Abstract
An important need of many cancer research projects is the availability of high-quality, appropriately selected tissue. Tissue biorepositories are organized to collect, process, store, and distribute samples of tumor and normal tissue for further use in fundamental and translational cancer research. This, in turn, provides investigators with an invaluable resource of appropriately examined and characterized tissue specimens and linked patient information. Human tissues, in particular, tumor tissues, are complex structures composed of heterogeneous mixtures of morphologically and functionally distinct cell types. It is essential to analyze specific cell types to identify and define accurately the biologically important processes in pathologic lesions. Laser capture microdissection (LCM) is state-of-the-art technology that provides the scientific community with a rapid and reliable method to isolate a homogeneous population of cells from heterogeneous tissue specimens, thus providing investigators with the ability to analyze DNA, RNA, and protein accurately from pure populations of cells. This is particularly well-suited for tumor cell isolation, which can be captured from complex tissue samples. The combination of LCM and a tissue biorepository offers a comprehensive means by which researchers can use valuable human biospecimens and cutting-edge technology to facilitate basic, translational, and clinical research. This review provides an overview of LCM technology with an emphasis on the applications of LCM in the setting of a tissue biorepository, based on the author's extensive experience in LCM procedures acquired at Fox Chase Cancer Center and Hollings Cancer Center.
Keywords: pathology, cancer biology, cells of interest
INTRODUCTION OVERVIEW OF LCM TECHNOLOGY
Laser capture microdissection (LCM) is a state-of-the-art technology for isolating pure cell populations from a heterogeneous tissue specimen. It can precisely target and capture the cells of interest for a wide range of downstream assays. In early 1986, the laser microbeam microdissection was first described.1 Following the first publication about LCM in 1996, which appeared in Science,2 LCM technology was rapidly commercialized by Arcturus (Molecular Devices, Inc., Sunnyvale, CA, USA). In the meantime, The PALM Microbeam (Carl Zeiss MicroImaging GmbH, Bernried, Germany) and Leica LMD6000 (Leica Microsystems Inc., Bannockburn, IL, USA) laser microdissection systems were also developed promptly and broadened its applications internationally. Today, thousands of researchers worldwide benefit from this technology, and thousands of publications involving LCM have appeared. As a result of the development of this technology, approaches to molecular analysis of pathologic processes have been enhanced significantly.
The LCM system is based on an inverted light microscope (with or without a fluorescent module), fitted with a laser device to facilitate the visualization and procurement of cells. Today, there are two general classes of laser microdissection: infrared (IR) laser-capture system and ultraviolet (UV) laser-cutting system. The laser-capture technique was developed in the 1990s by Dr. Emmert-Buck and colleagues at the National Institutes of Health (Bethesda, MD, USA).2 The principle of LCM consists of visualizing the cells of interest directly via microscopy, while having a thermoplastic transfer film attached to a plastic cap that overlies the cells. While viewing the cells, an IR, low-power laser is pulsed through the top of the cap, hitting the transfer film, which then melts and bonds to the cells or regions of interest. The film absorbs the laser radiation creating a gentle, nondamaging microdissection that preserves the integrity of the captured material. Laser impulses can be repeated multiple times across the whole cap; thus, up to 3000–5000 cells can be isolated onto a single cap. IR LCM platforms are available as PixCell, Veritas, and Arcturus XT systems (Molecular Devices, Inc.). The experience in my own research laboratory is with PixCell II, Veritas, and Arcturus XT systems. Distinct from LCM, laser-cutting microdissection uses a narrow-beam UV laser to draw around and excise cells of interest from surrounding cells and tissue. Variations of UV cutting systems include positioning and ablation with laser microbeam (PALM) systems (Carl Zeiss MicroImaging GmbH, http://www.zeiss.com) and UV laser-cutting systems (Leica LMD6000, Leica Microsystems, Inc., http://leica-microsystems.com). In contrast to LCM, the UV-microdissected cells or regions of interest are instead deposited directly into a collection tube by gravity or catapult. By ablating the adjacent rim of unwanted tissue, nonspecific adherence of tissue to the tube is avoided. Thus, these completely noncontact systems decrease the possibility of contamination from cells nonspecifically adhering to a plastic membrane cap. The solid-state IR laser applies a gentle capture technique, which preserves biomolecular integrity and is ideal for single cells and small numbers of cells. The UV laser delivers unprecedented speed and precision and is well-suited for microdissecting dense tissue structures and for capturing large numbers of cells. The Veritas and Arcurus XT systems are combined IR/UV systems for ultimate microdissection flexibility.
WORKFLOW OF LCM
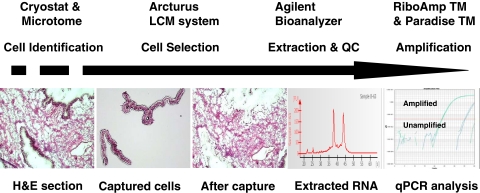
The process of LCM is straightforward, and there are a considerable number of commercially available kits that have aided in simplifying the process. The workflow of LCM is shown in Figure 1. Sample preparation for LCM is similar to routine slide preparation in the pathology laboratory. Optimal cutting temperature compound-embedded frozen tissue or formalin-fixed paraffin-embedded (FFPE) tissues are sectioned by cryostat or microtome. The optimal tissue section thickness for LCM is 4–15 μm, mounted onto glass slides, followed by dehydration after tissue staining. When using glass slides, they need to be uncharged and noncoated, as either feature may interfere with the transfer of tissue from the slide onto the cap. After staining tissue sections with H&E or by immunohistochemistry for specific cell identification, the LCM cap, comprised of a plastic support with thermolabile polymer film and dye, is then placed on top of the tissue section. The laser pulses through the cap and causes the thermoplastic film to form a thin protrusion that bridges the gap between the cap and tissue and adheres to the target cell. In this way, the dye on the film absorbs the laser energy and provides a means of visualizing where each laser pulse has targeted. The laser-activated film adheres tightly to the underlying cells, which are then removed selectively from the tissue section when the cap is lifted. After microdissection is completed, the cap with adhered target cells is placed into a tube that contains an appropriate buffer for DNA, RNA, or protein extraction, and the biomolecules are recovered for downstream analysis. The quality of the isolated DNA, RNA, and protein can be monitored and assessed with a bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The quality assessment of RNA, extracted from an entire tissue section, scraped from a slide, is usually necessary (and recommended) prior to LCM procedure. Two peaks of ribosomal RNA, 18S and 28S, and RNA integrity number (RIN) should be obtained in the bioanalyzer profile. High RINs represent successful RNA sample preparation (Fig. 2). For generating samples to be analyzed by cDNA microarray or comparative genomic hybridization, where large amounts of RNA or DNA are required, amplification of the captured and purified RNA/DNA can be performed to obtain sufficient starting material for these applications.
FIGURE 1.
The workflow of LCM. Tissue sections were cut using a cryostat or microtome and stained with hematoxylin and eosin (H&E) for cell identification. Pure epithelial cell populations from frozen ovarian tissue were captured using the Arcturus XT LCM system and visualized on the LCM cap. Total RNA was then extracted, and the quantity and quality of RNA were monitored on an Agilent bioanalyzer. Two peaks, 18S and 28S ribosomal RNA, were obtained in the RNA profile generated by the bioanalyzer, indicating successful RNA sample isolation. The RNA was amplified for downstream analysis. QC, quality control; qPCR, quantitative PCR.
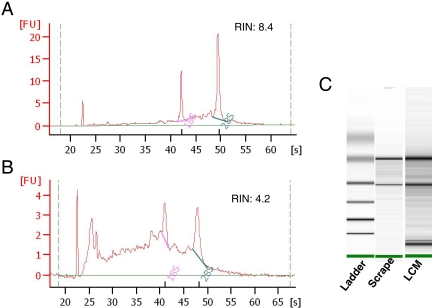
FIGURE 2.
RNA profile generated by Agilent Bioanalyzer. High-quality RNA was extracted from LCM-isolated cells. RNA size and quality were analyzed with the Agilent 2100 bioanalyzer. A pseudo gel image was created, and bands were sized and quantified. (A and B) The y-axes indicate fluorescence units (FU), and the x-axes indicate the length of the RNA in nucleotides. The peaks of 18S and 28S rRNA are clearly visible. Shown are the profiles of total RNA extracted from scrapes of the entire prostate section (A) and from LCM-isolated cells (B). (C) Gel electrophoresis analysis of the same RNA in A and B.
LCM has been applied to a wide range of tissues, including frozen and FFPE tissues. The ideal tissue preparation ensures that the morphology of tissue and biomolecules is preserved. Frozen tissue offers excellent preservation of RNA, DNA, and proteins and is optimal for RNA, DNA, or protein analysis. However, it lacks clear histological detail and is inconvenient for handling and storage. Frozen tissue sections need to be stored at –80°C prior to LCM. As RNA/protein quality will degrade rapidly after staining has occurred, once a tissue section has been stained, LCM must be performed and completed within 1 h. FFPE tissue is the standard for preservation of tissue morphology and has been used by most pathology laboratories for decades. However, it creates cross-links between nucleic acids and proteins and between different proteins. This cross-linking interferes with recovery of RNA and protein from FFPE tissue and is not appropriate for RNA or protein-based assays. DNA extraction from FFPE tissue generally requires a minimum of 16 h of proteinase K incubation. Table 1 shows the comparison of optimal material availability between frozen and FFPE tissues. Although proteins are not extractable from FFPE samples, RNA can be isolated from FFPE tissue for downstream applications such as RT-PCR and cDNA microarray.3 Consequently, ideal fixation protocols that maintain good histology and preserve biomolecules are being evaluated. A previous report showed that 70% ethanol is one candidate fixative that is optimal for histology and recovery of RNA.4
TABLE 1.
Comparison of the optimal yield of RNA, DNA, and protein between frozen and FFPE tissues
| Frozen | FFPE | |
|---|---|---|
| DNA | +++ | +++ |
| RNA | +++ | +∼++ |
| Protein | +++ | – |
IMPORTANCE OF LCM IN TISSUE BIOREPOSITORY
An important requirement of many cancer research projects is the availability of high-quality, appropriately selected tumor and normal tissues. The tissue biorepository serves as the central hub, providing investigators with an invaluable resource with appropriately examined and characterized tissue specimens and linked patient information. The goal of most tissue biorepositories is to collect, process, store, and distribute samples of tumor and normal tissue for use in basic and translational cancer research studies. Tissue is obtained fresh/snap-frozen and FFPE from patients who provided informed consent prior to undergoing surgery. As most solid tumors are comprised of a variety of mixed cell populations, direct experimental analysis of tumor samples usually reflects the major or predominant cell type within the specimen that may or may not be tumor cells. Similarly, analysis of protein, RNA, or DNA changes from such samples may not be biologically relevant to the tumor cells under investigation but rather, may reflect stromal cell type(s). It is essential to analyze specific cells from tissue specimens to identify and define accurately the biologically important processes in pathologic lesions. Thus, LCM is an important tool that enables investigators to analyze precisely specific cell types of interest and define biologically important processes therein. Moreover, it also enables investigators to isolate pure populations of cells representative of a disease or biological consequence of interest. Accordingly, LCM can be used to select specifically benign, premalignant cells, noninvasive cancer cells, or invasive cells in a given tissue section and provide cancer biologists the opportunity to study the fascinating evolution of cancer from early, premalignant stages to frankly invasive and metastatic disease. LCM can also differentially capture tumor stroma and epithelia to help define the signaling pathways involved in the communication between the different cellular compartments of the tumor microenvironment that contribute to the cancer phenotype. The development and application of LCM have revolutionized the molecular analysis of solid tumor tissue. The combination of LCM and the tissue biorepository offers a comprehensive means by which researchers can use valuable human biospecimens and cutting-edge technology to facilitate basic, translational, and clinical research.
APPLICATIONS, LIMITATIONS, AND PERSPECTIVE OF LCM
LCM is particularly applicable to the molecular profiling of cancer and noncaner tissue. LCM technology has been described in a number of publications2,5–14 and has been used in a wide variety of applications, including pathology,15,16 prefertilization genetic diagnosis,17 organ transplantation,18–20 psychiatric disorders,21 single cell mutation analysis,22,23 gene expression,24–28 tissue chimerism,18,19 and molecular characterization of cancer cells.29–31 The following briefly summarizes several studies with the intent of broadening the potential of LCM applications. Based on the presence of a multitude of different cell types, the application of LCM has become an important tool in malignant lymphoma research and essential in Hodgkin's lymphoma (HL) research. HL is unique in that the neoplastic Reed-Stenberg (RS) cells constitute a minority population (<1%) in the affected lymph nodes.32 In our previous work, LCM played an essential role in microdissecting pure, neoplastic RS cells to identify the molecular changes in these cells. We were able to detect alterations of the DNA damage-response gene, ATR, in HL by LCM following DNA sequencing analysis (Fig. 3A).33 Another difficulty in studying epithelial tumors is the inability to isolate pure tumor cells for DNA, RNA, and protein analysis from prostate cancer tissue as a result of its infiltrative nature. In the past, most molecular studies concentrated on the analysis of bulk tissue samples without careful dissection and removal of other tissue elements, such as supporting stroma and adjacent normal tissue. Thanks to LCM, however, we are now able to overcome these hurdles and precisely evaluate pure populations of benign or malignant epithelial cells or stromal cells in prostate cancer samples (Fig. 3B). The captured cells can be used for a wide range of downstream analyses, such as qRT-PCR, cDNA microarrays, DNA sequencing, Western blots, 2-dimensional gel electrophoresis, mass spectrometry, etc. As discussed, the benefit of analyzing pure cell populations is significantly more informative than the analysis of heterogeneous tissue samples. Currently, work from our tissue biorepository core laboratory shows that LCM significantly reduces tissue sample preanalytical variation. In addition, many forensic applications of LCM have been described in areas such as paternity testing,34–36 sperm isolation in sexual assault cases,5,37–39 and DNA mixtures,37,40 Such innovative applications for forensic purposes will likely have potential commercial applications for LCM, which has clearly emerged as a powerful tool in the effort to obtain pure targeted biosamples.
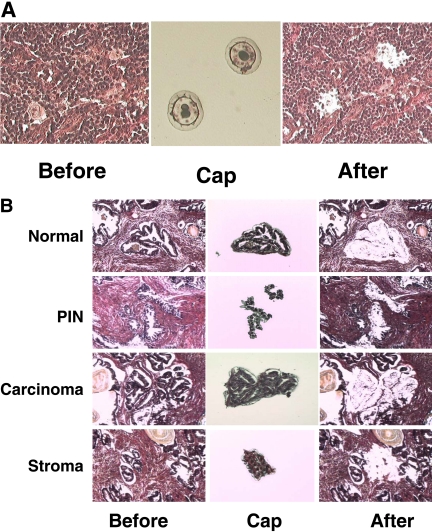
FIGURE 3.
Capture of pure cell populations by LCM. (A) LCM of HL and RS cells. A HL tissue section was stained with H&E (left), and the giant RS cells were captured successfully by LCM and visualized on the LCM cap (middle). (B) LCM of pure cell populations from a prostate cancer sample. A prostate adenocarcinoma tissue section was stained with H&E (left), and the benign, prostatic intraepithelial neoplasia (PIN), malignant, and stromal cells were isolated by LCM, respectively, and visualized on the LCM cap (middle). All human tissues were obtained from patients who provided informed consent and acquired through the Hollings Cancer Center Tissue Biorepository in accordance with an Institutional Review Board-approved protocol.
The major requirement for effective LCM is correct identification of cell subpopulations in a complex tissue structure by the operator. Therefore, a major limitation of LCM is the need to identify cells of interest based on morphologic characteristics, which in turn, requires a trained histologist or preferably, a pathologist. Combining an automatic imaging analyzer with LCM represents a future direction for expanding LCM applications. Although LCM can be used to analyze liver cell cultures, its strength and major advantage are the ability to isolate pure cell populations from heterogeneous tissue samples; this represents the advantage of LCM in the study of tissue-based projects.
In summary, LCM offers an effective approach to analyzing DNA, RNA, and protein from heterogeneous tissue samples by the isolation of pure cell populations. LCM can be applied to a wide range of cell and tissue preparations, thus providing an important advantage to tissue biorepositories engaged in translational and basic biomedical research. The development of novel, new applications of LCM is one of the subjects of ongoing investigation in our laboratory.
ACKNOWLEDGMENT
I thank Professor Steven A. Rosenzweig, Medical University of South Carolina, for his critical review and valuable advice.
Footnotes
All human tissues were obtained from patients who provided informed consent and acquired through the Hollings Cancer Center Tissue Biorepository in accordance with an Institutional Review Board-approved protocol.
REFERENCES
- 1.Monajembashi S, Cremer C, Cremer T, Wolfrum J, Greulich KO. Microdissection of human chromosomes by a laser microbeam. Exp Cell Res 1986;167:262–265 [DOI] [PubMed] [Google Scholar]
- 2.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science 1996;274:998–1001 [DOI] [PubMed] [Google Scholar]
- 3.Coudry RA, Meireles SI, Stoyanova R, et al. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn 2007;9:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su JM, Perlaky L, Li XN, et al. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol 2004;14:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anoruo B, van Oorschot R, Mitchell J, Howells D. Isolating cells from non-sperm cellular mixtures using the PALM microlaser micro dissection system. Forensic Sci Int 2007;173:93–96 [DOI] [PubMed] [Google Scholar]
- 6.Espina V, Heiby M, Pierobon M, Liotta LA. Laser capture microdissection technology. Expert Rev Mol Diagn 2007;7:647–657 [DOI] [PubMed] [Google Scholar]
- 7.Murray GI. An overview of laser microdissection technologies. Acta Histochem 2007;109:171–176 [DOI] [PubMed] [Google Scholar]
- 8.Espina V, Milia J, Wu G, Cowherd S, Liotta LA. Laser capture microdissection. Methods Mol Biol 2006;319:213–229 [DOI] [PubMed] [Google Scholar]
- 9.Hernández S, Lloreta J. Manual versus laser micro-dissection in molecular biology. Ultrastruct Pathol 2006;30:221–228 [DOI] [PubMed] [Google Scholar]
- 10.Ladanyi A, Sipos F, Szoke D, Galamb O, Molnar B, Tulassay Z. Laser microdissection in translational and clinical research. Cytometry A 2006;26:947–960 [DOI] [PubMed] [Google Scholar]
- 11.Chimenti C, Pieroni M, Russo A, et al. Laser microdissection in clinical cardiovascular research. Chest 2005;128:2876–2881 [DOI] [PubMed] [Google Scholar]
- 12.Burgemeister R. New aspects of laser microdissection in research and routine. J Histochem Cytochem 2005;53:409–412 [DOI] [PubMed] [Google Scholar]
- 13.Barisoni L, Star RA. Laser-capture microdissection. Methods Mol Med 2003;86:237–255 [DOI] [PubMed] [Google Scholar]
- 14.Fend F, Kremer M, Quintanilla-Martinez L. Laser capture microdissection: methodical aspects and applications with emphasis on immuno-laser capture microdissection. Pathobiology 2000;68:209–214 [DOI] [PubMed] [Google Scholar]
- 15.Agar NS, Halliday GM, Barnetson RS, Jones AM. A novel technique for the examination of skin biopsies by laser capture microdissection. J Cutan Pathol 2003;30:265–270 [DOI] [PubMed] [Google Scholar]
- 16.Okuducu AF, Hahne JC, Von Deimling A, Wernert N. Laser-assisted microdissection, techniques and applications in pathology. Int J Mol Med 2005;15:763–769 [PubMed] [Google Scholar]
- 17.Clement-Sengewald A, Buchholz T, Schütze K. Laser microdissection as a new approach to prefertilization genetic diagnosis. Pathobiology 2000;68:232–236 [DOI] [PubMed] [Google Scholar]
- 18.Kleeberger W, Rothämel T, Glöckner S, Lehmann U, Kreipe H. Laser-assisted microdissection and short tandem repeat PCR for the investigation of graft chimerism after solid organ transplantation. Pathobiology 2000;68:196–201 [DOI] [PubMed] [Google Scholar]
- 19.Kleeberger W, Rothämel T, Glöckner S, Flemming P, Lehmann U, Kreipe H. High frequency of epithelial chimerism in liver transplants demonstrated by microdissection and STR-analysis. Hepatology 2002;35:110–116 [DOI] [PubMed] [Google Scholar]
- 20.Lehmann U, Versmold A, Kreipe H. Combined laser-assisted microdissection and short tandem repeat analysis for detection of in situ microchimerism after solid organ transplantation. Methods Mol Biol 2005;293:113–123 [PubMed] [Google Scholar]
- 21.Burnet PW, Eastwood SL, Harrison PJ. Laser-assisted microdissection: methods for the molecular analysis of psychiatric disorders at a cellular resolution. Biol Psychiatry 2004;55:107–111 [DOI] [PubMed] [Google Scholar]
- 22.Persson A, Bäckvall H, Pontén F, Uhlén M, Lundeberg J. Single cell gene mutation analysis using laser-assisted microdissection of tissue sections. Methods Enzymol 2002;356:334–343 [DOI] [PubMed] [Google Scholar]
- 23.Nakamura N, Ruebel K, Jin L, Qian X, Zhang H, Lloyd RV. Laser capture microdissection for analysis of single cells. Methods Mol Med 2007;132:11–18 [DOI] [PubMed] [Google Scholar]
- 24.Esposito G. Complementary techniques: laser capture microdissection—increasing specificity of gene expression profiling of cancer specimens. Adv Exp Med Biol 2007;593:54–65 [DOI] [PubMed] [Google Scholar]
- 25.Kase M, Houtani T, Sakuma S, Tsutsumi T, Sugimoto T. Laser microdissection combined with immunohistochemistry on serial thin tissue sections: a method allowing efficient mRNA analysis. Histochem Cell Biol 2007;127:215–219 [DOI] [PubMed] [Google Scholar]
- 26.Manning CB, Mossman BT, Taatjes DJ. Analysis of asbestos-induced gene expression changes in bronchiolar epithelial cells using laser capture microdissection and quantitative reverse transcriptase-polymerase chain reaction. Methods Mol Biol 2006;319:231–236 [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Zhu JS, Song MQ, Chen GQ, Chen JL. Comparison of gene expression profiles between primary tumor and metastatic lesions in gastric cancer patients using laser microdissection and cDNA microarray. World J Gastroenterol 2006;12:6949–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikulowska-Mennis A, Taylor TB, Vishnu P, et al. High-quality RNA from cells isolated by laser capture microdissection. Biotechniques 2002;33:176–179 [DOI] [PubMed] [Google Scholar]
- 29.Huang C, Yang L, Li Z, et al. Detection of CCND1 amplification using laser capture microdissection coupled with real-time polymerase chain reaction in human esophageal squamous cell carcinoma. Cancer Genet Cytogenet 2007;175:19–25 [DOI] [PubMed] [Google Scholar]
- 30.Jensen LH, Cruger DG, Lindebjerg J, Byriel L, Bruun-Petersen G, Jakobsen A. Laser microdissection and microsatellite analysis of colorectal adenocarcinomas. Anticancer Res 2006;6:2069–2074 [PubMed] [Google Scholar]
- 31.Pai CY, Hsieh LL, Tsai CW, Chiou FS, Yang CH, Hsu BD. Allelic alterations at the STR markers in the buccal tissue cells of oral cancer patients and the oral epithelial cells of healthy betel quid-chewers: an evaluation of forensic applicability. Forensic Sci Int 2002;129:158–167 [DOI] [PubMed] [Google Scholar]
- 32.Mani H, Jaffe ES. Hodgkin lymphoma: an update on its biology with new insights into classification. Clin Lymphoma Myeloma 2009;9:206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu A, Takakuwa T, Fujita S, et al. ATR alterations in Hodgkin's lymphoma. Oncol Rep 2008;19:999–1005 [PubMed] [Google Scholar]
- 34.Budimlija ZM, Lechpammer M, Popiolek D, Fogt F, Prinz M, Bieber FR. Forensic applications of laser capture microdissection: use in DNA-based parentage testing and platform validation. Croat Med J 2005;46:549–555 [PubMed] [Google Scholar]
- 35.Delfin FC, Madrid BJ, Tan MP, De Ungria MC. Y-STR analysis for detection and objective confirmation of child sexual abuse. Int J Legal Med 2005;119:158–163 [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Kobilinsky L, Wolosin D, Shaler R, Baum H. A physical method for separating spermatozoa from epithelial cells in sexual assault evidence. J Forensic Sci 1998;43:114–118 [PubMed] [Google Scholar]
- 37.Vandewoestyne M, Van Hoofstat D, Van Nieuwerburgh F, Deforce D. Automatic detection of spermatozoa for laser capture microdissection. Int J Legal Med 2009;123:169–175 [DOI] [PubMed] [Google Scholar]
- 38.Horsman KM, Barker SL, Ferrance JP, Forrest KA, Koen KA, Landers JP. Separation of sperm and epithelial cells in a microfabricated device: potential application to forensic analysis of sexual assault evidence. Anal Chem 2005;77:742–749 [DOI] [PubMed] [Google Scholar]
- 39.Di Martino D, Giuffrè G, Staiti N, Simone A, Le Donne M, Saravo L. Single sperm cell isolation by laser microdissection. Forensic Sci Int 2004;146:151–153 [DOI] [PubMed] [Google Scholar]
- 40.Montesino M, Salas A, Crespillo M, et al. Analysis of body fluid mixtures by mtDNA sequencing: an inter-laboratory study of the GEP-ISFG working group. Forensic Sci Int 2007;168:42–56 [DOI] [PubMed] [Google Scholar]