Abstract
The origins of obligate pollination mutualisms, such as the classic yucca-yucca moth association, appear to require extensive trait evolution and specialization. To understand the extent to which traits truly evolved as part of establishing the mutualistic relationship, rather than being preadaptations, we used an expanded phylogenetic estimate with improved sampling of deeply-diverged groups to perform the first formal reconstruction of trait evolution in pollinating yucca moths and their non-pollinating relatives. Our analysis demonstrates that key life history traits of yucca moths, including larval feeding in the floral ovary and the associated specialized cutting ovipositor, as well as colonization of woody monocots in xeric habitats, may have been established before the obligate mutualism with yuccas. Given these preexisting traits, novel traits in the mutualist moths are limited to the active pollination behaviors and the tentacular appendages that facilitate pollen collection and deposition. These results suggest that a highly specialized obligate mutualism was built on the foundation of preexisting interactions between early Prodoxidae and their host plants, and arose with minimal trait evolution.
Suggested keywords: Prodoxidae, yucca moth, active pollination, obligate pollination mutualism, preadaptation
INTRODUCTION
Uncovering the origins of complex morphology and behavior offers insight into both the ecology and evolutionary dynamics of individual study organisms and the role of trait evolution in speciation and adaptive diversification (Britten & Davidson, 1971). An important question in this field is whether evolutionary novelties arise contemporaneously with an organism’s entry into a new adaptive zone(adaptation), or whether they are assembled via neofunctionalization of traits that predate the ecological transition (preadaptation or exaptation - Darwin, 1859; Armbruster, 1997; Gould, 2002; Lynch, 2007). Determining how particular traits achieved their present function can shed light on the selective pressures responsible for other adaptations (Armbruster, 1997), on the ecological opportunities that spur adaptive radiation (Schluter, 2000; Blount, Borland & Lenski, 2008), and on the relative roles of selection and contingency in the history of life (Lynch, 2007).
Tracing the evolutionary origins of complex traits has been a particular challenge in the case of obligate pollination mutualism, a class of plant-pollinator interactions in which the pollinator actively applies pollen to its host’s flowers so that its larvae may feed on a subset of the seeds produced. Examples include figs and fig wasps (Janzen, 1979; Herre, Jander & Machado, 2008), phyllanthaceous shrubs and Epicephala moths (Kawakita et al., 2004; Kawakita & Kato, 2006; Kawakita & Kato, 2009), Pachycereus (Lophocereus) cacti and the moth Upiga virescens (Holland & Rice, 1999; Holland, Buchanan & Loubeau, 2004), and yuccas and yucca moths (Pellmyr, 2003). These interactions are supported by complex arrays of traits in both partners, including specialized pollinator anatomy and behaviors for pollination and oviposition (Riley, 1872; Kawakita & Kato, 2006), and plant traits that prevent pollinators from overexploiting the interaction (Pellmyr & Huth, 1994) or otherwise manipulate pollinator behavior (Janzen, 1979).
The complexity and rarity of obligate pollination interactions point to the question of how they arise in the first place: to what extent are less-specialized ancestors adapted for obligate pollination after a transition to their present host plant, and to what extent do they shift to the host with some necessary traits already available to be exapted for the interaction? One way to answer this question is to estimate ancestral states using well-resolved phylogenetic relationships between the partners in an obligate mutualism and proximal lineages. This approach has been applied extensively in studies of insect-host associations (Armbruster, 1993; Kelley & Farrell, 1998; Nosil & Mooers, 2005; Stireman, 2005). However, because of a lack of phylogenetic resolution for relationships among lineages surrounding obligate mutualists, such an analysis has yet to be accomplished for any obligate pollination interaction.
One of the most classic such relationships is that between yuccas and yucca moths, which has been extensively described over more than a century of study (Riley, 1872; Pellmyr, 2003). A female yucca moth uses uniquely derived, tentacle-like mouthparts to collect pollen from one yucca flower and carry it to another, where she oviposits into the ovary and actively applies pollen to the stigma. Yuccas have no other pollinators, and yucca moth larvae feed only on developing yucca seeds or, in a few cases, other fruit tissue (Powell & Mackie, 1966; Davis, 1967; Powell, 1984; Pellmyr, 2003). Systematic and ecological studies of this model system have uncovered substantial diversity within the monophyletic group comprising the three yucca moth genera Tegeticula, Parategeticula, and Prodoxus (Table 1; Pellmyr, 1999; Pellmyr et al., 2005; Pellmyr et al., 2008). At least 22 species of “bogus yucca moths” in the genus Prodoxus oviposit on yuccas or Agave without pollinating, five species in Parategeticula are obligate pollinators, and 18 species of Tegeticula are obligate pollinators (Table 1; Davis, 1967; Pellmyr et al., 2005; Pellmyr et al., 2008). Two Tegeticula species are independently derived, non-pollinating “cheater yucca moths” (Pellmyr, Leebens-Mack & Huth, 1996a; Althoff et al., 2006). All feed on woody monocots in the family Agavaceae, and the vast majority use species in the genus Yucca. The sister group to the yucca-feeding moths is the genus Mesepiola, which feeds on woody monocots in the Ruscaceae (Table 1; Force & Thompson, 1984; Force, 1989).
TABLE 1.
Diversity of host associations and larval feeding habit in the Prodoxidae
| Genus (species sampled) | Pollination | Host family associations | Larvae feed in |
|---|---|---|---|
| Yucca moths | |||
| Tegeticula (20) | Active, obligate; two species non-pollinating | Agavaceae | Floral ovaries, mature fruit1 |
| Parategeticula (5) | Active, obligate | Agavaceae | Floral ovaries |
| Prodoxus (21) | Non-pollinating | Agavaceae | Floral stems, mature fruit, leaves |
| Basal Prodoxidae | |||
| Mesepiola (2) | Non-pollinating | Ruscaceae | Floral ovaries |
| Lampronia (7) | Non-pollinating | Betulaceae, Geraniaceae, Grossulariacae, Rosaceae, Saxifragaceae | Floral ovaries, twigs |
| Greya (5) | Non-pollinating; some passive, facultative | Saxifragaceae, Apiaceae | Floral ovaries, floral stems |
| Tetragma (1) | Non-pollinating | Rosaceae | Floral ovaries |
oviposition into mature fruit is associated with the two species of non-pollinating “late cheaters” in Tegeticula (Pellmyr & Leebens-Mack, 2000)
Previous studies have used molecular clock methods to estimate the age of the yucca-yucca moth mutualism at 35.6 ± 9.0 million years (Pellmyr & Leebens-Mack, 1999) and described the origins of the pollen-carrying tentacles unique to yucca moths (Pellmyr & Krenn, 2002), but none have reconstructed the evolution of the ecological traits that set the context in which the mutualism arose. This is due to the limitations of ancestral state reconstruction methods available when the first phylogeny of the Prodoxidae was published, a lack of phylogenetic resolution for the sister group to the woody monocot-feeding Prodoxidae, and poor sampling of deeply diverged lineages that left much of the group’s ecological diversity unrepresented. These include some 30 species in the genus Lampronia, one in Tetragma, and some 20 in Greya, which use an array of host plants and larval feeding habits (Table 1; Nielsen & Davis, 1985; Davis, Pellmyr & Thompson, 1992; Davis, 1999). In the species-rich Lampronia (for those species whose larval biology is known) larvae feed inside dicotyledonous host plants in the families Betulaceae, Geraniaceae, Grossulariacae, Rosaceae, and Saxifragaceae. The single species in Tetragma oviposits and feeds in the floral ovaries of Geum triflorum (Rosaceae; Davis et al., 1992). Finally, members of the genus Greya lay eggs on members of the Saxifragaceae and Apiaceae; alone among the basal Prodoxidae, some Greya species are passive, facultative pollinators of their host plants (Thompson & Pellmyr, 1992; Pellmyr et al., 1996b; Thompson & Fernandez, 2006).
The diversity of the Prodoxidae means that ancestral state reconstruction of the lineages leading to the yucca moths requires complete resolution of the relationships among Lampronia, Tetragma, Greya, and the monocot feeders. Two early phylogenetic estimates reported conflicting results about the identity of the sister group to the monocot-feeding Prodoxidae (Brown et al., 1994; Pellmyr & Leebens-Mack, 1999); however, both of these analyses included only a single representative of the deeply-diverged Lampronia. New collections now allow us to address these issue and develop the first rigorous ancestral state reconstruction for the Prodoxidae, using expanded sampling of Lampronia. We reconstructed the mitochondrial data set used in Pellmyr and Leebens-Mack’s (1999) analysis with existing sequence data (Table S1), and added newly-acquired sequences from a representative sample of the diverse genus Lampronia. We confirmed the robustness of the mitochondrial phylogeny by acquiring sequence data from the nuclear locus coding for arginine kinase (Arg-K) in a subset of taxa including representatives of each Prodoxid genus (Table S1) and testing for phylogenetic conflict between the mitochondrial and nuclear loci. We then used the mitochondrial phylogenetic estimate to reconstruct ancestral ecological states in the prodoxid moths that first colonized woody monocots and eventually established the obligate mutualism with yuccas. Our results show that the seemingly complex yucca moth life habit may have required few novel traits in its evolution from non-pollinating ancestors, and that the diversification of yucca moths may be linked not to the origin of obligate pollination but to dramatic habitat expansion into arid regions.
MATERIALS AND METHODS
We assembled specimens of the basal prodoxid genera Lampronia, Greya, and Mesepiola, and the outgroup Adela septentrionella (Lepdioptera: Adelidae) from sites in the United States and Europe (Table S1). We extracted whole genomic DNA from each specimen, and sequenced the mitochondrial COI-II and the nuclear Arg-K loci. We assessed phylogenetic conflict between the loci using PAUP* (v4.0b, Swofford, 2002), and estimated the mitochondrial phylogeny using MRBAYES (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003). We then used the resulting posterior sample of equally-credible trees for ancestral trait reconstruction in BayesTraits (Pagel, Meade & Barker, 2004). Genomic DNAacquisition, PCR, and sequence data collection were conducted using standard reagents and protocols; for details see the Supplementary Material.
Phylogenetic analysis
Sequences from each locus were automatically aligned and combined into contigs using CodonCode Aligner version 2.0.6 (CodonCode Corporation, Dedham, MA, USA), and inspected for read quality and accuracy. We assessed conflict between the mitochondrial and nuclear loci using a partition homogeneity test conducted in PAUP* (v4.0b, Swofford, 2002), which did not detect significant heterogeneity (p=0.97). We combined the newly collected mtDNA sequence data with previously published COI-COII sequences from Prodoxus, Tegeticula, Greya, and Lampronia (Table S1), automatically aligned them in CodonCode Aligner, and checked for conservation of amino acid sequences in MacClade (version 4.08; Maddison & Maddison, 2005). We chose a model of nucleotide substitution for the mitochondrial dataset using jModelTest version 0.0.1 (Posada & Crandall, 1998; Guindon & Gascuel, 2003; Posada, 2008) which selected a general-time reversible model with gamma-distributed rate variation and a proportion of invariant sites (GTR+I+Γ). We performed Bayesian phylogenetic reconstruction using MRBAYES version 3.1.2 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003), as described in the Supplementary Material.
Ancestral state reconstruction
Following phylogenetic estimation, we reconstructed the evolution of host association and larval feeding habit in the history of the Prodoxidae using the Multistate module of BayesTraits (www.evolution.rdg.ac.uk; Pagel et al., 2004). BayesTraits accounts for phylogenetic uncertainty in its ancestral state reconstructions by sampling the posterior distribution of equally credible phylogenetic trees—in this case, 706 post-burn-in trees from the above MRBAYES analysis. Ancestral states are estimated not for a given node in a consensus tree, but for the most recent common ancestor (MRCA) of selected taxa regardless of whether they form a monophyletic group in a particular topology, which allows BayesTraits to incorporate information about trait evolution from all trees in the posterior distribution. For simplicity, we report ancestral state reconstructions with reference to specific nodes on the consensus topology. Parameter settings and details of the BayesTraits analysis are given in the Supplementary Material.
Specific hypothesis testing
In some cases, BayesTraits could not clearly resolve ancestral states at nodes of particular interest in the history of the yucca moths, so that the most probable state was assigned with mean probability < 0.5. In these cases, we fixed the node in question at a possible character state using the BayesTraits fossil command and performed MCMC estimation of trait evolution as above, repeating this analysis for each possible character state that was assigned with mean probability > 0.1. We averaged the harmonic mean likelihood (HML) of the Markov chain Monte Carlo estimates across five independent replicate runs for each state, and determined the relative support for each possible character state using the Bayesian Information Criterion (BIC) test statistic, as recommended by the BayesTraits manual.
RESULTS
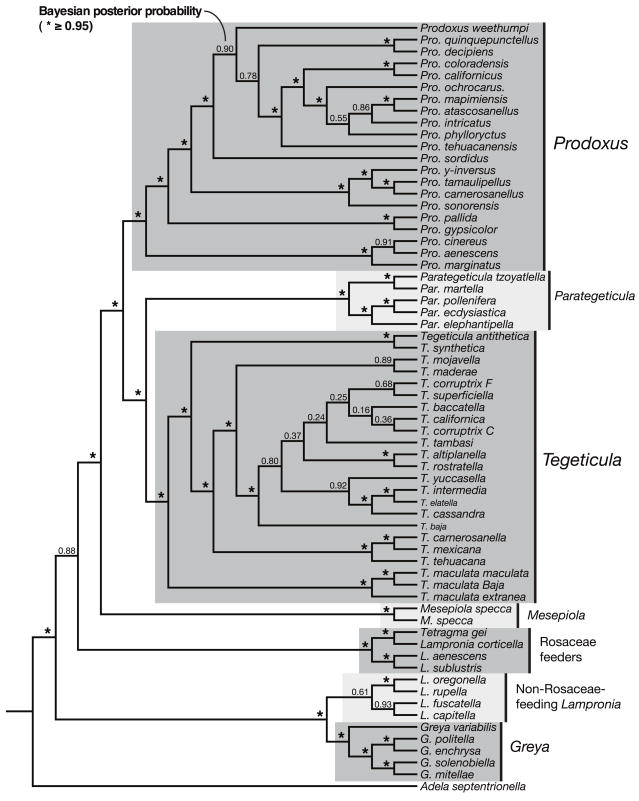
The final Bayesian consensus tree of the Prodoxidae is pictured in Figure 1. Within the monocot-feeding group composed of Mesepiola, Tegeticula, Parategeticula, and Prodoxus, genus-level relationships were consistent with the results presented by Pellmyr and Leebens-Mack (1999). The genera Lampronia, Tetragma, and Greya were resolved into two clades: one, the sister to the woody monocot feeders, consisted of all known Rosaceae-feeding members of Lampronia + Tetragma gei. The other, more basal clade consisted of the remainder of the Lampronia species and the genus Greya, which remained a monophyletic entity. In general, species on the tree formed monophyletic entities with common host associations (Figure 1).
FIGURE 1.
MRBAYES consensus topology for the Prodoxidae, showing all groupings compatible with the post-burn-in sample of equally credible trees. Bayesian posterior probabilities are printed at nodes unless greater than or equal to 0.95. The consensus topology matches that proposed by Pellmyr and Leebens-Mack (1999), placing a subset of Lampronia and Tetragma gei as sister group to the woody monocot feeders Mesepiola, Prodoxus, Tegeticula, and Parategeticula.
Ancestral host associations
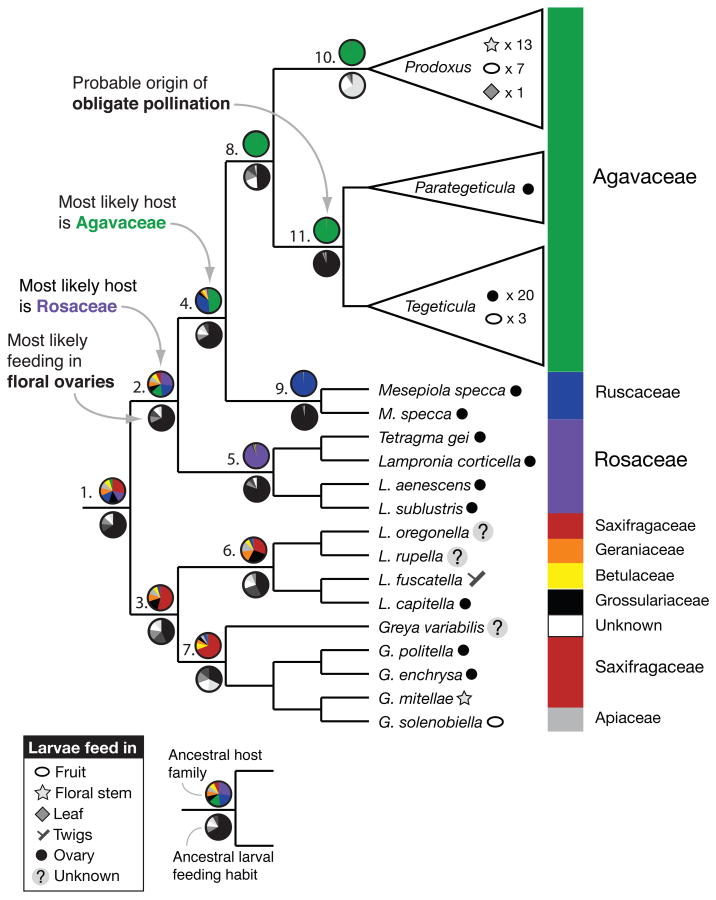
We reconstructed ancestral host associations for the most recent common ancestors of clades of interest in the phylogeny of the Prodoxidae (Figure 2, Table S4, Table S5). The MRCA of the woody monocot feeders and the rose feeders most likely fed on members of the Rosaceae in an unrestricted ancestral state estimation (mean probability = 0.2730±0.0048; Table S4), and hypothesis testing using the fossil command found significantly better support for Rosaceae at this node than for Ruscaceae (delta BIC=2.152) or Agavaceae (delta BIC=4.532). The MRCA of the woody monocot feeders was found to feed on woody monocots (mean probability for Ruscaceae or Agavaceae = 0.8414; Table S4); but while the probability of assignment from an unrestricted estimate favored Agavaceae (mean probability = 0.4872 ± 0.0079; Table S4), specific hypothesis testing gave Ruscaceae better support than Agavaceae (delta BIC= 2.279). Ancestral nodes within the yucca moth clade are strongly expected to have fed on members of the Agavaceae (mean probabilities all >0.95; Table S4). This suggests that the common ancestor of the woody monocot feeders transitioned to this host group from an earlier habit of oviposition and feeding in members of the Rosaceae.
FIGURE 2.
Simplified phylogeny of the Prodoxidae, showing host plant family associations and larval feeding habits for extant taxa, and the relative probabilities of host association and larval feeding habits for selected ancestral nodes, calculated as the mean of the posterior probabilities of possible states estimated by BayesTraits. Posterior probabilities of possible ancestral states for each numbered node are given in Table S4 (host association) and Table S5 (larval feeding site). Because BayesTraits estimates ancestral states not for nodes, but for the MRCA of selected taxa, these reconstructions are not for the nodes indicated per se, but for the common ancestor of the taxa arising from those nodes, regardless of whether they form a monophyletic group.
Ancestral larval feeding habit
BayesTraits reconstructions indicated that, for the deepest nodes, larval feeding primarily occurred in host plants’ floral ovaries, with transitions to feeding in other host tissues arising later (Figure 2; Table S5). The floral ovary was the most probable larval feeding site for the MRCA of the rose feeders and the woody monocot feeders (mean probability = 0.6824 ± 0.0069; Table S5), for the MRCA of the woody monocot feeders (mean probability = 0.6711 ± 0.0077; Table S5), and for the MRCA of the yucca moths Prodoxus + Tegeticula + Parategeticula (mean probability = 0.3886 ± 0.0075; delta BIC ≥ 5.708 versus all other possible states). The MRCA of the pollinating yucca moths Tegeticula + Parategeticula—which represents the probable origin of the obligate mutualism—is strongly expected to have fed inside floral ovaries (mean probability= 0.9415 ± 0.0022), whereas the MRCA of the “bogus yucca moths” in genus Prodoxus is estimated to have fed in floral stems (mean probability = 0.6605 ± 0.0076). This suggests that use of the floral ovary pre-dates the emergence of the yucca-yucca moth mutualism, and that the use of floral stem tissue or mature fruits by Prodoxus is the derived state.
DISCUSSION
Phylogenetic reconstruction based on a single locus may not capture the true species tree (Nichols, 2001; Degnan & Rosenberg, 2006; Edwards, Liu & Pearl, 2007). However, lacking nuclear loci that have been successfully amplified in an exhaustive sample of the Prodoxidae, the COI-COII locus remains the best available basis for phylogenetic estimation in this family. The poorly-resolved node joining the monocot-feeding clade to the rose-feeding Lampronia clade on the mitochondrial phylogeny is associated with relatively short branch lengths (Figure 1), suggesting that diversification occurred too rapidly for informative mutations to accumulate at this point in the tree. This problem will likely apply to nuclear loci as well as the mitochondrial genome, so that collection of further nuclear sequence data is unlikely to substantially improve phylogenetic resolution of this node. Nevertheless, the lack of phylogenetic conflict between the COI-II locus and the Arg-K locus corroborates the topology of the mitochondrial phylogeny.
Estimates of ancestral states may be directly impacted by the frequency of states among in the terminal taxa (Nosil & Mooers, 2005). While our analysis includes better sampling of the deeply-diverged genus Lampronia than any previous study, and represents much of the diversity of host associations and larval feeding habits within the group, it does not include every recognized Lampronia species. However, the finding that the sister to the monocot feeders is a clade of species that all feed in the floral ovaries of rosaceous plants suggests that our conclusions about the ecology of the Prodoxids that first colonized woody monocots would be robust to broader sampling.
In the two earlier attempts to resolve deep phylogenetic relationships in the Prodoxidae, only a single species of the large genus Lampronia (Davis, 1999) was available for analysis. Expanded sampling of deeply diverged lineages and formal reconstruction of the suites of traits that determined the ecological context in which the yucca-yucca moth mutualism arose reveals previously undetected structure in the diverse genus Lampronia and confirms earlier results indicating that obligate mutualism between the yuccas and yucca moths does not have a common origin with the facultative pollination mutualism seen in some members of Greya. More importantly, our results provide key insights to understanding the assembly of the yucca-yucca moth mutualism, indicating that the first monocot-feeding prodoxids arose from ancestral species using members of the Rosaceae, in a transition marked by minimal change in moth oviposition behavior and associated anatomy even as it required adjustment to dramatically different ecological conditions, moving from hosts associated with mesic conditions to hosts found in xeric or semi-xeric communities. This transition set the stage for the emergence of the classic obligate pollination mutualism, for which, our results suggest, the ancestral prodoxids were substantially preadapted.
Evolution of host association and larval feeding habits
Given a known pattern of specialization on specific plant tissues among the yucca moths (Pellmyr et al., 2005) and the strong mechanistic association between larval feeding on floral ovaries and active pollination (Pellmyr & Leebens-Mack, 2000), the evolution of larval feeding habits and host association in the deeper nodes of the Prodoxidae is central to understanding the origins of the yucca-yucca moth mutualism. All analyzed members of the sister group to the monocot feeders oviposit into and feed in floral ovaries, and this habit is conserved at the transition to woody monocots (Figure 2), as well as in the obligate pollinators in Tegeticula and Parategeticula (with the caveat that the latter oviposits in the floral pedicels or, rarely, petal tissue, and the larvae subsequently enter the ovary to feed).
These results have important implications for our understanding of how complex pollination mutualisms evolve. Key life habits within the lineage are conserved during the transition to obligate mutualism, particularly larval consumption of developing seeds and associated traits such as oviposition into the host ovary with a cutting ovipositor. The only significant morphological novelty in the yucca moths necessary for obligate mutualism, then, is the tentacular mouthparts used for pollen manipulation, which are suggested to have arisen through duplication of the galeae (Pellmyr & Krenn, 2002). These mouthparts provide for more efficient pollen manipulation, and would have been a highly adaptive innovation for ancestral yucca moths already dependent on developing seeds as a larval food source.
Ecological expansion with pollinator specialization
The transition from Rosaceae-feeding to woody monocots is associated not only with limited morphological change, but a distinct habitat expansion from originally mesic or humid habitats into semi-xeric and xeric areas. Although the colonization of the woody monocots—estimated to have occurred 44.1 ± 10.5 Mya (Pellmyr & Leebens-Mack 1999)—predates most estimates for the origin of the North American deserts(Hutchison, 1992; Becerra, 2005), this transition may have set the stage for subsequent diversification as arid environments expanded in western North America. This may explain the more than fivefold increase in prodoxid diversity in North America; of the ~57 North American prodoxid species, 48 feed on yuccas or other monocots (Pellmyr et al., 2005; Pellmyr et al., 2008; D.R. Davis, pers. comm.). Colonization of novel habitats is widely associated with diversification events such as that seen in the fivefold increase in Prodoxid diversity following colonization of woody monocots (Simpson, 1953), and diversification following colonization of arid habitats in particular has been observed in other specialized phytophagous insects (e.g., McLeish, Chapman & Schwarz, 2007).
To better understand the pattern of macroevolutionary change leading to this obligate pollination mutualism, we should then focus on factors leading to the colonization of arid habitats, which coincided with the stepwise colonization of two separate lineages of woody monocots. It is not until after these transitions that the morphologically and behaviorally minor, but ecologically major, changes appeared that led to the origin of the obligate mutualism. Thus it is reasonable to conclude that the obligate mutualism between yuccas and yucca moths arose at least in part because the early Prodoxidae were pre-adapted for the more specialized interaction.
Supplementary Material
Acknowledgments
The authors thank M. Barrlund, B.Å. Bengtsson, S. Hellqvist, and D. Quicke for providing critical specimens for this study. Members of the Lancashire & Cheshire Entomological Society provided helpful clues in tracking down the L. fuscatella sample. W.K.W. Godsoe and L.J. Harmon gave invaluable comments on earlier drafts of this paper. This project was funded by grants from NSF (DEB 0516841 to OP and CIS), and NIH (NCRR P20RR1644 and P20RR016454 to the IBEST Bioinformatics Core at the University of Idaho).
References
- Althoff DM, Segraves KA, Leebens-Mack J, Pellmyr O. Patterns of speciation in yucca moths: Parallel species radiations within the Tegeticula yuccasella species complex. Syst Biol. 2006;55:398–410. doi: 10.1080/10635150600697325. [DOI] [PubMed] [Google Scholar]
- Armbruster WS. Evolution of plant pollination systems: Hypotheses and tests with the neotropical vine Dalechampia. Evolution. 1993;47:1480–1505. doi: 10.1111/j.1558-5646.1993.tb02170.x. [DOI] [PubMed] [Google Scholar]
- Armbruster WS. Exaptations link evolution of plant-herbivore and plant-pollinator interactions: a phlyogenetic inquiry. Ecology. 1997;78:1661–1672. [Google Scholar]
- Becerra JX. Timing the origin and expansion of the Mexican tropical dry forest. Proc Nat Acad Sci USA. 2005;102:10919. doi: 10.1073/pnas.0409127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Nat Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Quarterly Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Brown JM, Pellmyr O, Thompson JN, Harrison RG. Phylogeny of Greya (Lepidoptera: Prodoxidae), based on nucleotide sequence variation in mitochondrial cytochrome oxidase I and II: congruence with morphological data. Mol Biol Evol. 1994;11:128–141. doi: 10.1093/oxfordjournals.molbev.a040087. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species. John Murray: London; 1859. [Google Scholar]
- Davis DR. A revision of the moths of the sub-family Prodoxidae (Lepidoptera: Incurvariidae) Bull US Natl Mus. 1967;255:1–170. [Google Scholar]
- Davis DR. The Monotrysian Heteroneura. In: Kristensen NP, editor. Handbook of Zoology, Part 35: Lepidoptera. Berlin, New York: W. de Gruyter; 1999. pp. 65–90. [Google Scholar]
- Davis DR, Pellmyr O, Thompson JN. Biology and systematics of Greya Busck and Tetragma n. gen. (Lepidoptera: Prodoxidae) Smithsonian Contrib Zool. 1992;524:1–88. [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genetics. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK. High-resolution species trees without concatenation. Proc Nat Acad Sci USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force DC. Observations on the parasitoids of Mesepiola specca Davis (Lepidoptera: Incurvariidae) Pan-Pacific Entomol. 1989;65:436–439. [Google Scholar]
- Force DC, Thompson ML. Parasitoids of the immature stages of several southwestern yucca moths. Southwestern Nat. 1984;29:45–56. [Google Scholar]
- Gould SJ. The Structure of Evolutionary Theory. Bellknap Press of Harvard; Cambridge: 2002. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Herre EA, Jander KC, Machado CA. Evolutionary ecology of figs and their associates: Recent progress and outstanding puzzles. Annu Rev Ecol Evol Syst. 2008;39:439–458. [Google Scholar]
- Holland B, Rice WR. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc Nat Acad Sci USA. 1999;96:5083–5088. doi: 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JN, Buchanan AL, Loubeau R. Oviposition choice and larval survival of an obligately pollinating granivorous moth. Evol Ecol Res. 2004;6:607–618. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hutchison JH. Western North American reptile and amphibian record across the Eocene/Oligocene boundary and its climatic implications. In: Prothero DR, Berggen WA, editors. Eocene-Oligocene Climatic and Biotic Evolution. Princeton, NJ: Princeton University Press; 1992. pp. 451–463. [Google Scholar]
- Janzen DH. How to be a fig. Annu Rev Ecol Syst. 1979;10:13–51. [Google Scholar]
- Kawakita A, Kato M. Assessment of the diversity and species specificity of the mutualistic association between Epicephala moths and Glochidion trees. Mol Ecol. 2006;15:3567–3581. doi: 10.1111/j.1365-294X.2006.03037.x. [DOI] [PubMed] [Google Scholar]
- Kawakita A, Kato M. Repeated independent evolution of obligate pollination mutualism in the Phyllantheae–Epicephala association. Proc R Soc B. 2009;276:417–426. doi: 10.1098/rspb.2008.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A, Sota T, Ito M, Ascher JS, Tanaka H, Kato M, Roubik DW. Phylogeny, historical biogeography, and character evolution in bumble bees (Bombus: Apidae) based on simultaneous analysis of three nuclear gene sequences. Mol Phylogenetics and Evol. 2004;31:799–804. doi: 10.1016/j.ympev.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Kelley ST, Farrell BD. Is specialization a dead end? The phylogeny of host use in Dendroctonus bark beetles (Scolytidae) Evolution. 1998;52:1731–1743. doi: 10.1111/j.1558-5646.1998.tb02253.x. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Nat Acad Sci USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade. 4.08. Sunderland, MA: Sinauer Associates, Inc; 2005. [Google Scholar]
- McLeish M, Chapman T, Schwarz M. Host-driven diversification of gall-inducing Acacia thrips and the aridification of Australia. BMC Biology. 2007;5:3. doi: 10.1186/1741-7007-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R. Gene trees and species trees are not the same. Trends Ecol Evol. 2001;16:358–364. doi: 10.1016/s0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- Nielsen ES, Davis DR. The first southern hemisphere prodoxid and the phylogeny of the Incurvarioidea (Lepidoptera) Syst Entomol. 1985;10:307–322. [Google Scholar]
- Nosil P, Mooers AO. Testing hypotheses about ecological specialization using phylogenetic trees. Evolution. 2005;59:2256–2263. [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Pellmyr O. Systematic revision of the yucca moths in the Tegeticula yucasella complex (Lepidoptera: Prodoxidae) north of Mexico. Syst Entomol. 1999;24:243–271. [Google Scholar]
- Pellmyr O. Yuccas, yucca moths, and coevolution: A review. Ann Missouri Bot Gard. 2003;90:35–55. [Google Scholar]
- Pellmyr O, Balcazar-Lara M, Althoff DM, Segraves KA, Leebens-Mack J. Phylogeny and life history evolution of Prodoxus yucca moths (Lepidoptera: Prodoxidae) Syst Entomol. 2005;31:1–20. [Google Scholar]
- Pellmyr O, Balcazar-Lara M, Segraves KA, Althoff DM, Littlefield RJ. Phylogeny of the pollinating yucca moths, with revision of Mexican species (Tegeticula and Parategeticula; Lepidoptera, Prodoxidae) Zool J Linnean Soc. 2008;152:297–314. [Google Scholar]
- Pellmyr O, Huth CJ. Evolutionary stability of mutualism between yuccas and yucca moths. Nature. 1994;372:257–260. [Google Scholar]
- Pellmyr O, Krenn HW. Origin of a complex key innovation in an obligate insect-plant mutualism. Proc Nat Acad Sci USA. 2002;99:5498–5502. doi: 10.1073/pnas.072588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J. Forty million years of mutualism: Evidence for Eocene origin of the yucca-yucca moth association. Proc Nat Acad Sci USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J. Reversal of mutualism as a mechanism for adaptive radiation in yucca moths. Am Nat. 2000;156:S62–S76. doi: 10.1086/303416. [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J, Huth CJ. Non-mutualistic yucca moths and their evolutionary consequences. Nature. 1996a;380:256–257. doi: 10.1038/380155a0. [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Thompson JN, Brown JM, Harrison RG. Evolution of pollination and mutualism in the yucca moth lineage. Am Nat. 1996b;148:827–848. [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. ModelTest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Powell JA. Biological interrelationships of moths and Yucca schottii. Univ Calif Publ Entomol. 1984;100:1–93. [Google Scholar]
- Powell JA, Mackie RA. Biological interrelationships of moths and Yucca whipplei (Lepidoptera: Gelechiidae, Blastobasidae, Prodoxidae) University of California Press; 1966. [Google Scholar]
- Riley CV. The fertilization of the yucca plant by Pronuba yuccasella. Can Entomol. 1872;4:182. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford Univ. Press; Oxford: 2000. [Google Scholar]
- Simpson GG. The Major Features of Evolution. Columbia University Press; New York: 1953. [Google Scholar]
- Stireman JO. The evolution of generalization? Parasitoid flies and the perils of inferring host range evolution from phylogenies. J Evol Biol. 2005;18:325–336. doi: 10.1111/j.1420-9101.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0. Sunderland, Mass: Sinauer Associates; 2002. [Google Scholar]
- Thompson JN, Fernandez CC. Temporal dynamics of antagonism and mutualism in a geographically variable plant-insect interaction. Ecology. 2006;87:103–112. doi: 10.1890/05-0123. [DOI] [PubMed] [Google Scholar]
- Thompson JN, Pellmyr O. Mutualism with pollinating seed parasites amid co-pollinators: constraints on specialization. Ecology. 1992;73:1780–1791. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.