Abstract
The filoviruses, Ebola and Marburg, utilize a multifaceted mechanism for assembly and budding of infectious virions from mammalian cells. Growing evidence not only demonstrates the importance of multiple viral proteins for efficient assembly and budding, but also the exploitation of various host proteins/pathways by the virus during this late stage of filovirus replication, including endocytic compartments, vacuolar protein sorting pathways, ubiquitination machinery, lipid rafts and cytoskeletal components. Continued elucidation of these complex and orchestrated virus-host interactions will provide a fundamental understanding of the molecular mechanisms of filovirus assembly/budding and ultimately lead to the development of novel viral- and/or host-oriented therapeutics to inhibit filovirus egress and spread. This article will focus on the most recent studies on host interactions and modulation of filovirus budding and summarize the key findings from these investigations.
Keywords: budding, Ebola, filovirus, Marburg, VLP
The Filoviridae family consists of two genera, Ebola virus (EBOV) and Marburg virus (MARV), which cause highly lethal, hemorrhagic disease in humans and nonhuman primates. Currently, there are no approved vaccines, or specific antivirals to prevent or treat filovirus infections. The high mortality rates, lack of therapeutic measures, potential use as weapons of bioterrorism and continuing outbreaks of deadly filovirus infections in various regions of the world contribute to the overall significance of filoviruses as worldwide public health concerns [1,2]. Filovirus outbreaks are typically sporadic and unpredictable, often interspersed by years without apparent disease activity, only to re-emerge again unexpectedly and with potentially high transmissibility [2–4]. Interestingly, fruit bats may represent the long sought-after reservoir for both EBOV and MARV [5–7].
The filoviruses are classified as biosafety level 4 and National Institute of Allergy and Infectious Diseases (NIAID) category A priority pathogens. Case fatality rates range between 20 and 90% depending on the virus species [8]. To date, five species of EBOV have been identified, including Zaire, Sudan, Reston, Cote d’Ivoire and Bundibugyo, whereas only one species of MARV has been described [2–4,9,10]. The filoviruses continue to represent significant public and global health concerns owing to the possible emergence of new species of filoviruses and their potential to spread and cause disease as zoonotic pathogens [11,12].
Filoviruses are enveloped, negative-sense RNA viruses belonging to the order Mononegavirales. The approximately 19.0-kb genome of the filoviruses encodes for the nucleoprotein (NP), VP35, matrix protein VP40, attachment glycoprotein (GP), VP30, VP24 and RNA polymerase protein. EBOV synthesizes an additional small/secreted nonstructural GP (sGP) generated by translation of unedited GP mRNA [13,14]. The VP40 matrix protein is the most abundant structural protein within the virion and plays a central role in directing virion assembly and egress [10,15–17]. In doing so, VP40 interacts with both viral and host proteins to promote filovirus budding [15,18–23]. This article will highlight the most recent findings regarding VP40 function and interactions that positively or negatively affect filovirus budding.
Functional budding domains within filovirus VP40
It is well documented that filovirus VP40, as well as the matrix proteins of other negative-sense RNA viruses, is the driving force of virion assembly and egress. We, and others, demonstrated that Zaire EBOV (ZEBOV) VP40 was necessary and sufficient to bud from mammalian cells as a viruslike particle (VLP) in the absence of any other viral proteins [10,15,16]. Remarkably, expression of either ZEBOV or MARV VP40 alone in mammalian cells led to the production and release of VLPs that were identical in overall morphology to authentic, infectious virions [16–18,24–27]. Filovirus VLP budding assays, which are performed under biosafety level 2, have served as the foundation for a plethora of studies aimed at elucidating the molecular mechanisms and host interactions required for efficient virion egress.
One of the first functional domains identified in ZEBOV VP40 that was important for VLP budding was termed the late (L) budding domain (for a recent review see [28]). In general, the core motifs that have been identified as functional L-domains in a wide array of RNA viruses include: P(T/S)AP, PPxY, YxxL, FPIV and LRSLF, where ‘x’ represents any amino acid. It has been well documented that the viral L-domain motifs P(T/S)AP, PPxY and YxxL interact with the host proteins: tumor susceptibility gene (Tsg)101, Nedd4.1, and ALG2 interacting protein X, respectively [15,18,21,29]. These three host proteins are known to play important roles in the vacuolar protein sorting (vps) pathway. Thus, the current model is that viral L-domains usurp or hijack these host proteins and exploit the vps machinery to help drive efficient release of the infectious virus.
For ZEBOV VP40, the L-domain is unique, in that it possesses two overlapping core motifs, 7PTAPPEY13 (7PTAP10 and 10PPEY13), both of which can function independently as L-domains [15,19,30]. By contrast, MARV (Musoke strain) VP40 contains a single 16PPxY19 motif [18,31]. Filovirus L-domains and their roles in budding have been the subject of several recent review articles [28,32–34]. Next, we will highlight recent findings regarding both filovirus L-domain activity and host interactions, as well as discuss other domains of VP40 that are important for budding.
Clearly, the L-domain motif represents only one component of VP40 that is important for efficient release of VLPs and virions. For example, L-domain deletion mutants of ZEBOV VP40 are still capable of budding as VLPs, albeit at significantly reduced levels compared with those of wild-type ZEBOV VP40 [10,15]. Similarly, although ZEBOV L-domains were found to enhance replication of infectious ZEBOV, they were not essential for virus replication in cell culture [35]. Thus, identification of additional sequences and functional domains of filovirus VP40 proteins remains an active area of investigation [36].
Towards this end, several recent studies aimed at further dissecting the functional domains of VP40 have yielded new findings. For example, Yamayoshi and Kawaoka generated a series of deletion and alanine-scanning mutants of ZEBOV VP40 in search of sequences that allowed for the budding of L-domain mutants of VP40 [37]. Yamayoshi and Kawaoka identified amino acids centered around Proline-53 of VP40 as being important for egress of VLPs and intracellular trafficking of VP40 to the site of VLP release at the plasma membrane [37].
McCarthy et al. [38] used a targeted mutagenesis approach based on the known crystal structure of ZEBOV VP40 [39] to mutagenize residues of VP40 predicted to be exposed on the surface and capable of interacting with lipid bilayers and/or proteins. In this study, the authors targeted three amino acids (212KLR214) in the C-terminal region of ZEBOV VP40 [38]. These residues formed part of a loop connecting two β-sheets in the C-terminal region of VP40 [39] and were predicted to be important for VP40 structure and/or oligomerization [38]. A series of alanine substitution mutants of the 212KLR214 region were generated and used to demonstrate that Leucine-213, in particular, was crucial for VP40 oligomerization, intracellular localization and subsequent release of VLPs [38].
More recently, the 96LPLGVA101 sequence of ZEBOV VP40 and the corresponding 84LPLGIM89 sequence of MARV VP40 were targeted for mutagenesis [36]. These filovirus VP40 sequences closely resembled the previously identified YPLGVG functional sequence of Nipah virus (NiV) matrix (M) protein [40]. Indeed, Patch and colleagues used site-directed mutagenesis to demonstrate that the NiV M sequence YPLGVG was required for M budding [40]. Although the YPLGVG was important for NiV M VLP budding, it did not appear to function as an L-domain, as budding of NiV M VLPs was insensitive to dominant-negative vps4 proteins [40]. Similarly, the 96LPLGVA101 sequence of ZEBOV VP40 and the 84LPLGIM89 sequence of MARV VP40 were found to be important for efficient egress of ZEBOV VP40 and MARV VP40 VLPs, respectively. However, these sequences are unlikely to function as L-domains [36]. Rather, we found that deletion of these motifs resulted in mislocalization, instability and, probably, misfolding of these VP40 deletion mutants, which subsequently led to their deficient release as VLPs [36].
Lastly, the ability of VP40 to oligomerize and adopt multiple conformations is crucial for its role in virion assembly and egress. Several key findings illustrating the importance of VP40 oligomerization for virion assembly and egress include:
The finding that the C-terminal domain of ZEBOV VP40 is required for membrane association, while the N-terminal region induces oligomerization [41];
ZEBOV VP40 can adopt the form of hexamers [42–44] and octamers that bind RNA and are incorporated into virions [45];
Octamers of ZEBOV VP40 are required for the release of infectious virions [46].
Taken together, these findings illustrate the fact that multiple domains of VP40 contribute to its overall structure and function, and emphasize the need for continued exploration of the VP40 sequences and structure to fully understand how this multifaceted protein promotes virus budding.
Role of additional viral proteins in budding
The VP40 proteins of ZEBOV and MARV are capable of budding independently from mammalian cells as VLPs; however, coexpression of additional filovirus proteins has been shown to facilitate VLP production (Table 1) [25–27,47–49]. This was not unexpected since coexpression of additional viral proteins should allow for essential protein–protein interactions to take place and the subsequent production of VLPs that more closely resemble the authentic, infectious virions. A few years ago, we demonstrated that coexpression of ZEBOV GP with VP40 resulted in a five- to eightfold enhancement of VLP budding as measured by quantification of VP40 in VLPs [49]. This finding has been confirmed by several other groups, and the mechanism of GP-mediated enhancement of VLP budding appears to be caused, at least in part, by its ability to function as an antagonist of tetherin (discussed later). We found that the most efficient release of VP40 VLPs occurred when VP40 was coexpressed with both GP and NP [49]. The addition of VP24 and VP35 did not significantly boost release of VLPs compared with that detected in the presence of VP40, GP and NP. In a follow-up study, Johnson et al. used electron microscopic analysis of ZEBOV VLPs to conclude that NP, GP and VP35 did not significantly alter the density, length or diameter of ZEBOV VP40 VLPs [50]. However, some structural changes were observed by electron microscopy within the core of the VLPs that contained both VP40 and NP, most likely reflecting the interactions between these two viral proteins [25].
Table 1.
Proteins that influence budding of filoviruses.
| Virus | Budding protein | Viral proteins that affect budding |
Host proteins that promote budding |
Host proteins that inhibit budding |
Ref. |
|---|---|---|---|---|---|
| Ebola | VP40 (PTAPPEY) |
GP | Tsg101 | ISG15 | [15–17,19,20,24,49,66–68,70] |
| NP | Nedd4 | Tetherin | [15,21,31,49,82,84,92,93] | ||
| VP24 | VPS4A/B | [49,94–96] | |||
| VP35 | Sec 24 | [25,70] | |||
| Marburg | VP40 (PPPY) |
GP NP VP24 VP35 |
Tsg101 Nedd4 VPS4A/B |
Tetherin | [18,19,26,27,31,47,48,57, 78,79,82,83,88] |
GP: Glycoprotein; ISG: Interferon-stimulated gene; NP: Nucleoprotein; Tsg: Tumor susceptibility gene; VPS: Vacuolar protein sorting.
As with EBOV, coexpression of MARV GP and NP with VP40 was demonstrated to enhance release of MARV VLPs by approximately tenfold compared with VP40 alone [18]. More recently, Wenigenrath et al. established an infectious VLP system for MARV and investigated the contributions of individual MARV proteins to the infectious VLP system [27]. Interestingly, the authors found that the infectivity of the VLPs was influenced by their length and morphology, and that the appropriate ratio or balance of the viral proteins NP and VP35 were crucial to the production of infectious VLPs [27]. As expected, GP was readily incorporated into budding VLPs and was essential for infectivity of the newly made VLPs [27]. By contrast, the viral polymerase protein (L) and VP24 were demonstrated to have no effect on the release of VLPs [27,48]. Importantly, this comprehensive analysis of the MARV VLP system underscores the importance of a balanced expression of viral proteins for the efficient production of morphologically authentic and infectious VLPs.
Host proteins that promote filovirus budding
A growing number of host proteins that can influence budding of filoviruses and VLPs continue to be identified. Several of the best characterized and most-recently identified host proteins implicated in either promoting or inhibiting budding of filoviruses are summarized in this article (see Table 1).
Tumor susceptibility gene 101
Tumor susceptibility gene 101 is an inactive E2 enzyme that functions as a component of the host vps pathway involved in protein sorting and targeting [51,52]. Tsg101 was first implicated in facilitating egress of HIV-1 and EBOV in an L-domain-dependent manner [19,53–56]. Since then, a plethora of studies have confirmed that expression of Tsg101 is important for efficient budding of ZEBOV VLPs, and that Tsg101-mediated enhancement of budding requires the ZEBOV VP40 PTAP-type L-domain (for review see [28]). Surprisingly, Tsg101 has also been reported to promote budding of MARV VP40 VLPs, which does not possess a PTAP-type L-domain [18]. Indeed, depletion of Tsg101 by an siRNA approach reduced budding of MARV VP40 VLPs compared with control siRNA [18]. Moreover, Tsg101 was packaged into VP40-WT VLPs; however, incorporation of Tsg101 into VP40-PPPA mutant VLPs was greatly reduced [18]. These findings, along with results of a GST-pulldown assay, suggest that the PPxY-type L-domain of MARV VP40 was required for Tsg101 interaction; however, whether Tsg101 interacts directly with the PPPY motif of MARV VP40 or indirectly, via another cell protein, was not determined.
Kolesnikova and colleagues also investigated the role of the host vps pathway in release of MARV VP40 VLPs [57]. An important finding from this report was that there were morphologically different populations of MARV VLPs released from cells expressing VP40. For example, the filamentous VLPs were composed primarily of VP40 with few host proteins, whereas the more spherical VLPs were composed of VP40 and considerable amounts of host proteins [57]. Kolesnikova and colleagues confirmed the importance of the PPxY-type L-domain of MARV VP40 for efficient egress of VLPs. However, in a yeast two-hybrid screen, the MARV VP40 protein failed to interact with host Tsg101 [57]. Instead, interactions between MARV VP40 and ubiquitin ligases AIP2, −4 and −5 were evident, as was the expected interaction between EBOV VP40 and Tsg101 [57]. Detection of VP40–host interactions, such as VP40–Tsg101, in mammalian cells has been difficult if not impossible. Although methods such as GST pulldowns and yeast two-hybrid assays have been useful, a method to detect and visualize these virus–host interactions in the natural environment of the cell in real time has yet to be developed [18,20,57]. One possible explanation to explain the difficulty is that the interactions between filovirus VP40 proteins and host proteins such as Tsg101 may be transient or have low binding affinities [20], making them difficult to detect in mammalian cells.
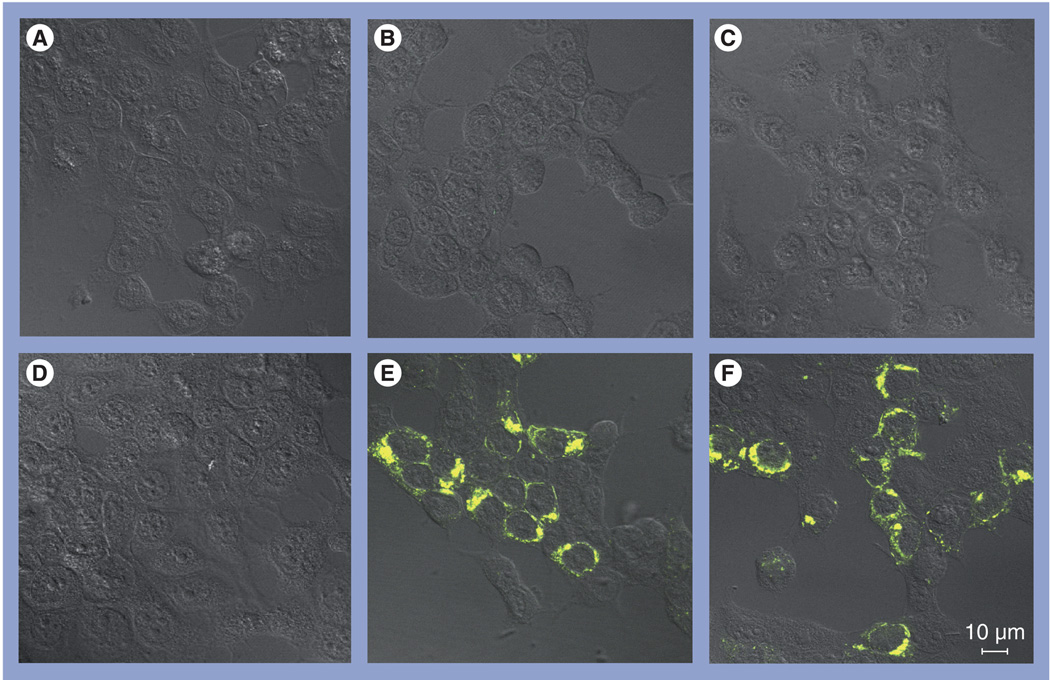
In an attempt to overcome this difficulty, we have recently established a bimolecular complementation (BiMC) assay using enhanced yellow fluorescent protein (EYFP) to detect and monitor VP40–host interactions in cell culture (Figure 1) [LIU Y, HARTY RN, UNPUBLISHED DATA]. To do this, the EYFP protein is split into its N- and C-terminal halves, which are then fused to the two proteins of interest. If the two proteins of interest interact in transfected cells, then the two YFP halves will be brought into close enough contact (5–10 nm) to restore fluorescent activity, which can be detected by confocal microscopy. Since EYFP activity caused by complementation is irreversible, this assay is ideal for detecting weak and/or transient protein–protein interactions in cell culture [58,59]. As expected, we readily detected an interaction between N-terminal YFP (NYFP)-Tsg101 and C-terminal YFP (CYFP)-EBOV VP40 as judged by fluorescence activity (Figure 1E). However, the EBOV VP40 L-domain deletion mutant (CYFP-EBOV VP40-ΔPT/PY) failed to interact with Tsg101 (Figure 1D). Interestingly, a similar level of fluorescent activity was detected in cells coexpressing NYFP-Tsg101 and CYFP-MARV VP40 (Figure 1F). These initial findings suggest that MARV VP40 interacts with Tsg101 in mammalian cells. However, a role for the PPPY motif in mediating this interaction remains to be determined. As negative controls, no background fluorescence activity was detected in cells expressing each of the plasmids alone (Figure 1A–C). Fluorescent activity generated by NYFP-Tsg101 and CYFP-EBOV VP40 could be detected in the cytoplasm as early as 3 h post-transfection and, overtime, the fluorescence accumulated at the plasma membrane of the cell [LIU Y, HARTY RN, UNPUBLISHED DATA]. We are currently using this assay to not only identify the protein domains required for interaction, but also to localize the intracellular sites of interaction and trafficking in real time [LIU Y, HARTY RN, UNPUBLISHED DATA]. In addition, our BiMC EYFP assay will be useful for screening small-molecule inhibitors of L-domain activity, VLP budding and overall VP40 function.
Figure 1. Bimolecular complementation assay.
Human 293T cells were transfected with (A) N-terminal yellow fluorescent protein (NYFP)-tumor susceptibility gene (Tsg)101 alone, (B) C-terminal YFP (CYFP)-Ebola virus (EBOV) VP40 alone, (C) CYFP-Marburg virus VP40 alone, (D) NYFP-Tsg101 plus CYFP-EBOV VP40-ΔPT/PY, (E) NYFP-Tsg101 plus CYFP-EBOV VP40 or (F) NYFP-Tsg101 plus CYFP-Marburg virus VP40. Cells were examined at 24 h post-transfection for YFP fluorescence activity by confocal microscopy using a Zeiss LSM-510 Meta system.
Along with other host proteins that interact physically and functionally with filovirus VP40, Tsg101 is of great interest as a potential target for therapeutic intervention. Antivirals capable of inhibiting virus budding by blocking L-domain interactions would be extremely attractive therapeutic candidates, since they probably possess a broad spectrum of activity against a wide array of human pathogens. Indeed, recent investigations into the development and use of host-oriented therapeutics have yielded preliminary, yet promising results [60–62]. Aman et al. identified a small-molecule therapeutic, FGI-106, which exhibits potent, broad-spectrum inhibition of EBOV, Rift Valley and Dengue Fever viruses in cell-based assays [61]. The authors speculate that FGI-106 interferes with a common cellular pathway utilized by different viruses [61]. In a similar study, Kinch and colleagues focused on host Tsg101 as the target for identification of a broad-spectrum, small-molecule inhibitor [60]. The authors identified FGI-104; a small-molecule inhibitor with broad-spectrum activity against multiple blood-borne pathogens (HCV, HBV and HIV-1) and emerging pathogens (Ebola, Venezuelan equine encephalitis virus, Cowpox virus and porcine reproductive and respiratory syndrome virus) [60]. The authors reasoned that by targeting the host, the pathogen can be prevented from causing disease [60]. In addition, the pathogen’s ability to become drug resistant will be minimized as selective pressure on the viral pathogen is eliminated. Overall, FGI-104 exhibited efficacy in a variety of cell-based assays against an array of human pathogens, and this efficacy was demonstrated in mouse-adapted ZEBOV [60].
Nedd4 E3 ubiquitin ligase
Nedd4.1 and −4.2 are membrane-localized E3 ubiquitin ligases that belong to the HECT family of E3 ligases (for review see [63]). Nedd4.1 contains an amino-terminal C2 domain that directs membrane binding and localization, four WW domains and a C-terminal HECT ubiquitin ligase catalytic domain that can covalently modify substrates through the addition of ubiquitin. Nedd4.1 has four hydrophobic WW domains within its central region, which are 35–40 amino acids in length and contain two conserved Trp residues spaced 21 amino acids apart [63]. It has been well documented that the PPxY-type L-domains present within matrix proteins of filoviruses, rhabdoviruses and retrovirus can interact physically and functionally with one or more WW domains of Nedd4, and these interactions are critical for efficient budding [15,20,21,31,64,65]. Ubiquitination of ZEBOV VP40 by Nedd4 and its yeast ortholog, Rsp5, has been documented [15,66,67]; although the mechanisms by which ubiquitination of VP40 enhances VLP budding remains unclear. Recently, Urata and Yasuda expanded on their findings that Nedd4 facilitated egress of MARV VP40 [21] by identifying the WW-domain of Nedd4 that interacts with the PPPY motif of MARV VP40 [31]. The authors reported that WW-domain 1 interacts with the PPPY motif of MARV VP40 [31], whereas WW-domain 3 predominantly interacts with ZEBOV PPEY [20]. These findings may reflect the differences in the nonconserved flanking sequences surrounding the PPxY L-domains of ZEBOV and MARV VP40, since the flanking residues and surrounding context of the L-domain core motif can influence host interactions [65,68]. It should also be noted that although many data have implicated Nedd4 as the primary ubiquitin ligase that interacts with both ZEBOV and MARV VP40, it is possible that other HECT domain E3 ligase family members may also play a role in mediating VP40 budding in different environments (e.g., different cell types or in vivo) [69].
Sec24C & the COPII transport system
A gap in our understanding of filovirus budding is the mechanism by which VP40 traffics to the plasma membrane to initiate the budding process. Recent findings by Yamayoshi et al. have shed some light on the intracellular transport of ZEBOV and MARV VP40 [70]. The authors used coimmunoprecipitation and mass spectrometric analyses to search for host proteins that interacted with VP40. They identified Sec24C, a member of the COPII vesicular transport system, and demonstrated that Sec24C interacted with amino acids 303–307 of ZEBOV VP40 [70]. A functional role for the COPII transport system in localizing VP40 to the plasma membrane was revealed using coimmunoprecipitation and dominant-negative mutants [70]. These findings have revealed a new area of investigation and will pave the way for future studies to elucidate the interplay between filovirus VP40 trafficking and function and the COPII transport system.
Cytoskeletal proteins & filovirus budding
Several studies have reported that the filoviruses exploit the cytoskeleton of host cells for protein transport and budding [71–77]. However, the exact role of host cytoskeletal proteins in filovirus budding remains to be defined. For example, actin was found to be present in both MARV and ZEBOV particles [76,77]. In addition to the possible role of cellular actin, Ruthel et al. demonstrated that EBOV associates with microtubules via the VP40 protein, which colocalizes with microtubule bundles [74]. Indeed, the authors reported that VP40 promotes tubulin polymerization, suggesting that microtubules may play an important role in the lifecycle of EBOV [74]. More recently, Kolesnikova et al. demonstrated that inhibition of actin polymerization in MARV-infected cells dramatically impaired the formation of MARV particles [71]. The authors also demonstrated that MARV VP40 VLP budding was strongly influenced by overexpression or inhibition of myosin 10 and Cdc42, host proteins important for filopodia formation and function [71]. Last, in other studies by Han et al., the authors found that alterations of cellular calcium/calmodulin influenced the efficiency of EBOV VP40 VLP budding through a mechanism that may involve the Ras/Raf/MEK/ERK signaling pathway [78]. Overall, these findings strongly implicate cytoskeletal elements as important for filovirus replication and budding.
Host proteins that inhibit filovirus budding
In contrast to the host proteins/machinery described previously, recent studies have identified host proteins/pathways that inhibit budding of filovirus particles, as well as other human viruses. Next, we summarize recent studies identifying two interferon-induced host proteins that are components of the innate immune antiviral defense and inhibit filovirus egress.
Tetherin
Second only to VP40, the filovirus surface-expressed GP contributes significantly to the budding process of filoviruses. As mentioned previously, coexpression of GP with VP40 leads to enhanced production of ZEBOV and MARV VLPs, which are now infectious owing to the presence of GP on their surface. One mechanism by which GP is thought to contribute to the budding process is by localizing viral components to lipid raft domains; the preferred site of budding [24,79–81]. More recently, a role for filovirus GP as an antagonist of the host membrane protein, tetherin, involved in tethering viruses to the plasma membrane to prevent their release, is gaining momentum [82–85]. Tetherin, also known as bone marrow stromal cell antigen 2, CD317 or HM1.24, is an interferon-induced protein that was first shown to block release of retroviruses and HIV-1 by tethering the virus particles to the plasma membrane [86]. The HIV-1 Vpu protein was found to function as an antagonist of tetherin activity, and expression of Vpu allowed for efficient and enhanced release of HIV-1 from infected cells [86,87]. Similarly, the filovirus GP protein appears to function in a manner similar to that of Vpu by relieving the inhibitory effect of tetherin and allowing for enhanced release of filovirus particles [84–86,88]; however, a role for tetherin during ZEBOV and MARV infection remains to be determined. Indeed, ZEBOV GP could substitute for Vpu to promote egress of HIV-1 from tetherin-expressing cells, and Vpu could substitute for ZEBOV GP to promote release of ZEBOV VLPs [84]. It should be noted that ZEBOV small/secreted GP was unable to rescue budding of ZEBOV VLPs from tetherin restriction [84]. While the mechanism by which tetherin restricts virus budding remains unclear, it appears that the C-terminal glycosylphosphatidylinositol-anchor region of tetherin is required for virus restriction [83,86]. Kaletsky et al. determined that ZEBOV GP colocalizes and interacts with tetherin, which may prevent tetherin activity [84]. Alternatively, it was recently demonstrated in Vpu-expressing HIV-infected cells that downregulation of tetherin may represent another mechanism to counteract its activity [87]. Finally, VLP production induced by MARV VP40 and the Z protein of Lassa virus was markedly inhibited by tetherin [88]. In a follow-up study, Sakuma et al. went on to demonstrate that dimerization of tetherin is not essential for its antiviral activity against MARV VP40 or Lassa virus Z protein [83]. In summary, tetherin is believed to be an antiviral factor with a broad-spectrum of activity to restrict the spread of numerous, unrelated enveloped viruses [82]. Interestingly, tetherin has been demonstrated to localize to cholesterol-rich plasma membrane domains, and, thus, it is not surprising that viruses, such as ZEBOV and HIV-1, that bud preferentially from these raft-like domains would need to possess a means of counteracting the antiviral activity of tetherin.
Interferon-stimulated gene 15
Interferon-stimulated gene (ISG)15 was one of the first of several hundred interferon-stimulated genes to be identified more than 20 years ago [89]. ISG15 is a small ubiquitin-like protein that can be conjugated (ISGylated) to specific target proteins, and may then alter their stability/function/localization. ISG15 and ISGylation have been demonstrated to possess a broad-spectrum of antiviral activity against both DNA and RNA viruses (reviewed recently in [34]). For example, new findings on the antiviral activity of ISG15 against both influenza A and B viruses have recently been reported [90,91].
Similar to ubiquitination pathways, ISGylation pathways involve a cascade of enzymatic reactions involving E1, −2 and −3 enzymes. In contrast to ubiquitination, which appears to promote budding of a wide array of RNA viruses including filoviruses, ISGylation was recently demonstrated to inhibit budding of ZEBOV by a previously undescribed mechanism [66,67]. Our laboratory [66] and Malakhova and Zhang [67], independently reported that ISG15 was able to inhibit ZEBOV VP40 VLP budding by disrupting Nedd4-mediated ubiquitination of the ZEBOV VP40 protein [66,67]. We found that free ISG15 was sufficient to negatively regulate Nedd4 ubiquitin ligase activity, and thus prevent the PPxY-dependent ubiquitination of ZEBOV VP40 required for optimal budding activity [66]. Accordingly, we found that budding of our ZEBOV VP40 L-domain deletion mutant was insensitive to ISG15-mediated inhibition, since it is also insensitive to ubiquitination by Nedd4 [66]. To address the molecular basis of ISG15 antiviral activity, Malakhova and Zhang reported that free ISG15 bound specifically to Nedd4, thus blocking its ability to interact with the corresponding E2 protein and preventing ubiquitination transfer from E2 to Nedd4 [67]. Since MARV VP40 also possesses a PPPY-type L-domain, it will be of interest to determine whether ISG15 has a similar inhibitory effect of MARV VP40 budding. It is also possible that direct ISGylation of EBOV or MARV VP40, or of host proteins, may contribute to the overall mechanism of inhibition of filovirus budding. In summary, the contradictory effects of ubiquitination (probudding) and ISGylation (antibudding) that relate to filovirus budding remain an intriguing area of investigation. Further investigation into the antiviral properties of ISG15 and tetherin will aid in our understanding of innate immune defenses to EBOV and MARV and prove useful in the pursuit of novel therapeutics.
Future perspective
While tremendous progress has been made in elucidating many of the molecular aspects of filovirus–host interactions and egress, many gaps and questions remain to be addressed to more completely understand the budding process. Some areas that await further investigation include: trafficking and intracellular transport of VP40 and interacting host proteins, the role of host cytoskeletal proteins, both during early and late stages of filovirus budding, and remodeling of cell membranes, via viral and/or host proteins, to initiate and complete the budding process. Lastly, as we continue to make progress in deciphering the events that are required for budding, we continue to inch closer to the identification of bona fide inhibitors of budding. Budding inhibitors remain attractive candidates for antiviral therapeutics, in part, owing to their potential broad-spectrum activity. The initial findings with FGI-104 and −106 are promising, as are the recent developments [27] and modifications to filovirus VLP budding assays (BiMC assay [LIU Y, HARTY RN, UNPUBLISHED DATA]) that will help facilitate screening and identification of new antivirals.
Executive summary
Domains of the filovirus VP40 protein
Multiple domains are required for efficient virion egress, including L-domains, membrane-binding domains and oligomerization domains.
L-domains of VP40 recruit members of the host endosomal sorting complex required for transport/vacuolar protein sorting pathways to facilitate budding.
Additional viral proteins & filovirus budding
Filovirus glycoproteins and nucleoproteins positively contribute to VP40-directed budding of virions and virus-like particles.
Host proteins & filovirus budding
Host proteins tumor susceptibility gene 101 and Nedd4 are recruited by L-domains of VP40 to enhance filovirus budding.
Cytoskeletal actin and microtubules are important for virion assembly and egress.
Small-molecule inhibitors targeting these VP40–host interactions may impair budding and have therapeutic potential.
Tetherin and interferon-stimulated gene 15 are interferon-induced, host innate immune proteins that disrupt budding of filovirus particles.
Acknowledgements
RN Harty would like to thank past and present members of his laboratory who contributed to this work, including: J Licata, T Irie, Z Han, R Johnson, S McCarthy and A Okumura. The authors would also like to thank A Stout, Director of the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania (PA, USA), for her assistance with confocal microscopy and the bimolecular complementation assay.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported in part by NIH grants to RN Harty. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Ascenzi P, Bocedi A, Heptonstall J, et al. Ebolavirus and Marburgvirus: insight the Filoviridae family. Mol. Aspects. Med. 2008;29:151–185. doi: 10.1016/j.mam.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Groseth A, Feldmann H, Strong JE. The ecology of Ebola virus. Trends Microbiol. 2007;15:408–416. doi: 10.1016/j.tim.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Martina BE, Osterhaus AD. “Filoviruses”: a real pandemic threat? EMBO Mol. Med. 2009;1:10–18. doi: 10.1002/emmm.200900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Towner JS, Sealy TK, Khristova ML, et al. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. ▪ First identification of the new Bundibugyo strain of Ebola virus (EBOV)
- 5.Leroy EM, Kumulungui B, Pourrut X, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 6.Swanepoel R, Smit SB, Rollin PE, et al. Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 2007;13:1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Towner JS, Amman BR, Sealy TK, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann H, Jones S, Klenk HD, Schnittler HJ. Ebola virus: from discovery to vaccine. Nat. Rev. Immunol. 2003;3:677–685. doi: 10.1038/nri1154. [DOI] [PubMed] [Google Scholar]
- 9.Diamond MS, Fremont DH. Ebola images emerge from the cave. Cell Host Microbe. 2008;4:87–89. doi: 10.1016/j.chom.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warfield KL, Deal EM, Bavari S. Filovirus infections. J. Am. Vet. Med. Assoc. 2009;234:1130–1139. doi: 10.2460/javma.234.9.1130. [DOI] [PubMed] [Google Scholar]
- 12. Barrette RW, Metwally SA, Rowland JM, et al. Discovery of swine as a host for the Reston Ebolavirus. Science. 2009;325:204–246. doi: 10.1126/science.1172705. ▪ Discovery of Reston EBOV in the human food chain in domestic swine.
- 13.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl Acad. Sci. USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volchkov VE, Becker S, Vochkova VA, et al. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 15. Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl Acad. Sci. USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. ▪ First report demonstrating that Zaire EBOV (ZEBOV) VP40 can bud as a virus-like particle (VLP) in an L-domain-dependent manner.
- 16.Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of Ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 17.Noda T, Sagara H, Suzuki E, et al. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J. Virol. 2007;81:4895–4899. doi: 10.1128/JVI.02829-06. ▪ Host Tsg101 is implicated in budding of Marburg virus VP40 VLPs.
- 19.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 20.Timmins J, Schoehn G, Ricard-Blum S, et al. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 2003;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 21.Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolesnikova L, Bugany H, Klenk HD, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 2002;76:1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolesnikova L, Bamberg S, Berghofer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J. Virol. 2004;78:2382–2393. doi: 10.1128/JVI.78.5.2382-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bavari S, Bosio CM, Wiegand E, et al. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. ▪ Budding of filoviruses occurs at lipid raft microdomains.
- 25.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E–5E minigenome RNA into virus-like particles. J. Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swenson DL, Warfield KL, Kuehl K, et al. Generation of Marburg virus-like particles by coexpression of glycoprotein and matrix protein. FEMS Immunol. Med. Microbiol. 2004;40:27–31. doi: 10.1016/S0928-8244(03)00273-6. [DOI] [PubMed] [Google Scholar]
- 27. Wenigenrath J, Kolesnikova L, Hoenen T, Mittler E, Becker S. Establishment and application of an infectious virus-like particle system for Marburg virus. J. Gen. Virol. 2010;91(Pt 5):1325–1334. doi: 10.1099/vir.0.018226-0. ▪ Development of an infectious VLP system to investigate multiple stages of the Marburg virus lifecycle.
- 28. Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. ▪ Comprehensive review of enveloped virus budding and host interactions.
- 29.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 Functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 30. Licata JM, Simpson-Holley M, Wright NT, Han ZY, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. ▪ Functional dissection of the overlapping L-domain motifs of ZEBOV VP40.
- 31.Urata S, Yasuda J. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J. Gen. Virol. 2010;91:228–234. doi: 10.1099/vir.0.015495-0. [DOI] [PubMed] [Google Scholar]
- 32.Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Jasenosky LD, Kawaoka Y. Filovirus budding. Virus Res. 2004;106:181–188. doi: 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Harty RN, Pitha PM, Okumura A. Antiviral activity of innate immune protein ISG15. J. Innate. Immun. 2009;1:397–404. doi: 10.1159/000226245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neumann G, Ebihara H, Takada A, et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J. Virol. 2005;79:10300–10307. doi: 10.1128/JVI.79.16.10300-10307.2005. ▪ Incorporation of L-domain mutations into infectious EBOV. The EBOV VP40 L-domains were found to enhance virus replication, but were not absolutely required for virus replication in cell culture.
- 36.Liu YL, Cocka L, Okumura A, Zhang YA, Sunyer JO, Harty RN. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J. Virol. 2010;84:2294–2303. doi: 10.1128/JVI.02034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamayoshi S, Kawaoka Y. Mapping of a region of Ebola virus VP40 that is important in the production of virus-like particles. J. Infect. Dis. 2007;196 Suppl. 2:S291–S295. doi: 10.1086/520595. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy SE, Johnson RF, Zhang YA, Sunyer JO, Harty RN. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J. Virol. 2007;81:11452–11460. doi: 10.1128/JVI.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. ▪ First report to provide important structural information and imaging of ZEBOV VP40.
- 40.Patch JR, Han ZY, McCarthy SE, et al. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol. J. 2008;5:137. doi: 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruigrok RW, Schoehn G, Dessen A, et al. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 42.Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, Weissenhorn W. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 2000;19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;312:59–68. doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen TL, Schoehn G, Weissenhorn W, et al. An all-atom model of the pore-like structure of hexameric VP40 from Ebola: structural insights into the monomer-hexamer transition. J. Struct. Biol. 2005;151:30–40. doi: 10.1016/j.jsb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Gomis-Rüth FX, Dessen A, Timmins J, et al. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11:423–433. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoenen T, Volchkov V, Kolesnikova L, et al. VP40 octamers are essential for Ebola virus replication. J. Virol. 2005;79:1898–1905. doi: 10.1128/JVI.79.3.1898-1905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittler E, Kolesnikova L, Strecker T, Garten W, Becker S. Role of the transmembrane domain of marburg virus surface protein GP in assembly of the viral envelope. J. Virol. 2007;81:3942–3948. doi: 10.1128/JVI.02263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bamberg S, Kolesnikova L, Moller P, Klenk HD, Becker S. VP24 of Marburg virus influences formation of infectious particles. J. Virol. 2005;79:13421–13433. doi: 10.1128/JVI.79.21.13421-13433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licata JM, Johnson RF, Han ZY, Harty RN. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 2004;78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson RF, Bell P, Harty RN. Effect of Ebola virus proteins GP, NP and VP35 on VP40 VLP morphology. Virol. J. 2006;233:31. doi: 10.1186/1743-422X-3-31. ▪ Analysis of ZEBOV VP40 VLP morphology by electron microscopy.
- 51.Li L, Cohen SN. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 52.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev. Mol. Cell. Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 53.Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell. Biol. 2002;12:569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 54.VerPlank L, Bouamr F, LaGarassa TJ, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc. Natl Acad. Sci. USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrus JE, Schwedler UK, Pornillos OW, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 56.Carter CA. Tsg101: HIV-1’s ticket to ride. Trends Microbiol. 2002;10:203–205. doi: 10.1016/s0966-842x(02)02350-8. [DOI] [PubMed] [Google Scholar]
- 57. Kolesnikova L, Strecker T, Morita E, et al. Vacuolar protein sorting pathway contributes to the release of Marburg virus. J. Virol. 2009;83:2327–2337. doi: 10.1128/JVI.02184-08. ▪ Similar to EBOV, Marburg virus utilizes the host vacuolar protein sorting pathway for efficient egress.
- 58.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 59.Magliery TJ, Wilson CGM, Pan W, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 60.Kinch MS, Yunus A, Mao H, et al. FGI-104: a broad-spectrum small molecule inhibitor of viral infection. Am. J. Transl. Res. 2009;1:87–98. [PMC free article] [PubMed] [Google Scholar]
- 61.Aman MJ, Kinch MS, Warfield K, et al. Development of a broad-spectrum antiviral with activity against Ebola virus. Antiviral Res. 2009;83:245–251. doi: 10.1016/j.antiviral.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Chen H, Liu X, Li Z, Zhan P, De Clercq E. TSG101: A novel anti-HIV-1 drug target. Curr. Med. Chem. 2010;17(8):750–758. doi: 10.2174/092986710790514444. [DOI] [PubMed] [Google Scholar]
- 63.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 64.Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 2004;78:2657–2665. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl Acad. Sci. USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. ▪ Elucidation of the mechanism by which human interferon-stimulated gene 15 inhibits budding of ZEBOV VP40 VLPs.
- 67. Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. ▪ Elucidation of the mechanism by which human interferon-stimulated gene 15 inhibits budding of ZEBOV VP40 VLPs.
- 68. Irie T, Licata JM, Harty RN. Functional characterization of Ebola virus L-domains using VSV recombinants. Virology. 2005;336:291–298. doi: 10.1016/j.virol.2005.03.027. ▪ Ebola virus VP40 L-domains could replace those of vesicular stomatitis virus M protein and were functional in the context of a vesicular stomatitis virus infection in cell culture.
- 69.Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J. Virol. 2010;84:704–715. doi: 10.1128/JVI.01319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamayoshi S, Noda T, Ebihara H, et al. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe. 2008;3:168–177. doi: 10.1016/j.chom.2008.02.001. ▪ Sec24C, a component of the host COPII vesicular transport system, interacts with ZEBOV VP40 and plays a critical role in transport of VP40 to the site of budding.
- 71.Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9:939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 72.Smith GA, Enquist LW. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 2002;18:135–161. doi: 10.1146/annurev.cellbio.18.012502.105920. [DOI] [PubMed] [Google Scholar]
- 73.Noda T. Assembly and budding of Ebolavirus. PloS Pathog. 2006;2:864–872. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ruthel G, Demmin GL, Kallstrom G, et al. Association of Ebola virus matrix protein VP40 with microtubules. J. Virol. 2005;79:4709–4719. doi: 10.1128/JVI.79.8.4709-4719.2005. ▪ Demonstrates the importance of cytoskeletal elements for VP40 transport and egress.
- 75.Wahl-Jensen VM, Afanasieva TA, Seebach J, Stroher U, Feldmann H, Schnittler HJ. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J. Virol. 2005;79:10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolesnikova LH, Bugany H-D, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 2002;76:1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Z, Harty RN. Packaging of actin into Ebola virus VLPs. Virol. J. 2005;2:92. doi: 10.1186/1743-422X-2-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han ZY, Harty RN. Influence of calcium/calmodulin on budding of Ebola VLPs: implications for the involvement of the Ras/Raf/MEK/ERK pathway. Virus Genes. 2007;35:511–520. doi: 10.1007/s11262-007-0125-9. [DOI] [PubMed] [Google Scholar]
- 79.Panchal RG, Ruthel G, Kenny T, et al. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl Acad. Sci. USA. 2003;100:15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aman MJ, Bosio CM, Panchal RG, et al. Molecular mechanisms of filovirus cellular trafficking. Microbes Infect. 2003;5:639–649. doi: 10.1016/s1286-4579(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 81.Freitas MS, Gaspar LP, Lorenzoni M, et al. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J. Biol. Chem. 2007;282:27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- 82.Jouvenet N, Neil SJ, Zhadina M, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakuma T, Sakurai A, Yasuda J. Dimerization of tetherin is not essential for its antiviral activity against Lassa and Marburg viruses. PLoS One. 2009;4:e6934. doi: 10.1371/journal.pone.0006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl Acad. Sci. USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. ▪ Tetherin-mediated restriction of EBOV VLPs can be overcome by coexpression of EBOV glycoprotein.
- 85.Tokarev A, Skasko M, Fitzpatrick K, Guatelli J. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res. Hum. Retroviruses. 2009;25:1197–1210. doi: 10.1089/aid.2009.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 87.Van Damme N, Goff D, Katsura C, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 2009;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korant BD, Blomstrom DC, Jonak GJ, et al. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J. Biol. Chem. 1984;259:14835–14839. [PubMed] [Google Scholar]
- 90.Sridharan H, Zhao C, Krug RM. Species specificity of the NS1 protein of influenza B virus: It binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 2010;285(11):7852–7856. doi: 10.1074/jbc.C109.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao C, Hsiang TY, Kuo RL, Krug RM. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl Acad. Sci. USA. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kallstrom G, Warfield KL, Swenson DL, et al. Analysis of Ebola virus and VLP release using an immunocapture assay. J. Virol. Methods. 2005;127:1–9. doi: 10.1016/j.jviromet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 93.Shi W, Huang Y, Sutton-Smith M, et al. A filovirus-unique region of Ebola virus nucleoprotein confers aberrant migration and mediates its incorporation into virions. J. Virol. 2008;82:6190–6199. doi: 10.1128/JVI.02731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han ZY, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 2003;77:1793–1800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoenen T, Allison G, Larissa K, et al. Infection of naive target cells with virus-like particles: implications for the function of Ebola virus VP24. J. Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silvestri LS, Ruthel G, Kallstrom G, et al. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J. Infect. Dis. 2007;196 Suppl. 2:S264–S270. doi: 10.1086/520610. [DOI] [PubMed] [Google Scholar]