Abstract
The aim of this project was to identify the best method for the enrichment of plasma membrane (PM) proteins for proteomics experiments. Following tryptic digestion and extended liquid chromatography-tandem mass spectrometry acquisitions, data were processed using MaxQuant and Gene Ontology (GO) terms used to determine protein subcellular localization. The following techniques were examined for the total number and percentage purity of PM proteins identified: (a) whole cell lysate (total number, 84–112; percentage purity, 9–13%); (b) crude membrane preparation (104–111; 17–20%); (c) biotinylation of surface proteins with N-hydroxysulfosuccinimydyl-S,S-biotin and streptavidin pulldown (78–115; 27–31%); (d) biotinylation of surface glycoproteins with biocytin hydrazide and streptavidin pulldown (41–54; 59–85%); or (e) biotinylation of surface glycoproteins with amino-oxy-biotin (which labels the sialylated fraction of PM glycoproteins) and streptavidin pulldown (120; 65%). A two- to threefold increase in the overall number of proteins identified was achieved by using stop and go extraction tip (StageTip)-based anion exchange (SAX) fractionation. Combining technique (e) with SAX fractionation increased the number of proteins identified to 281 (54%). Analysis of GO terms describing these proteins identified a large subset of proteins integral to the membrane with no subcellular assignment. These are likely to be of PM location and bring the total PM protein identifications to 364 (68%). This study suggests that selective biotinylation of the cell surface using amino-oxy-biotin in combination with SAX fractionation is a useful method for identification of sialylated PM proteins.
Keywords: biotin, glycoprotein, NHS-SS-biotin, sialic acid, proteomics
INTRODUCTION
Proteins at the plasma membrane (PM) allow cells to communicate with each other and with the extracellular environment and are critical for the propagation of signaling cascades, vesicle trafficking, ion transport, and protein translocation/integration.1–3 Proteomic approaches to the study of PM proteins are complicated by the relatively low abundance of PM proteins, hydrophobicity, and problems associated with the separation of PM proteins from those of other organelles.4 To identify a reproducible method for the enrichment of PM proteins, we compared five existing methodologies and for each, determined the total number and percentage purity of PM proteins identified.
MATERIALS AND METHODS
Cell Culture
Sultan, THP-1, and Jurkat cells were grown in RPMI-1640 medium (Thermo Pierce, Cramlington, UK), supplemented with 10% FCS (PAA, Yeovil, UK), penicillin at 100 units/ml, and streptomycin at 0.1 mg/ml (Sigma, Dorset, UK).
Methods of PM Protein Preparation
Whole cell lysate
Whole cell lysates were prepared as described by Wisniewski et al.5 Briefly, cells were washed once in PBS and then solubilized at a concentration of 107 cells/100 μl in 4% (w/v) SDS, 0.1 M DTT, 0.1 M Tris-HCl, pH 7.6 (SDT buffer) at room temperature. Brief sonication was performed to reduce the viscosity of the lysate, which was centrifuged for 10 min at 16,000 g to remove debris and the supernatant heated for 20 min at 50°C.
Crude membrane preparation
Cells were resuspended in ice-cold hypotonic lysis buffer [25 mM imidazole, pH 7.0, supplemented with complete inhibitor cocktail without EDTA (Roche, Burgess Hill, UK)]. Cells were then lysed using a cell disruption bomb (Parr Instrument Co., Moline, IL, USA); the resultant mixture was centrifuged at 2800 g for 10 min at 4°C to remove nuclei. The postnuclear supernatant was centrifuged subsequently at 200,000 g for 1 h 30 min and the crude membrane pellet carefully washed in PBS. Two hundred microliters of 150 mM Tris-HCl, pH 7.6, was added, and the pellet was dispersed thoroughly using the plunger of a 1-ml syringe. SDS (20%), 1 M DTT, and water were added to a final volume of 300 μl, giving the same final composition as SDT buffer. The lysate was sonicated briefly and then spun for 10 min at 16,000 g to remove debris, and the supernatant was heated for 20 min at 50°C.
Preparation of PM proteins using N-hydroxysulfosuccinimydyl-S,S-biotin (NHS-SS-biotin)
Cells were washed once with ice-cold PBS, resuspended at a concentration of 107 cells/ml in ice-cold biotinylation mix [one 12 mg vial of NHS-SS-biotin (Thermo Pierce) dissolved in 12 ml ice-cold PBS, pH 8.0, immediately before addition to cells], and incubated for 30 min at 4°C. The biotinylation reaction was quenched by the addition of 1 ml quenching solution (cell surface protein isolation kit, Thermo Pierce). Labeled cells were washed twice in ice-cold, Tris-buffered saline, pH 7.4, and a crude membrane preparation was performed as described above. The pellet was dispersed in 100 μl 50 mM Tris-HCl, pH 7.6, using the plunger of a 1-ml syringe, and the preparation was made up to a final volume of 500 μ l at a final concentration of 0.5% (w/v) SDS, 1% (v/v) Triton, 150 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl, 1× protease inhibitor (Roche), and 0.1 mg/ml PMSF (Sigma; lysis buffer). Sonication was performed at medium power (Sonoprobe, MSE, London, UK) at 4°C, and the lysate was centrifuged for 10 min at 16,000 g. Biotinylated proteins were enriched using 500 μl Neutravidin agarose (Thermo Pierce) for 2 h at 4°C. Beads were washed three times with lysis buffer, then twice with lysis buffer containing 500 mM NaCl, and then once with salt-free lysis buffer. Biotinylated proteins were eluted from beads by incubation for 1 h at room temperature in 1% (w/v) SDS, 50 mM DTT, 100 mM Tris-HCl. Proteins were denatured subsequently by heating for 20 min at 50°C.
Methods used to improve the purity of recovered PM proteins are described in Results.
Processing of PM Proteins for Mass Spectrometry (MS)
Samples were processed using filter-aided sample preparation (FASP).5 Briefly, protein concentration was estimated using absorption at λ = 280 nm on a NanoDrop spectrophotometer (Thermo Pierce), and 200 μg lysate was mixed with UA buffer (8 M urea in 0.1 M Tris-HCl, pH 8.5) in a 30-kDa cut-off Microcon device (Millipore, Watford, UK). The device was centrifuged at 14,000 g at 20°C for 15 min. Successive washes or incubations were performed using: UA buffer, 50 mM iodoacetamide in UA buffer, UA buffer and then 50 mM NH4HCO3.5 The concentrate was then subjected to proteolytic digestion overnight at room temperature using 2 μg-modified sequencing grade trypsin (Promega, Southampton, UK) in 40 μl 50 mM NH4HCO3. Tryptic peptides were collected by centrifugation. For samples to be fractionated by strong anion exchange (SAX), the filter device was rinsed with a further 50-μl 50 mM NH4HCO3; for other samples, the device was rinsed with 50 μl 0.5 M NaCl and centrifuged. The peptide content was estimated by Nanodrop. Peptide was fractionated using SAX or desalted using C18 StageTips constructed with three membrane plugs, as per Rappsilber et al.6
Preparation of PM proteins using amino-oxy-biotin
Cells were biotinylated and PM proteins prepared as described by Zeng et al.7 Briefly, cells were washed twice in 50 ml ice-cold PBS, pH 7.4, with CaCl2 and MgCl2 (Sigma). Surface sialic acid residues were oxidized with 1 mM sodium meta-periodate for 20 min in the dark at 4°C. The oxidation mixture was quenched by addition of glycerol to a final concentration of 1 mM. Cells were washed twice in PBS, pH 7.4/5% (v/v) FCS, and then biotinylated in 100 mM amino-oxy-biotin (Biotium Inc., Hayward, CA, USA) and 10 mM aniline (Sigma) in PBS, pH 6.7/5% (v/v) FCS. After 1 h at 4°C, cells were washed once with PBS, pH 7.4/5% (v/v) FCS, and then once with PBS, pH 7.4. Biotinylated cells were spun at 400 g at 4°C, and the resulting cell pellet was resuspended and incubated at 4°C for 30 min in lysis buffer [1% Triton X-100 (high purity, Thermo Pierce), 150 mM NaCl, 1× protease inhibitor (complete, without EDTA, Roche), 5 mM iodoacetamide (Sigma), 0.1 mg/ml PMSF, and 10 mM Tris-HCl, pH 7.6]. Nuclei were removed by centrifugation at 4°C three times for 10 min at 2800 g and then 16,000 g. Biotinylated proteins were enriched by incubating for 2 h at 4°C with high-affinity streptavidin agarose (Thermo Pierce). Extensive washing was performed using a vacuum manifold and Snap Cap spin columns (Thermo Pierce) with intermittent centrifugation at 1000 g for 1 min to ensure complete removal of wash buffers. Beads were washed initially with lysis buffer and then PBS/0.5% (w/v) SDS. Beads were incubated next for 20 min at room temperature with PBS/0.5% (w/v) SDS/100 mM DTT. Further washing was performed using UC buffer (6 M urea, 100 mM Tris-HCl, pH 8.5), followed by incubation for 20 min at room temperature with UC buffer containing 50 mM iodoacetamide. Beads were washed with UC buffer, then PBS, and then water, and biotinylated glycoproteins were digested on-bead overnight in 400 μl 50 mM NH4HCO3 containing 4 μg-modified sequencing grade trypsin (Promega). Tryptic peptides were collected by centrifugation at 1000 g for 1 min. Beads were washed once with 50 mM NH4HCO3, and pooled tryptic fractions were desalted using StageTips6 or fractionated by SAX. To elute glycopeptides, beads were washed with PBS, then water, and then G7 buffer (New England Biolabs, Hitchin, UK). Beads were incubated overnight in 400 μl G7 buffer containing 30,000 units glycerol-free peptide N-glycanase (PNGase; New England Biolabs). Glycopeptides were collected by centrifugation at 1000 g for 1 min, beads were washed once with G7 buffer, and eluates were pooled and desalted using StageTips.6
Preparation of PM proteins using biocytin hydrazide
Cells were biotinylated and PM proteins prepared as described by Wollscheid et al.8 A method similar to that used for amino-oxy-biotin was used. Significant differences included using labeling buffer for all washes and incubations [PBS, pH 6.5, 0.1% (v/v) FCS], oxidation with 1.6 mM sodium meta-periodate for 10 min, and biotinylation with 5 mM biocytin hydrazide (Biotium Inc.). A crude membrane preparation, as described above, was performed instead of a 1% (v/v) Triton lysis. Membrane pellet was solubilized in SDT buffer, denatured, and sialylated glycoprotein-encriched using Neutravadin agarose resin (Thermo Pierce). Extensive washing of beads was performed as described by Wollscheid et al.8 Tryptic and PNGase fractions were generated and collected as described above.
SAX Fractionation
SAX was performed as described by Wisniewski et al.9 Briefly, 30–50 μg tryptic peptide was loaded at pH 11 on a tip-based anion exchanger constructed using six layers of Empore anion exchange disk (3M, Bracknell, UK). The column was equilibrated and fractions eluted using Britton & Robinson buffer (20 mM acetic acid, 20 mM phosphoric acid, 20 mM boric acid titrated with NaOH to the desired pH). Fractions were eluted subsequently with buffer solutions of pH 11, 8, 6, 5, 4, and 3 onto StageTips6 containing three layers of C18 membrane.
MS Analysis and Database Searching
Unfractionated samples were eluted from StageTips, dried almost to completion, and resuspended in 11 μl MS solvent [3% (v/v) acetonitrile (MeCN), 0.1% (v/v) formic acid]. For each liquid chromatography (LC)-tandem MS (MS/MS) run, 3 μl was injected onto a NanoAcquity Ultra Performance LC (Waters, Milford, MA, USA). Peptides were loaded onto a 180-μm/20 mm Symmetry C18 trap column (Waters) for 1 min at 15 μl/min and eluted to a 75-μm/150 mm BEH130 C18 analytical column. Peptides were eluted with a MeCN gradient rising from 3% to 25% by 130 min, 25% to 40% by 205 min, and to 85% by 210 min. The total run time was 240 min. Peptides were eluted into the LTQ OrbiTrap XL (Thermo Pierce) via 10 μm SilicaTip emitters (New Objectives, Woburn, MA, USA). Data were acquired by a Top 10 data-dependent acquisition method with survey scans acquired at 60,000 resolution (full width at half maximum at a mass-to-charge ratio of 430). Fractionated samples were analyzed as described above but with duplicate 240 min gradients. Raw MS files were processed using MaxQuant, Version 1.0.12.3110 with .msm output files searched against decoy International Protein Index human database (download 04/2009)11 containing forward and reverse protein sequences using MASCOT Daemon 2.2.0.12 Fragment ion tolerance was set to 0.5 Da, with a maximum of two missed tryptic cleavage sites. Carbamidomethyl cysteine was defined as a fixed modification, oxidized methionine, deamidation, and in the NHS-SS-biotin experiments, CAMthiopropanoyl (K and protein N-terminus) were selected as variable modifications. The false-discovery rate for peptides and proteins was set at 0.01. Gene Ontology (GO) information was annotated to data by MaxQuant.10
RESULTS
PM Protein Preparation Following NHS-SS-Biotin Cell Surface Labeling Allows Only A Modest Enrichment Above Crude Membrane Preparation. Only A Small Proportion of Proteins Identified in Whole Cell Lysates is of PM Origin
Three different methods were used to purify PM proteins (Table 1). For each preparation, 4 μg peptide was desalted on a StageTip. Using whole cell lysates, we identified 930–1060 total proteins (Table 1) with only a small proportion of these annotated as PM: 84–112 (9–13%). Crude membrane preparation led to a small increase in purity: 525–685 total proteins, of which 104–111 were annotated as PM (17–21%). Preparation of PM proteins using NHS-SS-biotin gave a modest enrichment above crude membrane preparation: 293–434 total proteins, of which 78–115 were annotated as PM (27–31%).
TABLE 1.
PM protein preparation following NHS-SS-biotin cell surface labeling allows only a modest enrichment above crude membrane preparation. Only a small proportion of proteins identified in whole cell lysates are of PM origin
| Whole cell lysate 1: Sultan cells | Whole cell lysate 2: Sultan cells | Whole cell lysate 3: THP-1 cells | Crude Prep 1: Sultan cells | Crude Prep 2: Jurkat cells | NHS-SS-Biotin Prep 1: Sultan cells | NHS-SS-Biotin Prep 2: THP-1 cells | |
|---|---|---|---|---|---|---|---|
| Total protein identifications (IDs) | 941 | 1060 | 931 | 685 | 525 | 293 | 434 |
| Membrane annotation | 192 | 224 | 228 | 272 | 336 | 145 | 223 |
| PM annotation | 84 | 103 | 112 | 111 | 104 | 78 | 115 |
| Integral to PM | 14 | 19 | 21 | 34 | 36 | 33 | 48 |
| % PM | 9% | 10% | 13% | 17% | 21% | 31% | 27% |
The total number of proteins with a PM annotation provided by GO is shown in bold in all tables. This includes a small subset of proteins annotated as “cell surface” (data not shown), most of which are also annotated as PM and the subset annotated as “Integral to PM”. % PM is calculated by (PM annotation) /(Total protein IDs–unidentified proteins). Data for numbers of unidentified proteins are not shown.
To determine whether NHS-SS-biotin also biotinylated intracellular proteins, we analyzed processed data to determine which types of protein had a residual CAMthiopropanoyl modification (residual modification left after cleavage of NHS-SS-biotin at the disulfide linker). We expected that this analysis would only identify a subset of all biotinylated proteins, as identification of this modification was dependent on identification of a modified peptide. For NHS-SS-biotin Prep 2, a total of 77/434 proteins were modified with CAMthiopropanoyl, of which 43% were annotated as PM (including proteins additionally annotated as “nucleus” or “cytoplasm”), 29% nucleus, 5% cytoplasm, 6% “cytoplasm and nucleus”, 12% “membrane” (but not any of the prior terms), and 5% other.
Various Methods Trialed to Improve the Purity of PM Protein Preparation Following NHS-SS-Biotinylation Lead to Only A Modest Improvement in Purity
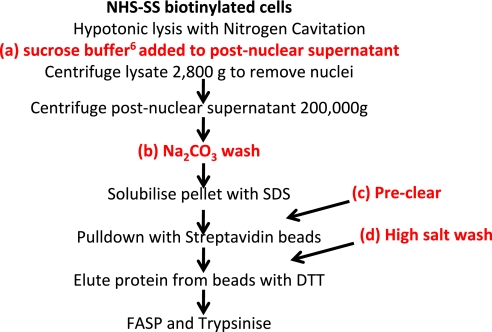
We examined in parallel four different modifications to improve the purity of PM proteins (Fig. 1 and Table 2). Hypertonic wash with sucrose buffer was used as described8; briefly, equal volumes of crude membrane in hypotonic lysis buffer and membrane prep buffer (containing 280 mM sucrose) were mixed and incubated 10 min on ice, and then nuclei were removed by centrifugation. Na2CO3 wash: crude membrane pellet was resuspended thoroughly in a small volume of 100 mM Na2CO3, followed by rotation in 12 ml Na2CO3 for 30 min at 4°C. A further spin at 200,000 g for 1.5 h was performed. The resulting pellet was washed carefully in 100 mM Tris-HCl, pH 7.6, and then solubilized in SDS. Preclear: prior to pulldown, the solubilized membrane pellet was incubated for 1 h with control agarose resin (Thermo Pierce) at 4°C. High salt wash: after pulldown, beads were washed using 5 M NaCl and 100 mM Na2CO3 in addition to standard washes.
FIGURE 1.
Workflow illustrating additional methods attempted to increase purity of PM preparation using NHS-SS-biotinylation of the cell surface.
TABLE 2.
Various methods trialed to improve the purity of PM protein preparation following NHS-SS-biotinylation lead to only a modest improvement in purity
| Normal | Sucrose wash | Na2CO3 wash | Preclear | High salt wash | |
|---|---|---|---|---|---|
| Total protein IDs | 544 | 555 | 527 | 492 | 515 |
| Nucleus annotation | 259 | 210 | 134 | 248 | 251 |
| Membrane annotation | 167 | 249 | 305 | 150 | 159 |
| PM annotation | 89 | 106 | 126 | 80 | 80 |
| Integral to PM | 24 | 38 | 46 | 20 | 18 |
| % PM | 17% | 20% | 25% | 17% | 16% |
5×107 input cells were used for each method apart from the Na2CO3 wash, where 1 × 108 cells were used in anticipation of significant losses. For each preparation, the total peptide yield was: normal, 38.5 μg; sucrose wash, 13.9 μg; Na2CO3 wash, 13.9 μg; preclear, 40.1 μg; high salt wash, 37.6 μg. For each preparation, 14 μg peptide was desalted on a StageTip. No improvement in purity of PM proteins was observed following a preclear or high salt wash (Table 2). A possible slight improvement in purity was seen with sucrose buffer. The most significant improvement in purity was observed using a Na2CO3 wash, chiefly as a result of a decrease in contamination from nuclear proteins (Table 2), which accounted for 50–53% of annotated proteins (normal, preclear, high salt wash preps) but only 39% of proteins after sucrose wash and 27% after Na2CO3 wash. Of note, a number of nuclear proteins were annotated additionally as PM—22 (Na2CO3 wash) and 16–18 (all other preps). However, the improved purity with the Na2CO3 wash coincided with significant losses in terms of final peptide yield, especially when considering that twice as many cells were used.
Use of Biocytin Hydrazide or Amino-oxy-Biotin to Isolate Cell Surface Proteins Leads to A Substantial Improvement in Purity. Protein Yield is Greater with Amino-Oxy-Biotin and Improved by Peptide Fractionation
To increase the specificity of biotin labeling of the PM, we used biocytin hydrazide first to label cell surface glycoproteins of THP-1 cells using a modified method based on that described.8 In two independent experiments, we identified a small number of PM proteins (Table 3): 41–54 (59–85%). Analysis of the PNGase fraction identified a smaller number of PM proteins at very high purity: 13 (76%). Seven of these 13 proteins were also identified in the corresponding tryptic fraction.
TABLE 3.
Use of biocytin hydrazide or amino-oxy-biotin to isolate cell surface proteins leads to a substantial improvement in purity. Protein yield is greater with amino-oxy-biotin and improved by peptide fractionation
| Prep 1 |
Prep 2 |
Prep 1 |
Prep 2 |
||||
|---|---|---|---|---|---|---|---|
| Tryptic peptides | Tryptic peptides | PNGase peptides | Tryptic peptides, no SAX | Tryptic peptides, six fractions | PNGase peptides | Combined data | |
| Total protein IDs | 49 | 95 | 21 | 188 | 541 | 155 | 539 |
| Membrane annotation | 44 | 63 | 14 | 158 | 396 | 150 | 398 |
| PM annotation | 41 | 54 | 13 | 120 | 276 | 113 | 281 |
| Integral to PM | 23 | 28 | 9 | 70 | 149 | 61 | 151 |
| % PM | 85% | 59% | 76% | 65% | 52% | 74% | 54% |
A second way of labeling glycoproteins by amino-oxy-biotin was used to label cell surface proteins from THP-1 cells, and after pulldown and tryptic digestion, the resulting peptides were analyzed using a single, 4-h LC-MSMS run (Prep 1) or duplicate, 4-h runs on six SAX fractions (Prep 2). For Prep 2, after elution of tryptic peptides, beads were washed and glycopeptides eluted from beads using PNGase. These peptides were analyzed in duplicate, 4-h LC-MSMS runs, and the data generated were combined with data from the SAX fractions (Table 3). Significantly more PM proteins were identified than by biocytin hydrazide, even without fractionation (120 proteins compared with 41–54). Purity of the PM preparation was similar for both methods (Table 3). SAX fractionation of tryptic peptides led to a substantial increase in PM protein identifications: 276 (52%). Combining trypsin and PNGase data resulted in a slight increase in the total number of PM protein identifications: 281 (54%; Table 3).
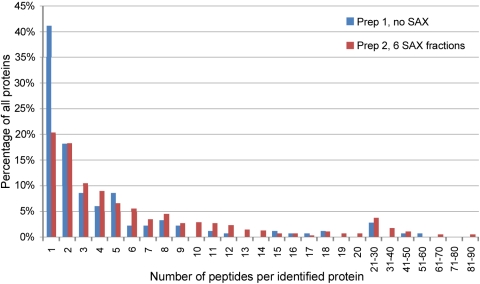
In addition to improving the total number of proteins isolated, we found that SAX fractionation of tryptic peptides increased substantially the number of peptides identified per protein (Fig. 2). Without fractionation, 41% of proteins were identified on the basis of a single peptide; with fractionation, this number decreased to only 20%. With fractionation, one protein was identified by 89 unique peptides, and without fractionation, the maximum number of peptides per protein was 57.
FIGURE 2.
SAX fractionation of peptides generated by using the amino-oxy-biotin technique leads to a substantial increase in the number of peptides per protein identified. We compared data for the two amino-oxy-biotin preparations shown in Table 3. Prep 1—single, 4-h MS run, no fractionation; Prep 2—duplicate, 4-h MS run on six peptide fractions.
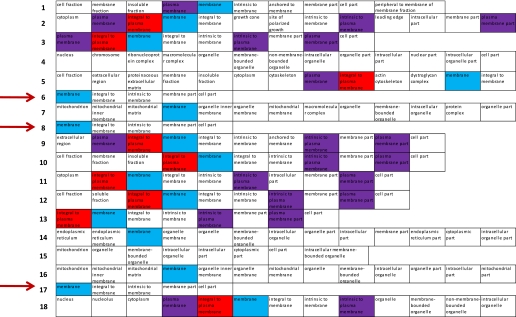
We analyzed further the full list of proteins identified in amino-oxy-biotin Prep 2 (Table 3) to try to identify additional PM proteins. By analysis of GO terms annotated to each protein, we identified a substantial subset of proteins that were likely to be PM-localized. A very short GO descriptor identified 71 out of 539 proteins (Fig. 3, arrows) as integral to membrane. Although these have no subcellular assignment, we suspect that the majority are likely to be PM proteins. In addition, 56 out of 539 proteins were identified as “extracellular” by GO; 12 of these were not included already in the total count of PM proteins, as they had no PM annotation. Including these two subsets brings the total PM protein identifications to 364 (68%).
FIGURE 3.
A substantial number of proteins isolated using amino-oxy-biotin are identified by GO as integral to the membrane but have no subcellular assignment. These are likely to be PM proteins. Separated GO terms are shown for a representative sample of the full list of proteins identified in amino-oxy-biotin Prep 2, Table 3. Seventy-one out of 539 total proteins had a very short GO descriptor (examples indicated by arrows), identifying the protein as integral to membrane but with no subcellular assignment. These are likely to be PM proteins.
We analyzed further the full list of proteins identified in the amino-oxy-biotin Prep 2 using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA, USA) to assess assignments of protein subcellular localization using a technique independent of GO. This identified: PM (277), extracellular space (19), nucleus (51), cytoplasm (131), unknown (50). Many of the proteins identified with an unknown localization were solute carriers, tetraspanins, lectins, or transmembrane proteins; a substantial number of these proteins were likely to be of PM localization. It is possible that some of the proteins labeled as nucleus or cytoplasm resided additionally in the PM. As Ingenuity Pathway Analysis provided single annotations for each protein to describe subcellular localization, this will not accommodate proteins that are located in more than one compartment.
DISCUSSION
We examined five different methods for enrichment of PM proteins and found that the combination of amino-oxy-biotinylation of sialylated glycoproteins with SAX fractionation of tryptic peptides provided the optimal PM protein identification (364) at the highest purity (68%). Other methods examined were in our hands inferior, identifying fewer PM proteins at considerably lower purity.
We identified 930–1060 total proteins from three whole cell lysates; 9–13% had a PM annotation (Table 1) and 1.1–1.6% of proteins had a cell surface annotation. This was in agreement with the published protocol we used for the generation and processing of whole cell lysates.5 In this study, 12% total proteins were annotated as PM (J.R. Wisniewski, personal communication) and 0.9–1.8% of proteins annotated as cell surface.5 The total number of proteins we identified using a single, 4-h LC-MSMS run was substantially lower than the number identified in a comparable experiment (2745±91).5 This likely reflects the use of different LC equipment and a nanoESI source or different acquisition parameters and data analysis tools.
When crude membrane was prepared by nitrogen cavitation, 17–21% of proteins identified were annotated as PM, comparable with other data relating to crude membrane extractions (for example, mouse hippocampi, 26% PM).9
Enrichment of PM proteins by cell surface labeling with NHS-SS-biotin gave disappointing results, with at best, a modest improvement in purity above crude membrane preparation alone (17–31% purity compared with 17–21% purity, Tables 1 and 2). Our attempts to improve the enrichment of PM proteins using this technique (Table 2) were not especially successful. Washing crude membrane with Na2CO3 improved only modestly the purity of PM proteins recovered compared with no wash (25% compared with 17% purity, Table 2). NHS-SS-biotin is at least partially membrane-permeable, as 57% of the proteins we identified in NHS-SS-biotin Prep 2 (Table 1) were not annotated PM and were chiefly nuclear or cytoplasmic in origin. Dead cells or cell fragments may have been labeled by NHS-SS-biotin, but they were not identified by Trypan blue staining. NHS-SS-biotin is a popular technique to demonstrate or confirm cell surface localization with the assumption that it is membrane-impermeable.13–16 However, other groups have also found labeling of intracellular proteins when using NHS-SS-biotin on intact cells.17 An alternative technique is to label PM proteins with NHS-SS-biotin and then homogenize labeled cells using a Dounce homogenizer.18 Membrane fragments that contain a biotinylated protein are enriched on streptavidin magnetic beads and washed at high salt and high pH. The theoretical advantage of this technique is the enrichment of both biotinylated proteins as well as other proteins embedded in the PM that have not been biotinylated. Using this technique, 781/898 proteins (87%) had a reported PM annotation.18 Our attempts to reproduce their data achieved a purity of <20% PM (data not shown). It has been suggested that NHS-LC-biotin may be less membrane-permeable than NHS-SS-biotin,19 although we did not examine this further.
Using biocytin hydrazide to label sialylated glycoproteins, 41–54 (59–85%) of identified proteins were annotated as PM, representing a considerable improvement in purity on the approaches described above. In the original description of the technique, biotinylated crude membrane was dissolved in Rapigest, digested with Lys-C and trypsin and biotinylated glycopeptides, and enriched with UltraLink Streptavidin Plus beads (Thermo Pierce) prior to release with PNGase F.8 Our modifications included dissolving biotinylated crude membrane in SDS, pulldown of labeled glycoproteins, and tryptic digestion on Neutravidin agarose beads combined with PNGase release of glycopeptides. Tryptic and PNGase fractions were analyzed by LC-MSMS. The reported increased number of identified glycoproteins (up to 292) may reflect improved PM protein digestion or reduced digestion of agarose-linked streptavidin. In our biocytin hydrazide experiments, we used Neutravidin agarose to enrich biotinylated proteins, from which we identified abundant streptavidin peptides that dominated mass spectra (data not shown), and Wollscheid et al.8 used UltraLink Streptavidin Plus beads (Thermo Pierce), which may be less susceptible to tryptic digestion. In our initial experiments with amino-oxy-biotin, we enriched biotinylated proteins using avidin, streptavidin (Thermo Pierce), or Captavidin (Invitrogen, Paisley, UK) beads and had problems as a result of the identification of abundant streptavidin peptides. We therefore changed to high-capacity streptavidin beads (Thermo Pierce; T.N. Ramya, personal communication),7 which were much less susceptible to tryptic digestion (data not shown).
Isolation of sialylated glycoproteins with amino-oxy-biotin was highly reproducible and identified PM proteins at high purity. The oxime ligation of amino-oxy-biotin to cell surface aldehyde generates a more stable product than the hydrazone ligation of biocytin hydrazide,20 and the use of aniline as a catalyst enables use of amino-oxy-biotin at a relatively low concentration (100 μM).7 Using SAX fractionation, the identification of 364 PM proteins, representing 68% of all annotated proteins, is comparable with the best data from other groups and our own using different techniques, including: colloidal silica [450 total proteins identified from lung tissue (81% PM annotation)21 and 157 proteins identified from an immortalized B-lymphocyte cell line (16%)]22; sucrose density gradient with two-phase partitioning (428 proteins identified from rat liver, 67% PM annotation)23; and high pH-proteinase K preparations (670 total proteins, 72% predicted integral membrane proteins).24
We conclude that selective biotinylation of the cell surface using amino-oxy-biotin in combination with SAX fractionation is a useful method for identification of sialylated PM proteins.
ACKNOWLEDGMENT
This work was supported by the Wellcome Trust. M.P.W. is a Next Generation Fellow at Cambridge Institute for Medical Research.
Footnotes
There are no conflicts of interest.
REFERENCES
- 1.Ott CM, Lingappa VR. Integral membrane protein biosynthesis: why topology is hard to predict. J Cell Sci 2002;115:2003–2009 [DOI] [PubMed] [Google Scholar]
- 2.Torres J, Stevens TJ, Samso M. Membrane proteins: the “Wild West” of structural biology. Trends Biochem Sci 2003;28:137–144 [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Zheng Y, Zhou Y. Structure prediction of membrane proteins. Genomics Proteomics Bioinformatics 2004;2:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speers AE, Wu CC. Proteomics of integral membrane proteins—theory and application. Chem Rev 2007;107:3687–3714 [DOI] [PubMed] [Google Scholar]
- 5.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359–362 [DOI] [PubMed] [Google Scholar]
- 6.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007;2:1896–1906 [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. High-efficiency labeling of sialylated glycoproteins on living cells. Nat Methods 2009:6:207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wollscheid B, Bausch-Fluck D, Henderson C, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol 2009;27:378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 2009;8:5674–5678 [DOI] [PubMed] [Google Scholar]
- 10.Cox J, Matic I, Hilger M, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc 2009;4:698–705 [DOI] [PubMed] [Google Scholar]
- 11.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics 2004;4:1985–1988 [DOI] [PubMed] [Google Scholar]
- 12.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999;20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 13.Bertinato J, Swist E, Plouffe LJ, Brooks SP, L'Abbe MR. Ctr2 is partially localized to the plasma membrane and stimulates copper uptake in COS-7 cells. Biochem J 2008;409:731–740 [DOI] [PubMed] [Google Scholar]
- 14.Ge Y, Rikihisa Y. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J Bacteriol 2007;189:7819–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge Y, Rikihisa Y. Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun 2007;75:3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SJ, Kim KH, Park JS, et al. Comparative analysis of cell surface proteins in chronic and acute leukemia cell lines. Biochem Biophys Res Commun 2007;357:620–626 [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, McDonald K, Hanrahan JW. Trafficking of immature DeltaF508-CFTR to the plasma membrane and its detection by biotinylation. Biochem J 2009;419:211–219 [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Zhang W, Kho Y. Proteomic analysis of integral plasma membrane proteins. Anal Chem 2004;76:1817–1823 [DOI] [PubMed] [Google Scholar]
- 19.Nunomura K, Nagano K, Itagaki C, et al. Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol Cell Proteomics 2005;4:1968–1976 [DOI] [PubMed] [Google Scholar]
- 20.Kalia J, Raines RT. Hydrolytic stability of hydrazones and oximes. Angew Chem Int Ed Engl 2008;47:7523–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durr E, Yu J, Krasinska KM, et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol 2004;22:985–992 [DOI] [PubMed] [Google Scholar]
- 22.Hor S, Ziv T, Admon A, Lehner PJ. Stable isotope labeling by amino acids in cell culture and differential plasma membrane proteome quantitation identify new substrates for the MARCH9 transmembrane E3 ligase. Mol Cell Proteomics 2009;8:1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao R, Li X, Liu Z, et al. Integration of a two-phase partition method into proteomics research on rat liver plasma membrane proteins. J Proteome Res 2006;5:634–642 [DOI] [PubMed] [Google Scholar]
- 24.Blackler AR, Speers AE, Ladinsky MS, Wu CC. A shotgun proteomic method for the identification of membrane-embedded proteins and peptides. J Proteome Res 2008;7:3028–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]