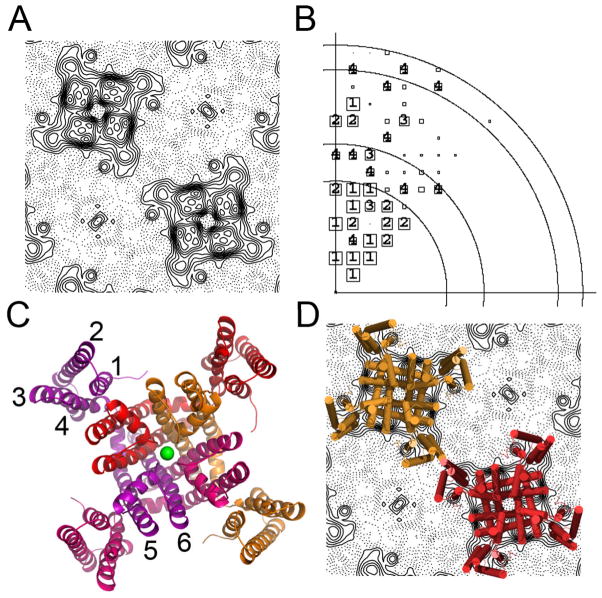
Despite severe difficulties in preparing vitreous samples (see methods) we were able to collect projection data from 2D frozen crystals grown in the absence of ligand and to calculate a 9Å projection map of the channel (Fig. 2A). The cryo data established without ambiguity (Fig. 2B, Table 1 and Supplemental Information) that the channel molecules in these crystals pack in space group P4212 (unit cell dimensions: 130 Å, 130 Å, 90°), with one channel subunit in the asymmetric cell, and channel molecules alternating in an up-and-down disposition. The recently reported 3.1 Å resolution structure of the MlotiK1 potassium channel (Fig 2C) (Clayton et al., 2008) was easily fitted into this projection map (Fig 2D). The channel structure has a four-fold symmetric central pore domain (formed by TMs 5–6 from four subunits) and four peripheral S1-S4 domains (TMs 1-4). In this structure the cytoplasmic cyclic nucleotide binding domains are disordered. The fit between the X-ray structure and the cryo-data projection map reveals that lattice contacts are established through the S1-S4 domains of neighbouring channel molecules, with possible participation of phospholipids.
Figure 2. 2D and 3D structures.
A) Cryo-data projection of the MlotiK1 channel showing 2 channel molecules in the lattice. B) Phase error plot of cryo-data. Resolution circles: 20Å, 15Å, 10Å, 9Å; phase errors are encoded as follows: 1<8°, 2<14°, 3<20°, 4<30°, 5<40°, 6<50°, 7<80°, 8<90° where 90° is random. The size of the plot symbols depends on the error of the reflection, with categories 1–4 individually labelled. C) Ribbon representation of the structure of the MlotiK1 channel (PDB code: 3BEH), subunits are shown in different colours; green sphere represents a potassium ion bound in the selectivity filter. Transmembrane helices are labelled 1 to 6. D) Superposition of the X-ray structure onto the cryo-data projection.
Table 1.
Statistics for Projection Data
| − cAMP P4 | + cAMP P4 | Cryo P4212 | |||||
|---|---|---|---|---|---|---|---|
| resolution | # | ERR | # | ERR | # | ERR | |
| 130–30.2 | 15 | 12.1 | 14 | 11.2 | 130–15.6 | 28 | 11.6 |
| 30.1-20.9- | 13 | 9.6 | 13 | 4.4 | 15.5–11.0 | 17 | 20.2 |
| 20.8–17.1 | 13 | 17.7 | 10 | 31.3 | 10.9–9.0 | 12 | 28.8 |
| OVERALL | 41 | 13.2 | 37 | 13.9 | OVERALL | 57 | 17.8 |
cAMP are with and without cAMP respectively. Data sets from negatively stained samples were obtained tilt-series as explained in the methods. In the case of the unstained samples, two images were used. The phase error (ERR) is expressed as the phase residual with respect to the target phase of 0° or 180°. In this comparison, 45° is random. Only reflections with an IQ of 6 (signal-to-noise ratio of at least 1.3) were included in the calculations.
number of unique reflections in the resolution bin indicated to the left.
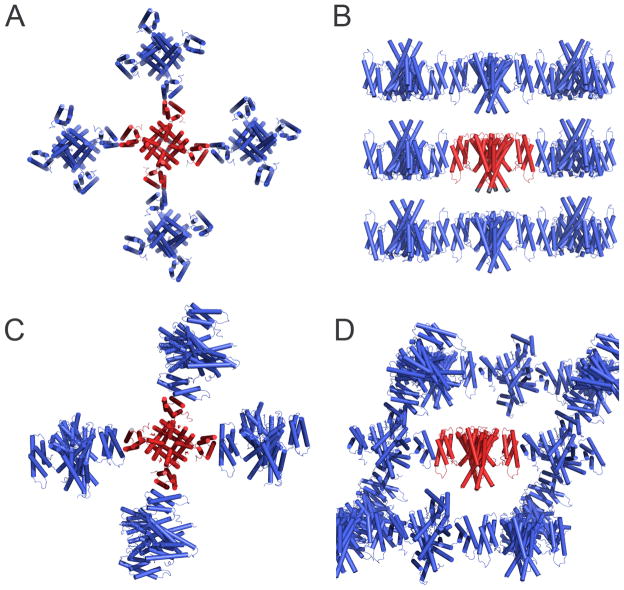
Using the X-ray model, determined from crystals in space group R3, we have also obtained molecular replacement solutions for the 3D crystal forms in space groups P21 and P4212. These models have not been fully refined but the analysis of their lattices provides interesting insights about the relationship between the 2D and 3D crystals. The 3D P4212 form is particularly interesting since its symmetry is identical to the symmetry the 2D crystals and the cell dimensions (125.1 Å, 125.1 Å, 81.8 Å, 90°, 90°, 90°) are very similar. The 3D packing of channel molecules is basically identical, with layers formed by molecules in an alternating up-and-down disposition (Fig 3A and B) and layer lattice contacts established through the S1-S4 domains. The higher resolution of the 3D crystals allowed us to determine that these contacts are established through TM2 and TM3, with a total buried surface between two neighbouring molecules of ~460 Å 2. Layers are stacked on top of each other to form the third dimension (Fig 3B); although the ligand binding domains are not defined in the electron-density maps, partly due to the resolution limits of the data (~6 Å), the contacts between layers are most likely established through these domains.
Figure 3. Three-dimensional crystal lattices.
A) Crystal lattice P4212. View along the axis of symmetry of central channel molecule. MlotiK1 channel molecules are shown in cartoon representation with central molecule in red and others in blue. B) Same as in A) but side-view of channel in red. C) Crystal lattice R3. View is along axis of symmetry of central channel molecule (red). Details as in A). D) Same as in C) but side-view of central channel molecule.
At first it appears that there is no relation between the 2D crystal lattice and the lattices of the 3D crystal forms, R3 and P21. Crystal symmetry is different and the number of molecules in the asymmetric units is also different: R3 crystals contain 1 channel molecule in the asymmetric unit, P21 crystals contain 3 channel molecules and P4212 crystals, both 2D and 3D, have ½ of a molecule or one channel subunit. However, in all of these crystal forms, each channel molecule establishes contacts with 4 neighbouring molecules through the S1-S4 domains (Fig. 2D and 3). More specifically, in the 3D crystals, and possibly also in the 2D, these contacts are established through TM2 and TM3 from each molecule. In the 2D crystals, the two molecules in contact are related by 180°; in R3, the molecules are no longer restricted by the lipid bilayer and the relation between contacting channels is ~45° (buried surface area ~340 Å2) or ~315° (buried surface area ~430 Å2), depending on the lattice direction (Fig 3C). In P21, the lattice is very similar to the one in R3 but there are small adjustments in the relative position of the molecules (not shown) that result in the symmetry change. In either R3 or P21 crystal forms, the non-180° angle between neighbouring molecules provides the possibility for the formation of three-dimensional lattices maintained solely by membrane buried regions (Fig 3D).
This analysis shows that there is a fundamental relationship between the lattices of the 2D and 3D MlotiK1 crystals, providing an explanation for the easy transfer of crystal quality improvements from one system to the other.
2D and 3D crystals
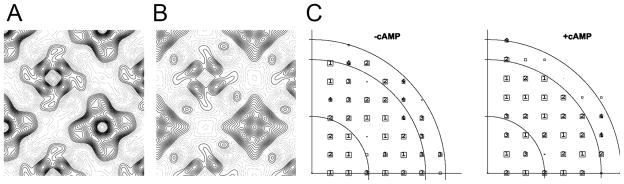
To understand the molecular details of the ligand activation mechanism of the MlotiK1 channel we require structures in the bound and unbound states. Unfortunately, the low stability of the detergent-solubilized MlotiK1 protein in the absence of ligand has precluded so far the study of ligand binding effects with the 3D crystals. However, due to the stabilizing effect of phospholipids, we could easily obtain 2D crystals of both cAMP-bound protein and apo-protein. The apo-protein 2D crystals were obtained by performing dialysis over 7 days against buffer without ligand. After multiple buffer changes the final ligand concentration was close to 1 pM, well below the channel affinity for cAMP (KD ~80 nM) (Cukkemane et al., 2007). We have collected data from negative stained crystals with and without cAMP and calculated experimental projection maps in space group P4 (Fig. 4 and Table 1). Both maps show two channel molecules per unit cell; strikingly, these two molecules appear very different in the negative stain. The variation in lattice symmetry between negatively stained and vitreous samples, as well as the presence of two apparently different molecules in each of the unit cells, had been previously determined to be a consequence of the up-and-down disposition of neighbouring molecules and their differential staining by the heavy atom compound (Chiu et al., 2007). A comparison between the two negative stain maps reveals clear differences between channel molecules with and without ligand. This demonstrates that the procedure for removal of cAMP works well and it also shows that the stabilizing effect of the lipid bilayer in 2D crystals extends the range of experimental conditions accessible for structural studies of this channel. Although the molecular nature of the structural change is not clear at this stage, we can gain some insights about the ligand binding effects on channel conformation. In particular, the peripheral regions of the channel appear to undergo major alterations suggesting that either the positions or the conformations of the ligand binding domains and/or of the S1-S4 domains are altered relative to the pore domain, upon ligand binding.
Figure 4. Negative stain projections.
A) View of the asymmetric unit in a negative stain projection of two-dimensional crystal grown without ligand. B) View of the asymmetric unit in a negative stain projection of two-dimensional crystal grown in the presence of cAMP. C) Phase error plots for the data collected from negative stained crystals grown with and without cAMP. Resolution circles: 40Å, 20Å, 17Å; phase errors are encoded as follows: 1<8°, 2<14°, 3<20°, 4<30°, 5<40°, 6<50°, 7<80°, 8<90° where 90° is random. The size of the plot symbols depends on the error of the reflection, with categories 1–4 individually labelled.
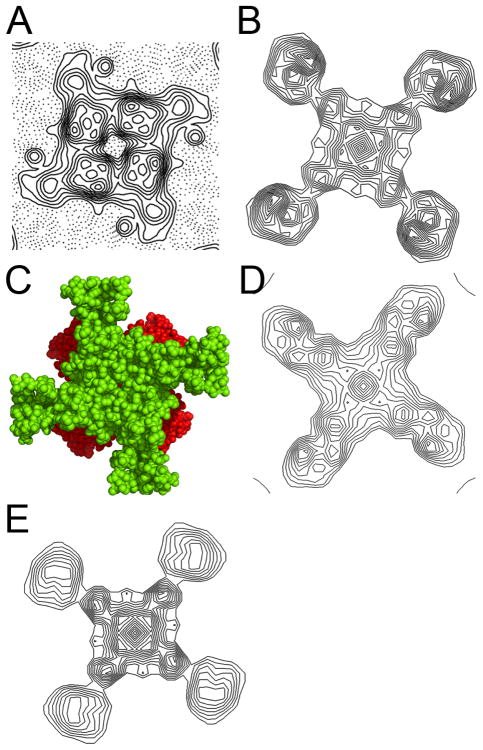
The effect of the lipid bilayer in 2D crystals is also revealed in another important aspect. Above we have shown that the X-ray model fits easily into the cryo-data projection map. A more detailed analysis of the experimental projection map shows that the electron density levels for the S1-S4 domains are much weaker than in the pore region (Fig. 5A). This is distinct from an equivalent map calculated from the X-ray model (Fig 5B), where the density levels are equivalent in the pore domain and S1-S4 domains. There are several possible explanations for this striking feature: 1) the cytoplasmic ligand binding domains are positioned below the pore domain resulting in stronger density in the central region of the projection. To test this hypothesis we constructed models of the full-length channel using the MlotiK1 channel structure and the ligand binding domain structure previously determined (Clayton et al., 2004). It became obvious that due to their dimensions it is not possible to arrange four globular ligand binding domains on the cytoplasmic face of the channel without having them stick out significantly under the “shadow” of the pore domain (Fig. 5C). Positioning the domain as in Figure 5A would create density regions protruding from the pore domain that are not observed in the experimental map and are not accounted by the channel X-ray structure alone. Alternatively, the domains can be positioned so that they are totally “covered” both by the pore and by the S1-S4 domains. This arrangement results in a model projection map (Fig. 5D) that still does not present the strong-weak density features of the cryo-data projection. We also considered the possibility that just two of the four cytoplasmic domains are positioned under the pore domain, while the other two are crystallographically disordered. However, in this arrangement the electron-density contribution from the ordered ligand binding domains is halved due to four-fold crystal symmetry averaging. As a consequence the calculated projection map does not present the strong-weak density feature (map not shown). 2) Another possible explanation for the weak-strong density feature is that in the lipid bilayer the S1-S4 domains adopt an orientation that is different from the one in the X-ray structure. We calculated projection maps after re-orienting the S1-S4 domain in our X-ray model; we tried many different positions, with the domains tilted up to 30° (which still keeps the helices in a transmembrane arrangement), but could not reproduce the features of the experimental density (calculated maps not shown). We also calculated maps where the helices of the S1-S4 domain were splayed apart like it has been observed in the voltage-sensor domain of voltage-gated potassium channels. This arrangement also did not reproduce the features of the experimental map (calculated maps not shown). 3) Finally, the difference in density levels may result from high mobility or from multiple orientations of the S1-S4 domains relative to the pore domain. Strikingly, a projection map calculated with a total of 14 superimposed models, where in each model the S1-S4 domain has been reorientated differently relative to the pore, recapitulates the general features of the experimental cryo data projection, with the density levels for the pore region much stronger than for S1-S4 domains (Fig. 5E).
Figure 5. Modelling the cryo-data projection.
A) Density for one MlotiK1 channel molecule in the cryo-data projection map. B) Calculated projection of MlotiK1 channel X-ray model. C) Extracellular view of the MlotiK1 channel structure (in green), with the oriented ligand binding domains in red. D) Calculated projection map of the MlotiK1 channel with the ligand binding domains positioned on the cytoplasmic face of the channel, under the S1-S4 and pore domains. E) Calculated projection map of 14 different models of the MlotiK1 channel structure superimposed through the pore domain. In each of the models the orientation and position of the S1-S4 domain was altered relative to the pore domain.
This analysis supports a model where the S1-S4 domain and the pore domain are not tightly attached and the domains are able to adopt multiple positions relative to each other.
Discussion
We have combined electron and X-ray crystallography to study the MlotiK1 cyclic nucleotide regulated potassium channel. At a first level we used electron crystallography to provide direct and rapid turn-over information on the factors that affect crystal quality. Crucially, the improvements in the 2D crystal quality were easily translated into growth of high quality 3D crystals. It may be the case that the similarities in channel packing in the 2D and 3D crystals explain the success of this strategy. However, we suggest that the combination of electron and X-ray crystallography could be explored as a general approach to define more rapidly the optimal conditions for crystal growth of membrane proteins.
We also combined electron and X-ray crystallographic information to better understand the properties of the MlotiK1 channel. Interestingly, the major hurdle we faced in the use of electron crystallography was not obtaining 2D crystals but coaxing them to adhere reliably onto grids in good form. In some instances the handling of the crystals resulted in their disappearance or drastic deformation. Further development or refinement of methods for handling these very fragile samples is required.
Electron crystallography projection maps have allowed us to uncover structural properties that are dependent on the natural environment of the MlotiK1 channel, the phospholipid bilayer. For example, we could only obtain crystals of MlotiK1 in the unliganded state with channel embedded in a bilayer. The differences in the negative stain projections of the channel with and without ligand suggest alterations in the S1-S4 domains and/or the cytoplasmic ligand binding domains. Importantly, a 9Å cryo-data projection map of the unliganded channel presents a striking density feature, the density levels in the pore domain are much higher than in the S1-S4 domain regions. We found that the best explanation for this characteristic is that the S1-S4 domains, equivalent to the voltage-sensor domains of voltage-gated channels, can adopt multiple orientations relative to the pore.
The fact that the 2D crystal contacts involve the S1-S4 domain is not an obstacle to this conclusion; there is the possibility that some of the contacts are mediated by lipids and our crystal lattice analysis showed that the helices of the S1-S4 domain involved in lattice contacts are able to maintain the same basic crystal contact while pivoting across the contact. Noticeably, the relative positions of the S1-S4 and pore domains are the same in the different 3D crystal forms of the MlotiK1 channel. Therefore, the loose arrangement between the pore and S1-S4 domains is only observed in the 2D crystals, when the protein is embedded in a lipid bilayer. This observation emphasizes the importance of phospholipid molecules in the structural and functional properties of 6 TM channels.
The structural relationship between the S1-S4 domain and the pore domain in 6 TM voltage gated channels has been a point of debate. The question has been whether the interaction between the two functionally important domains is tight or weak and whether the X-ray structures reveal all the structural properties of this interaction. A mutagenesis study of the interface between the voltage-sensor domains (equivalent to the S1-S4 domain) in the voltage gated Shaker potassium channel revealed a surface that is highly tolerant of drastic amino acid changes (Soler-Llavina et al., 2006). Electron-spin resonance (ESR) data measured from the voltage gated KvAP channel embedded in a lipid bilayer is not directly explained by the X-ray structures of voltage-gated channels (Chakrapani et al., 2008; Vamvouka et al., 2008). The ESR data suggest alternative relative dispositions of the domains. Interestingly, a superposition of the structures of the MlotiK1 channel and Kv1.2 voltage gated potassium channel, through the pore domains, shows the S1-S4 domains of the two channels shifted relative to each other (Clayton et al., 2008). Together with our 2D cryo-data projection data these studies support a model where, in the lipid bilayer, the S1-S4 domains (equivalent to the voltage-sensor domains in voltage-gated channels) are loosely attached to the pore domain and can adopt multiple orientations. The low resolution of our map requires further confirmation of the model but it is clear that these loose multiple orientations would result in a breakdown of 4-fold symmetry between the different S1-S4 domains. A symmetry breakdown will have a dramatic impact on many of the approaches presently used to study the voltage-sensor in voltage-gated channels since many assume and impose 4-fold symmetry on the S1-S4 domains during data interpretation.
Methods
The optimized purification protocol is as follows, with all solutions being prepared with deionized-distilled water. The cell pellets from 8 liters of culture were resuspended in 100 ml of PBS (phosphate buffered saline solution; prepared with only potassium salts) and were lysed by 2 passages in a cell-cracker Avestin C5. Cell debris was removed by a 10 minute spin at 4°C, 12000 rpm in a Beckman SS34 rotor; cell membranes were pelleted in a 3 hour, 50000 rpm spin in a Ti70 Beckman rotor at 4°C. Membrane protein was extracted by mixing the pellets with 270 ml of PBS with 25 mM decylmaltoside (DM) and 200 μM cAMP, followed by a 45 minute centrifugation at 17000 rpm. Supernatant was incubated with nickel-agarose beads for one hour; the membrane protein was eluted, pooled and concentrated to ~0.5 ml before being loaded into a size exclusion chromatography column Sephadex-200, equilibrated with PBS plus 5 mM DM, 200 μM cAMP. Pooled fractions were incubated overnight at 4°C with thrombin to remove the His-tag. The following day the protein was diluted in half with Tris pH 7.5, 150 mM KCl, 5 mM LDAO detergent, 200 μM cAMP and concentrated to ~0.5 ml. This solution was loaded into a size exclusion chromatography column Sephadex-200 equilibrated with 20 mM Tris buffer pH 7.5, 5 mM LDAO or 5 mM DM (for P4212 crystals), 150 mM KCl, 5 mM DTT, 200 μM cAMP. Crystal growth conditions are described in the results section.
The electron microscopy work was performed on a Philips Tecnai 12 transmission microscope equipped with a LaB6 filament, operating at 120kV. Carbon film was floated onto 400mesh Cu/Rh-grids on the day of use, and glow discharged for 15 seconds in air. An aliquot of the crystal suspension was briefly vortexed, 3μl were applied to the grid, and subsequently stained with 2% uranyl-acetate. Because the lattices had a strong tendency to stack, grids were screened at 500x magnification to identify suitable targets. For negatively stained samples, data were collected at 30,000x magnification, using a 1024×1024 GATAN CCD camera. The defocus values were adjusted for each image to place the first zero of the Contrast Transfer Function (CTF) beyond the 17Å cutoff used during data merging. This made it unnecessary to correct for CTF-dependent modulation of phase values beyond the first zero, and minimized the effect of the amplitude modulation. Indexing of the calculated transforms yielded unit cells dimensions of: a=b=124.2±2.0 Å, γ=89.6o±0.4 (crystals without cAMP; negative stain, n=4), a=b=120.3±2.7 Å, γ=89.8o±0.4 (with cAMP, negative stain, n=5). ALLSPACE suggested that the crystals exhibited P4212 plane group. However, this symmetry did not hold when single-axis tilt series from these crystals were analyzed. Therefore, the lower plane group symmetry (P4) was used for all calculations in case of the negatively stained samples. After merging the data from the tilt series, AVRGAMPHS was used to obtain projection data (z*=±0.005 cutoff, IQ6 cutoff), and phase statistics for the two data sets were calculated with FOMSTATS. Individual phase errors after adjustment of the average phase values to the closer target value of 0°/180° were plotted with PLOTALL. Projections were sharpened using a B-factor of −3000Å 2. Statistics in Table 1.
We encountered two significant problems while trying to collect data from vitreous samples, poor adherence of crystals and disruption of the lattice in adhered crystals. In some instances adherence was so poor that it led us to conclude incorrectly that protein crystals had not been formed. After experimenting with various approaches it appears that the ratio of hydrophobicity and hydrophilicity of the carbon support in the grids affects adherence of crystals and their lattice quality. Specifically, MlotiK1 2D crystals tended to adhere inconsistently and often became disordered or damaged when we used non-tuneable plasma glow dischargers, with pre-set discharge times. The best conditions for adherence were achieved in tuneable plasma glow dischargers with weak charging, such as 20 second at 40mA (with or without amylamine). Crystals were deposited by back blotting method (Nigel Unwin - personal communication; more details below) onto weakly charged molybdenum, gold or copper grids, followed by direct ethane plunging in high humidity (90%). In addition, we used freshly peeled mica and carbon support, prepared without sparking in an effort to reduce surface particles. The carbon was either baked or “cured” by keeping them in a vacuum dessicator for one week or two, to increase its hydrophobicity, prior to plasma glow discharge. In more detail: 2D crystals were pipetted onto the surface of the carbon side of the grid. On the opposite side of the grid, the mica side, filter paper was used to blot the grid, drawing the crystal sample towards the filter paper. Holes in the carbon, natural or designed, allow crystal buffer solution to flow onto the filter paper but are not generally large enough to let the crystals through. In this way excess buffer is removed onto the filter paper, the crystals are drawn towards the carbon and are protected from the metal surface of the grid. The grid is quickly plunged into ethane before the crystals can re-drift up off the carbon.
Data from vitrified crystals were collected on a Tecnai F20 Field Emission microscope at 200kV, nominal magnification of 50,000-fold and underfocus values of ~1–1.5μm. Micrographs were recorded on SO163-film using ~10e/Å 2. After development in full strength D19, two images were scanned on a SCAI image scanner at 7μm stepsize, and processed using standard 2D-MRC protocols. The defocus (~1.4μm underfocus) was determined with CTFFIND3, and CTFAPPLY was used for adjusting phases for the impact of the CTF beyond the first zero. Analysis of phase data obtained from both images of unstained crystals revealed that one of the crystals was untilted while the other contained ~13° of tilt when compared against a low resolution 3D-model generated from the corresponding negatively stained samples. For the data from the untilted crystal, ALLSPACE indicated P4212 plane group symmetry (Supplemental Information and Table 1). Using the appropriate phase origin, data from both images were merged, and AVRGAMPHS was used with a z*=±0.005 cutoff (IQ6 maximum) to obtain the averaged projection data. Phase errors of each reflection were plotted after adjustment of the phases to the closer target phase of 0/180°. Indexing of the transform of the nominally untilted crystal yielded cell dimensions of a=b=130.3±2.1, γ=89.9° (n=1). A B-factor of −350Å 2 and a nominal resolution cutoff of 9Å were used for the calculation of the projection map. Statistics in Table 1.
Supplementary Material
Acknowledgments
We would like to thank the staff of X25 and X29 beamlines at the National Synchrotron Light source and the Cornell High Energy Synchrotron Source (CHESS). This work was supported by grants from the American Heart Association (#0555822T) and National Institutes of Health (GM068585) awarded to JHM-C and from PHS grants GM66145, GM071590 awarded to VMU.
Footnotes
The authors declare they have no competing financial interests.
References
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu PL, Pagel MD, Evans J, Chou HT, Zeng X, Gipson B, Stahlberg H, Nimigean CM. The Structure of the Prokaryotic Cyclic Nucleotide-Modulated Potassium Channel MloK1 at 16 A Resolution. Structure. 2007;15:1053–1064. doi: 10.1016/j.str.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci U S A. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH. Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell. 2004;119:615–627. doi: 10.1016/j.cell.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Craven KB, Zagotta WN. CNG and HCN channels: two peas, one pod. Annu Rev Physiol. 2006;68:375–401. doi: 10.1146/annurev.physiol.68.040104.134728. [DOI] [PubMed] [Google Scholar]
- Cukkemane A, Gruter B, Novak K, Gensch T, Bonigk W, Gerharz T, Kaupp UB, Seifert R. Subunits act independently in a cyclic nucleotide-activated K(+) channel. EMBO Rep. 2007;8:749–755. doi: 10.1038/sj.embor.7401025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y. Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J Struct Biol. 2004;146:325–333. doi: 10.1016/j.jsb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean CM, Shane T, Miller C. A cyclic nucleotide modulated prokaryotic K+ channel. J Gen Physiol. 2004;124:203–210. doi: 10.1085/jgp.200409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Soler-Llavina GJ, Chang TH, Swartz KJ. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Vamvouka M, Cieslak J, Van Eps N, Hubbell W, Gross A. The structure of the lipid-embedded potassium channel voltage sensor determined by double-electron-electron resonance spectroscopy. Protein Sci. 2008;17:506–517. doi: 10.1110/ps.073310008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.