Abstract
The establishment and maintenance of cellular identity are ultimately dependent upon the accurate regulation of gene expression, the process by which genetic information is used to synthesize functional gene products. The post-transcriptional, pre-translational regulation of RNA constitutes RNA processing, which plays a prominent role in the modulation of gene expression in differentiated animal cells. The multi-protein Exon Junction Complex (EJC) serves as a critical signaling hub within the network that underlies many RNA processing events. Here, we identify a requirement for the EJC during early vertebrate embryogenesis. Knockdown of the EJC component Eukaryotic initiation factor 4a3 (Eif4a3) in embryos of the frog Xenopus laevis results in full-body paralysis, with defects in sensory neuron, pigment cell, and cardiac development; similar phenotypes are seen following knockdown of other “core” EJC protein constituents. Our studies point to an essential role for the EJC in the development of neural plate border derivatives.
Keywords: Eif4a3, EJC, Xenopus laevis, neural crest, melanophore, RNA processing
Introduction
Gene expression during vertebrate embryogenesis is regulated at many steps, from transcription through post-translational modification. Until recently, relatively little has been known about the role of RNA processing, defined here as the regulatory steps following transcription and preceding translation, during development of the early vertebrate embryo. Over the past several years, biochemical studies have established that the multi-protein Exon Junction Complex (EJC) functions as a major signaling locus within a large protein network underlying eukaryotic RNA processing (Moore and Proudfoot, 2009).
The EJC comprises four “core” factors, Eif4a3, Y14, Magoh, and MLN51, and several accessory proteins; the complex binds RNA in a sequence-independent fashion, 25-50 nucleotides upstream of exon-exon splice junctions (Le Hir et al., 2000). The EJC has been implicated in a number of RNA processing events, including translational regulation, subcellular mRNA localization, and nonsense-mediated decay (NMD) (Tange et al., 2004). Eif4a3 (eukaryotic translation initiation factor 4a3) is the EJC core component thought to directly contact RNA (Shibuya et al., 2004; Ballut et al., 2005). Eif4a3 was named for its high sequence homology to Eif4a1 and Eif4a2; these latter genes encode components of the cap-binding translation initiation complex Eif4F (Owttrim et al., 1991; Weinstein et al., 1997). Eif4a3 cannot, however, functionally substitute for Eif4a1/2 during translation initiation (Li et al., 1999), and appears to function primarily in the context of the EJC. Although gain-of-function studies have implicated Eif4a3 in the patterning of the embryonic ectoderm in the frog Xenopus laevis, little is known about the potential requirement for Eif4a3, or the EJC, during vertebrate development (Weinstein et al., 1997).
Here, we describe the analysis of Eif4a3 loss-of-function during Xenopus development. We find that morpholino-mediated Eif4a3 knockdown results in a suite of phenotypes that include 1) defects in sensory neuron development; 2) complete paralysis and lack of response to contact stimuli; 3) defects in the neural crest-derived melanophores; 4) failure of heart looping and edema in the region surrounding the embryonic heart. Our data support a model in which Eif4a3 activity is required for the development of the neural-epidermal border region, via its role as a core component of the EJC.
Results
Eif4a3 is required for melanophore development, cardiac looping, and the embryonic touch response
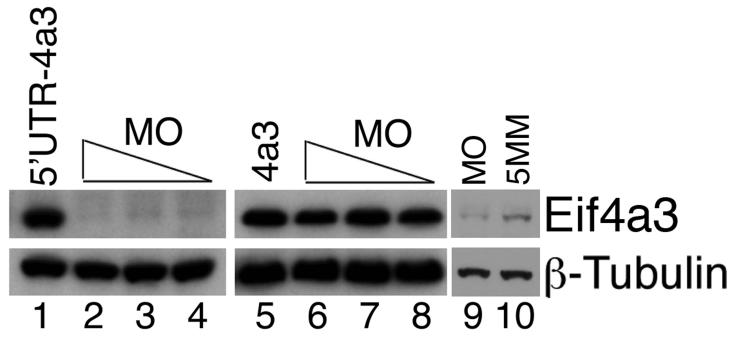
To investigate a potential requirement for Eif4a3 in early development, we designed translation-blocking, antisense morpholino oligonucleotides against Eif4a3 (4a3MO) for use in knockdown assays in Xenopus embryos. Injection of 4a3MO effectively inhibits translation of exogenous Xenopus eif4a3 RNA, as detected with an antibody against mouse Eif4a3 (Fig. 1, lanes 1-4); injection of RNA synthesized from an Eif4a3 construct that lacks the 4a3MO-binding site (found in the 5′-untranslated region of the eif4a3 transcript) is not inhibited by injection of the 4a3MO (Fig. 1, lanes 5-8). Injection of 4a3MO also inhibits translation of native eif4a3 RNA (Fig. 1, lanes 9-10). 4a3MO is thus a useful reagent for generating Eif4a3 knockdown morphants.
Fig. 1.
Activity of Eif4a3 translation-blocking morpholinos. Western blot analysis of exogenous (lanes 1-8) and endogenous (lanes 9 and 10) Eif4a3 protein, visualized using a mouse monoclonal anti-Eif4a3 antibody. Eif4a3MO inhibits translation of eif4a3 RNA containing the Eif4a3 morpholino binding site (5′UTR-4a3); translation of an RNA that lacks the Eif4a3MO binding site (4a3) is not inhibited. 21ng, 10.5ng, or 5.25ng Eif4a3 morpholino (MO) and/or 2ng 5′UTR-4a3 or 4a3 RNA were injected, as listed; higher morpholino doses did not lead to an appreciable increase in the degree of translational block (data not shown). Longer exposure times and 21ng morpholino (MO or 5 base pair mismatch, 5MM) were used for lanes 9 and 10. Whole-embryo lysates from stage 14 embryos were used for this study; β-Tubulin levels were used as a loading control.
Xenopus eif4a3 is first detected as a maternal transcript; it is strongly expressed following the start of zygotic transcription at the midblastula transition (Weinstein et al., 1997). Development of Eif4a3 morphants appears normal through stage 24 (data not shown). Injection of 21ng 4a3MO into early cleavage stage embryos results in the following phenotypes:
Defects in pigment forming cells
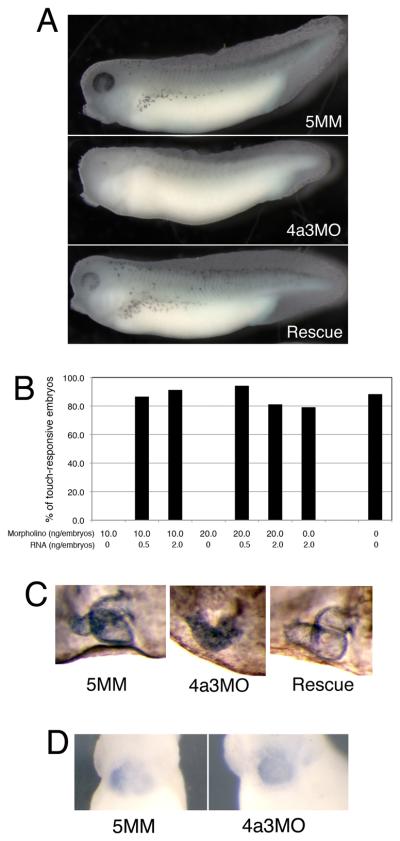
During normal development, the pigment-containing melanophores are clearly apparent by tadpole stages 33/34; in marked contrast, Eif4a3 morphants show a dramatic reduction in overall eye and body pigment, and in the number of pigment-containing melanophores (Fig. 2A, middle panel; Table I). Reduced pigmentation is observed in the Eif4a3 morphants at all stage examined (33-42), suggesting that this defect is not due to developmental delay. Embryos injected with a morpholino that differs from Eif4a3 in five base pairs (5 base pair mismatch, 5MM), resemble uninjected siblings in this and other respects (Fig. 2A, top panel, and see below). Embryos used in these studies were derived from albino eggs fertilized with sperm from wild-type males; loss of melanophores was also observed in morphants generated from pigmented sperm and egg (data not shown). The number of pigment-positive cells is partially but consistently rescued by co-expression of eif4a3 RNA that lacks the 4a3MO binding site (4a3RNA; Fig. 2A, bottom panel; Table I); injection of eif4a3 RNA alone has a modest and variable effect on pigmentation, with occasional reduction in melanophore number (Table I, and data not shown).
Fig. 2.
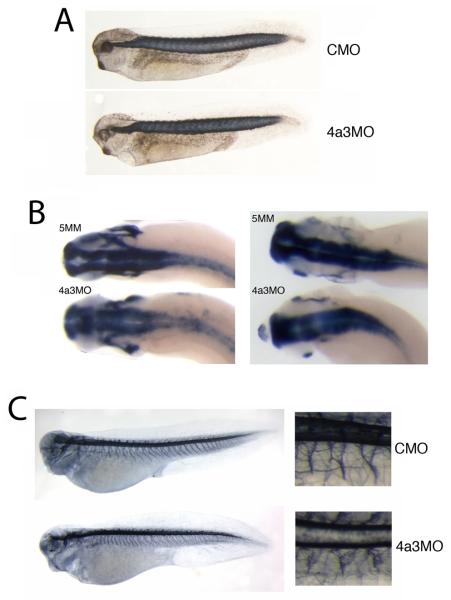
The Eif4a3 morphant phenotype. A: Loss of pigment-forming cells following Eif4a3 knockdown. 21ng of each morpholino (“4a3MO”: Eif4a3 morpholino; “5MM”: 5 base pair mismatch Eif4a3 morpholino) was injected, as listed, in this and subsequent figures. “Rescue”: Eif4a3MO+2ng eif4a3 RNA. Embryos were derived from albino eggs fertilized with sperm from wild-type males. B: Loss of touch response behavior in Eif4a3 morphants. Graph depicting percentage of touch-responsive embryos injected with listed doses of eif4a3 RNA and/or morpholino (4a3MO), as shown. C: Defects of heart formation in Eif4a3 morphants. Troponin T staining of cardiac tissue in stage 37/38 embryos injected with 4a3MO, 5MM, or 4a3MO+Eif4a3 RNA (rescue). Note lack of heart looping in 4a3MO-injected embryo. D: Whole-mount in situ hybridization with antisense probes against Xnkx-2.5 in stage 28 embryos injected with Eif4a3MO (4a3MO) or Eif4a3MM (5MM), as listed. No significant differences were seen in the expression of this marker at neurula or tailbud stages (data not shown).
Table I.
Effects of morpholino-mediated knockdown of EJC core components Eif4a3, Magoh, and Y14, scored at stage 32 (touch response) and stage 37 (melanophore and heart defects). nd=not determined. The Y14 morpholino was injected at lower concentration (2.6ng/embryo) than were the Eif4a3 or Magoh morpholinos: weak Y14 RNA-mediated rescue was achieved when 10ng/embryo or 20ng/embryo Y14 morpholino/embryo was used.
| Melanophore (%) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Morpholino (ng/embryo) |
RNA (ng/embryo) | Touch- responsive (%) |
normal | reduced | none | Heart defect (%) |
n | |
| Eif4A3 | 10 | 0 | 0 | 13.3 | 55.6 | 31.1 | 88.9 | 46 |
| 10 | 0.5 | 86.4 | 85.7 | 12.7 | 1.6 | 26.2 | 66 | |
| 10 | 2 | 91.1 | 82.1 | 14.3 | 3.6 | 14.8 | 56 | |
| 20 | 0 | 0 | 3.3 | 73.3 | 23.3 | 70 | 36 | |
| 20 | 0.5 | 94 | 78.8 | 17.3 | 3.8 | 34.6 | 50 | |
| 20 | 2 | 81 | 61.9 | 23.8 | 14.3 | 40 | 21 | |
| 0 | 2 | 78.9 | 52.6 | 34.2 | 13.2 | 23.7 | 38 | |
| Magoh | 10 | 0 | 0 | 0 | 78.8 | 21.2 | nd | 83 |
| 10 | 0.5 | 94.3 | 40 | 60 | 0 | nd | 31 | |
| 10 | 2 | 97.3 | 29.7 | 70.3 | 0 | nd | 74 | |
| 20 | 0 | 0 | 6 | 71.1 | 22.9 | 83.6 | 33 | |
| 20 | 0.5 | 100 | 64.5 | 35.5 | 0 | nd | 35 | |
| 20 | 2 | 98.6 | 75.7 | 21.6 | 2.7 | 10.2 | 37 | |
| 0 | 2 | 100 | 78.7 | 21.3 | 0 | 0 | 61 | |
| Y14 | 2.6 | 0 | 8.0 | 40.0 | 52.0 | 8.0 | 100.0 | 25 |
| 2.6 | 0.5 | 100.0 | 91.9 | 8.1 | 0.0 | 2.7 | 37 | |
| 2.6 | 2 | 96.9 | 79.2 | 20.8 | 0.0 | 0.0 | 32 | |
| 0 | 2 | 82.6 | 72.7 | 13.6 | 13.6 | 34.8 | 23 | |
| uninjected | 88.1 | 87.5 | 7.5 | 5.0 | 13.1 | 40 | ||
Paralysis and lack of touch response
Late neurula and tadpole stage embryos respond to tactile stimulation by twitching or swimming away from the stimulus, respectively; Eif4a3 morphants, however, are completely unresponsive to touch (Fig. 2B; Table I). No touch response behavior is observed in the Eif4a3 morphants at any stage examined (25-42); moreover, Eif4a3 morphants display no spontaneous movement whatsoever, suggesting that the lack of touch response is secondary to Eif4a3 knockdown-mediated paralysis (data not shown). Movement and touch response is partially rescued by co-expression of eif4a3 RNA: RNA injection alone has minimal effect on embryonic movement (Fig. 2B, Table I, and data not shown).
Defects in cardiac development
The Eif4a3 morphants exhibit a failure of heart looping, often accompanied by edema in the pericardial coelom; heart defects are rescued by co-injection of eif4a3 RNA, which alone has little effect on heart development (Fig. 2C; Table I, and data not shown). Despite defects in looping, Eif4a3 morphant hearts show normal levels of expression of the cardiac enzyme Troponin-T (Kolker et al., 2000); moreover, expression of Xnkx-2.5, a marker of cardiac progenitor cells and later, the myocardium, appears unaffected in Eif4a3 morphants at tailbud stages (Fig. 2D) (Tonissen et al., 1994). A heartbeat was observed in Eif4a3 morphants, albeit at a lower rate than in controls (roughly 50%, data not shown), providing further evidence for the presence of differentiated cardiomyocytes. Heart looping defects are more pronounced in embryos injected vegetally at the eight-cell stage; conversely, pigment and movement defects are seen when embryos are injected animally, but not vegetally, at the eight-cell stage (data not shown). These data suggest that loss of Eif4a3 in the mesendoderm inhibits normal cardiac development, whereas loss of Eif4a3 in the ectoderm underlies the melanophore and paralysis phenotypes. All developmental defects are observed following animal pole injection at the two-cell stage, likely owing to the dispersal of the small morpholino oligonucleotides throughout the blastomeres at early cleavage stages. All phenotypes were also observed following injection of a second morpholino against Eif4a3 (data not shown).
Knockdown of other Exon Junction Complex (EJC) components phenocopy Eif4a3 loss-of-function
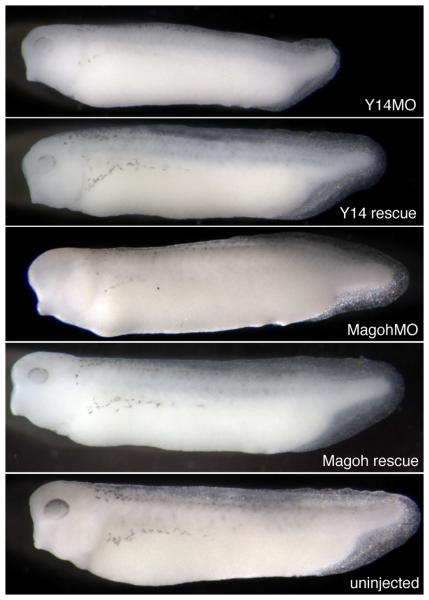
Eif4a3 is known to function as a component of a multi-protein structure called the Exon Junction Complex (EJC) (Tange et al., 2004). The requirement for Eif4a3 during embryonic development, however, may be due to distinct function(s) of this factor. For example, Eif4a3 is highly homologous to Eif4a1 and Eif4a2, components of the eukaryotic translation initiation machinery (Gingras et al., 1999); although Eif4a3 cannot itself functionally substitute for Eif4a1/2, its ability to complex with the translation initiation machinery suggests that it could function as a dominant inhibitor of translation (Li et al., 1999). To begin to address the mechanisms by which Eif4a3 regulates development, we used antisense morpholino oligonucleotides to knock down the “core” EJC components Y14 and Magoh. Knockdown of either Y14 or Magoh results in a phenotype virtually identical to that seen in the Eif4a3 morphants; moreover, these defects are generally rescued by co-expression, respectively, of Y14 and magoh RNA constructs engineered to lack the morpholino binding sites (Figure 3 and Table I). A previous study had noted embryonic paralysis, along with neural tube defects, following Magoh knockdown in Xenopus tropicalis (Kenwrick et al., 2004); RNA-mediated rescue of the morpholino effect was not shown in that earlier work. Overall, these data suggest that the requirement for Eif4a3 during early development is mediated primarily through its function as a component of the EJC.
Fig. 3.
The Y14 and Magoh morphant phenotypes. Loss of pigment-forming cells following Y14 or Magoh knockdown. 2.6ng of Y14 morpholino (“Y14MO”) or 10ng of Magoh morpholino (“Magoh MO”) was injected, as listed. “Rescue”: MO+2ng corresponding RNA. Embryos were derived from albino eggs fertilized with sperm from wild-type males.
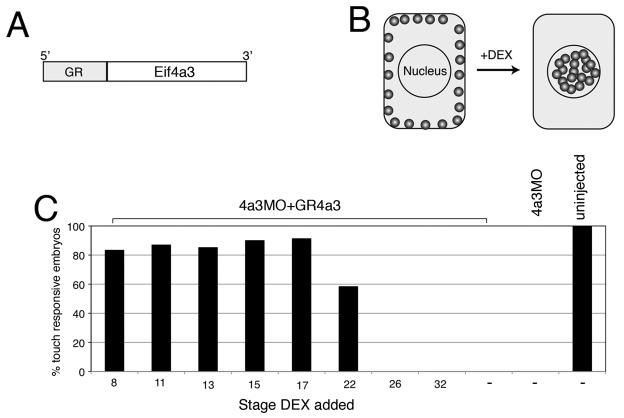
Loss of Eif4a3 at neurula stages leads to embryonic paralysis
In an attempt to determine the stage(s) at which Eif4a3 is required for normal development, we designed a glucocorticoid receptor-Eif4a3 (GR-Eif4a3) fusion construct (Fig. 4A). Protein generated from this construct is predicted to remain associated with the cytoplasm in embryonic Xenopus cells; addition of the hormone dexamethasone (DEX) will drive translocation of the fusion protein to the nucleus, postulated to be the initial site of EJC complex association (Fig. 4B) (Chang et al., 2007). For this study, we have focused on the rescue of embryonic paralysis, as this is the earliest detectable phenotype in the Eif4a3 morphants. We find that DEX addition partially rescues movement in Eif4a3 morphants up to neurula stage 22, with DEX addition between early cleavage stages and stage 17 eliciting the same degree of rescue; embryos injected with GR-Eif4a3 RNA but not treated with DEX show no rescue (Fig. 4C and data not shown). These studies indicate that Eif4a3 activity is required by neurula stages, but not before, for normal embryonic movement.
Fig. 4.
Eif4a3 is required by neurula stages for movement. A: Glucocorticoid receptor-Eif4a3 fusion construct. B: Addition of dexamethasone (DEX) allows nuclear entry of GR-Eif4a3. C: Addition of DEX at or before stage 17 to embryos injected with Eif4a3MO+GR-Eif4a3 RNA rescues the embryonic touch response, scored at stage 32; diminished rescue or lack of rescue is seen with DEX addition at later stages. DEX addition prior to stage 8 showed similar rescue to that seen from stages 8-17 (data not shown).
Molecular characterization of the Eif4a3 morphant phenotype
The paralysis of the Eif4a3 morphants could be due to a variety of factors, including defects in skeletal muscle, sensory and/or motor neuron development. We do not find marked changes in somite structure following Eif4a3 knockdown (Fig. 5A). As reported for the Magoh morphant, the brains of Eif4a3 morphants appear slightly wider than in control embryos, as visualized after staining with antisense elrC or sox3 probes (Kenwrick et al., 2004)(Fig. 5B, left and right panels, respectively); we have not observed, however, a loss of expression of the pan-neural marker elrC, as reported in the Magoh morphants (Kenwrick et al., 2004). Neuroanatomy, as visualized by staining with the monoclonal antibody 3A10 seems relatively unaffected by Eif4a3 loss-of-function, with the exception of less-uniformly bundled motor neurons (Ericson et al., 1996)(Fig. 5C). These data suggest that Eif4a3 is not required for neural induction, neurogenesis, or the gross development of the neural tube.
Fig. 5.
Somite and neuronal development in Eif4a3 morphants. A: 12/101 antibody staining of somitic mesoderm in embryos injected with Eif4a3MO (4a3MO) or a control (scrambled) morpholino (CMO). No differences in somite staining were observed between the two populations. B: Whole-mount in situ hybridization with antisense probes against elrC (left panels) or sox3 (right panels) in embryos injected with Eif4a3MO (4a3MO) or Eif4a3MM (5MM). Note slight expansion of anterior neural tube in Eif4a3 morphants. C: Left panels: 3A10 antibody staining of neurons in embryos injected with Eif4a3MO (4a3MO) or a control (scrambled) morpholino (CMO). Right panels: high magnification of trunk detail of embryos at left. Note disorganization of “Y”-shaped motor neuron bundles in Eif4a3 morphants.
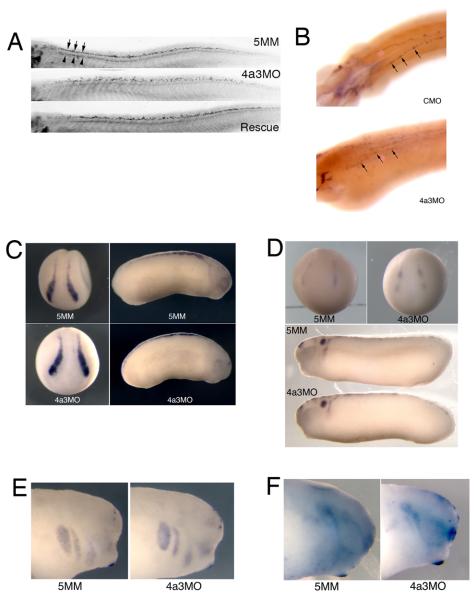
The Islet-1 transcription factor is expressed in the nuclei of both motor and Rohon-Beard sensory neurons during tadpole stages of Xenopus development (Chen et al., 2007). An antibody against the Islet-1 protein shows a reduction in Islet-1 signal in motor neurons of Eif4a3 morphants; the localization of these cells, however, appears unaffected (Fig. 6A, top panel). We find, however, that the dorsal spinal population of Islet-1-positive nuclei, corresponding to the Rohon-Beard neurons, occupy a wider dorsoventral zone than is seen in control embryos (Fig. 6A top and middle panels, respectively) (Chen et al., 2007). Both Islet-1 signal intensity and Islet-1-positive cell localization are rescued by co-injection of eif4a3 RNA (Fig. 6A). Consistently, we also observe a disorganization of cells expressing hox-11L2, a marker of Rohon-Beard sensory neurons and cranial neural crest, in Eif4a3 morphants, as well as an overall reduction in hox-11L2 signal intensity (Fig. 6B) (Patterson and Krieg, 1999). As the Rohon-Beard cells have been shown to be required for the embryonic touch response, these data suggest that the lack of touch response following Eif4a3 knockdown could be mediated by an effect on these neurons (Clarke et al., 1984; Roberts, 2000). The lack of touch response is, however, likely secondary to the complete paralysis observed in the Eif4a3 morphants; thus, motor neuron defects, indicated by improper bundling and decreased Islet-1 signal, rather than sensory neuron defects, may underlie the paralysis phenotype.
Fig. 6.
Eif4a3 morphants display late defects in sensory neuron and neural crest development. A: Islet-1 antibody staining of Rohon-Beard (top band of nuclei in top panel, arrows) and motor (bottom band of nuclei in top panel, arrowheads) neurons. Increased dorsoventral spread of Islet-1 positive Rohon-Beard cells, as well as a reduction in intensity of Islet-1 staining in motor neuron nuclei, was observed in Eif4a3 morphants; both defects were partially rescued by co-injection of eif4a3 RNA. B: Whole-mount in situ hybridization with an antisense probe against hox11L2 in embryos injected with Eif4a3MO (4a3MO) or a control (scrambled) morpholino (CMO). Note disorganization of hox11L2-positive Rohon-Beard cells (arrows) in the Eif4a3 morphant embryo. C, D, E, F: Whole-mount in situ hybridization with antisense probes against slug/snail2 (C), sox10 (D), Xtwist (E), or Xsix-1 (F) in embryos injected with Eif4a3MO (4a3MO) or Eif4a3MM (5MM) morpholinos, as listed. No consistent differences were seen in the expression of these markers at neurula or tailbud stages.
Rohon-Beard cells arise from the same progenitor population that gives rise to the cranial neural crest (Artinger et al., 1999; Cornell and Eisen, 2000; Cornell and Eisen, 2002). In order to determine whether neural crest development is broadly disrupted in the Eif4a3 morphants, we examined the expression of slug/snail2, one of the earliest markers of neural crest differentiation (Linker et al., 2000). We find no change in intensity or localization of slug/snail2 expression in Eif4a3 morphants during neural plate or tailbud stages (Fig. 6C). The expression of the neural crest marker sox-10 appears similarly unaffected in Eif4a3 morphants (Fig. 6D). Furthermore, we observe no change in the localization of Xtwist, a marker of cranial migratory neural crest, following Eif4a3 knockdown (Fig. 6E) (Hopwood et al., 1989; Linker et al., 2000)). Lastly, we examined the expression of Xsix-1, a marker of neurogenic cranial placodes derived, like the neural crest, from the border between the neural plate and the epidermis (Pandur and Moody, 2000; Schlosser, 2006); Xsix-1 expression appears normal following Eif4a3 knockdown (Fig. 6F). These data suggest that Eif4a3 is neither required for the initial differentiation of the neural crest nor for the migration or differentiation of at least some neural crest and other neural plate border derivatives (Linker et al., 2000).
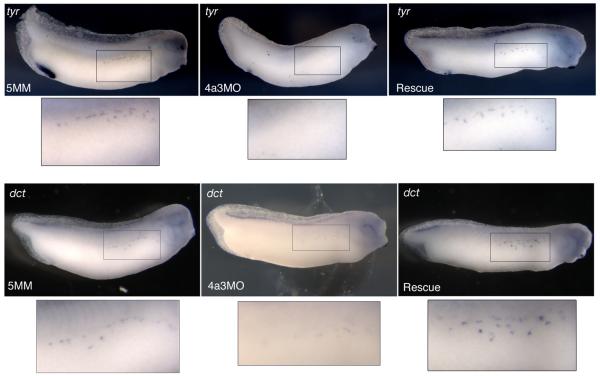
The loss of melanophores in the Eif4a3 morphants prompted us to examine the neural crest cells that give rise to this population. The tyrosinase family genes code for enzymes that catalyze melanin production and are expressed in Xenopus melanophores by late tailbud stages (Kumasaka et al., 2003). We find that the expression of both tyrosinase (tyr) and tyrosinase-related protein-2 (dct) are reduced in the Eif4a3 morphants at stage 32; this effect is rescued by co-expression of eif4a3 RNA (Fig. 7). These data confirm that Eif4a3 is necessary for normal melanophore development, and may be required for expression of the melanogenic enzymes, themselves.
Fig. 7.
Eif4a3 is required for expression of tyrosinase genes. Whole-mount in situ hybridization with antisense probes against tyrosinase (tyr) (top panels) or tyrosinase-related protein-2 (dct) (bottom panels) in embryos injected with Eif4a3MO (4a3MO), Eif4a3MM (5MM), or Eif4a3MO+2ng eif4a3 RNA (Rescue). With both probes, intensity of signal and number of positive cells were reduced in Eif4a3 morphants at tailbud stages; this effect was rescued by co-expression of eif4a3 RNA.
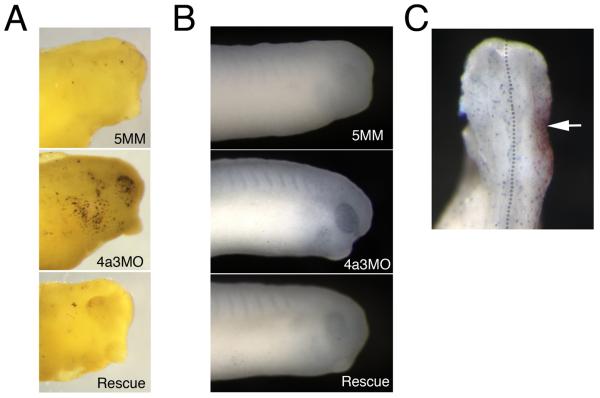
The reduction of Rohon-Beard cells and melanophores at tadpole stages in the Eif4a3 morphants could be due to increased rates of apoptotic death among these cell types. To address this possibility, we performed the Terminal deoxynucleotidyl transferase dUTP nick end labeling assay (TUNEL), to identify cells undergoing apoptosis. We found a dramatic increase in TUNEL-positive cells in the head region of tadpole stage embryos (Fig. 8A); we do not observe this increase in other regions of the morphant embryos (data not shown). A slight increase in TUNEL-positive cells is initially apparent in the eyes of Eif4a3 morphants beginning at late neurula stages (data not shown); more prominently, the eyes of late neurula stage embryos also displayed increased expression of the pro-apoptotic protease Caspase 3 (Fig. 8B). These data suggest that the loss of anterior pigmentation observed following Eif4a3 knockdown results from, or in, apoptotic cell death. The anterior restriction of TUNEL-positive cells also suggests that apoptosis does not mediate defects in Rohon-Beard sensory neurons or the loss of trunk pigmentation seen in the Eif4a3 morphants; these latter phenotypes may result from inappropriate differentiation or migration of subpopulations of trunk neural crest.
Fig. 8.
Apoptotic cell death is increased in Eif4a3 morphants. A: TUNEL assay of embryos injected with Eif4a3MO (4a3MO), Eif4a3MM (5MM), or Eif4a3MO+2ng eif4a3 RNA (Rescue) at stage 32. Strong staining observed in head only. B: Immunohistochemistry using Caspase 3 antibodies in embryos injected with Eif4a3MO (4a3MO), Eif4a3MM (5MM), or Eif4a3MO+2ng eif4a3 RNA (Rescue); signal was observed in the eyes of Eif4a3 morphants. C: Phosphohistone H3 antibody staining (blue) of embryos injected on one side (arrowhead) with Eif4a3MO (4a3MO) and 500pg β-gal RNA as a lineage trace (red) at stage 32. Phosphohistone H3 signal was not significantly affected by Eif4a3 knockdown.
The loss of neural crest and neuronal cell populations in the Eif4a3 morphants could also result from decreased proliferation of these cell types. We used an antibody to examine the levels of phosphorylated histone H3, the presence of which is a hallmark of dividing cells, in wild-type and Eif4a3 morphant embryos (Saka and Smith, 2001). We did not detect any appreciable difference between the number of phospho-histone H3-positive cells in Eif4a3 morphants and control embryos examined between neurula and tadpole stages (Fig. 8C and data not shown). This study suggests that Eif4a3 loss-of-function does not result in altered cell proliferation rates.
Discussion
Eif4a3 is a critical component of the multi-protein exon junction complex (EJC), implicated in the regulation of a number of RNA processing events. Here, we demonstrate that Eif4a3 is required for embryonic movement, and for melanophore and cardiac development, in Xenopus laevis. Loss-of-function of the other “core” EJC components gives rise to similar phenotypes, suggesting that the embryonic requirement for Eif4a3 is via its role in the EJC.
Our studies suggest that Eif4a3 may play a particularly important role in the development of a subset of cell types that emerge from the ectoderm at the border between the neural plate and epidermis. Eif4a3 activity is required for the development of the Rohon-Beard sensory neurons; these cells are formed during Xenopus primary neurogenesis, and are derived from the neural plate border cells that give rise to the melanophores and other derivatives of the migratory neural crest (Artinger et al., 1999; Cornell and Eisen, 2000; Cornell and Eisen, 2002). In addition, Eif4a3 knockdown results in defects in the neural crest-derived melanophores, themselves. Finally, while the cardiac abnormalities observed following Eif4a3 knockdown could result from a wide range of embryonic defects, the demonstration that ablation of the cardiac neural crest leads to a loss of heart looping in Xenopus suggest that Eif4a3 may regulate cardiac development via actions on the neural crest (Martinsen et al., 2004); studies to determine the underlying basis of the heart defects in the Eif4a3 morphants are in progress. The apparently normal Xsix-1 expression pattern in, and head structure of, the Eif4a3 morphants suggests that the development of at least some derivatives of the neural plate border, including the cranial placodes and the cartilage derived from the cranial neural crest, respectively, are independent of regulation by Eif4a3 or the EJC. Conversely, the apparent defects in motor neurons in the Eif4a3 morphants suggests that cells derived from regions other than the neural plate border also require EJC function for their proper development.
Precise regulation of BMP signaling is critical during several steps of neural crest development, including induction and maintenance of the premigratory neural crest cells, when both gastrula stage inhibition and neurula stage activation of BMP signaling, respectively, are required (Steventon et al., 2009). Our earlier work suggested that Eif4a3 gain-of-function is sufficient to enrich BMP activity in the early embryo via activation of an extracellular BMP signaling component (Weinstein et al., 1997). We have not observed outcomes associated with BMP signal attenuation following Eif4a3 loss-of-function: neither increased neuralization nor decreased levels of phosphorylated Smad1 protein have been observed in Eif4a3 morphants or in ectodermal explants derived from morphant embryos (data not shown). Thus, Eif4a3 does not appear to be required for regulating epidermal induction/neuralization during early Xenopus development. The lack of global alteration of BMP signaling following Eif4a3 loss-of-function could be due to the dynamic and at times highly localized expression of the zygotic component of Eif4a3 (Weinstein et al., 1997); i.e., Eif4a3 may only regulate BMP signaling within the Eif4a3 expression domain, perhaps including the neural plate/neural tube border region. The additional requirement for Y14 and Magoh for normal movement and pigmentation further suggests that regulation of BMP signaling can only occur in regions of the embryo where expression of the core EJC components overlap. Studies are in progress to establish the embryonic expression domains of Y14, Magoh, and MLN 51; there appears to be a high degree of overlap, in the tadpole stage head, of Eif4a3 and Magoh (Weinstein et al., 1997; Kenwrick et al., 2004) (data not shown).
To date, EJC function is perhaps best understood in the context of nonsense-mediated decay (NMD), a post-transcriptional surveillance mechanism by which eukaryotic cells detect and eliminate transcripts containing premature termination codons (PTCs) (Chang et al., 2007). Historically, NMD has been regarded as a surveillance mechanism by which aberrant transcripts, induced by mutation, are identified and degraded. More recent studies, however, have revealed that some wild-type transcripts, derived from genes with a particular structural organization, can also trigger NMD activation, suggesting a more comprehensive role for this pathway in the regulation of gene expression (Giorgi et al., 2007). The identification of nearly 200 candidate targets of NMD in human cells suggest that a major function of NMD is to suppress excess production, and thus regulate homeostatic gene expression, of these wild-type transcripts (Khajavi et al., 2006). The EJC has been shown to be essential for recognition and eventual degradation, via NMD, of RNA containing premature termination codons (PTCs) (Chang et al., 2007). As Eif4a3 is an essential component of the EJC, the developmental defects seen following Eif4a3 knockdown may arise as a result of defects in NMD.
At present, we cannot rule out the possibility that loss of EJC components leads to a general alteration of gene processing, with observed phenotypes seen in those systems that are most sensitive to defects in an as-yet undetermined, EJC-mediated global RNA processing event. Microarray data from our laboratory suggests that this is not the case, however: the level of most transcripts are unaffected by Eif4a3 knockdown, with a small subset (approximately 0.1%) of transcripts consistently down- or upregulated (data not shown). We are in the process of determining whether these changes reflect alterations in cells of a particular subtype, and/or whether dysregulated transcripts mediate the Eif4a3 morphant phenotype.
Experimental Procedures
Eif4a3 and GR-Eif4a3 fusion construct
A construct containing only the coding sequence of Xenopus Eif4a3 was described previously (Weinstein et al., 1997); this construct lacks the Eif4a3MO binding site. For 5′UTR-Eif4a3, the Eif4a3 coding sequence was amplified by PCR using T3 and the following primer, which includes 5′UTR sequence (AGATCTcttgcacattcgctaagggaaaatggcggccgcagctgttgc). The amplification product was cloned into the BglII and StuI sites of pCS2++. For the glucocorticoid receptor-Eif4a3 (GR-Eif4a3) fusion construct, the coding region of Eif4a3, minus the initiating Methionine, was amplified by PCR and cloned into the StuI and XhoI sites of pCS2-hGRN.
Xenopus Y14 constructs were generated from IMAGE clone 8318419. Wild-type Y14 was subcloned into the pCS2++ vector using the following primers: Y14ClaI: 5′-ATCGATGTTGTTGTTTAGCGCTAC; Y14XhoI: 5′-CTCGAGCTAACGTCGTCTTCTTTC.
Xenopus Magoh constructs were generated from IMAGE clone 6873436. Wild-type Magoh was subcloned into the pCS2++ vector using the following primers: MagohF Eco: 5′-GAATTCCACCGCGACCGGTAAC; MagohR Xho: 5′-CTCGAGTTAGATGGGCTTAATTTTG.
Silent mutations were introduced by PCR using KOD Hot Start Polymerase (Novagen). A Y14 morpholino-insensitive silent mutation was generated using the following primers: Y14smtF: 5′- ACTTTGCtGCaAACATGGCaGATGTgCTcGATCT; Y14smtR: 5′-AGATCgAGcACATCtGCCATGTTtGCaGCAAAGT.
A Magoh morpholino-insensitive silent mutation was generated using the following primers: MagohMT-F: 5′- CCGGTAACATGGGctcgGAcTTCTaCCTCCGCTATTATGTTGG; MagohMT-R: 5′-CCAACATAATAGCGGAGGtAGAAgTCcgagCCCATGTTACCGG.
RNA preparation and embryo culture
RNA was synthesized in vitro in the presence of cap analog using the mMessage mMachine kit (Ambion). Microinjection and embryo culture were performed as described (Hemmati-Brivanlou and Melton, 1994; Wilson and Hemmati-Brivanlou, 1995).
Morpholinos
Morpholino oligos (Gene Tools) were heated for five minutes at 65 degrees C, then quenched on ice, prior to injection at the 2- or 4-cell stage.
4a3MO- GGCCGCCATTTTCCCTTAGCGAATG; 4a3MO#2- TCCCTTAGCGAATGTGCAAGAGCCC; 4a3MM- GGaCGgCATTTTCCgTTAcCGAtTG; Y14- CCAGTACATCCGCCATGTTCGCTGC; Magoh- GCGGAGATAGAAATCGCTCCCCATG; CMO- CCTCTTACCTCAGTTACAATTTATA.
Immunohistochemistry, TUNEL assay, and in situ hybridization
Whole-mount antibody staining and in situ hybridization was performed as described (Suri et al., 2005). An antibody against mouse Eif4a3 (2256C1; Abcam ab50741) was used for Western blot analysis. The 12/101, 3A10, 40.2D6 (Islet-1) and CT3 (Troponin T) monoclonal antibodies were used at a 1:1, 1:10, 1:3, and 1:1 dilution, respectively. These antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology. Anti-phosphohistone H3 (Cell Signaling 9701) and anti-Caspase3 (Abcam ab13847) rabbit polyclonal antibodies were used at 1:100 and 1:1000 dilutions, respectively. Secondary antibodies, donkey anti-mouse and/or anti-rabbit IgG coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratories), were used at 1:1,000 dilution. Color reactions were performed using the Vector SG kit (Vector Laboratories). The TUNEL assay was performed as described in (Hensey and Gautier, 1998).
Acknowledgments
We wish to thank A. Brivanlou, P. Krieg, A. Kulozik, C. LaBonne, N. Papalopulu, P. Wilson, and H. Yamamoto for gifts of reagents. This work is supported by PHS grant R01-GM61671 (DCW) and funds from Queens College of the City University of New York.
Grant Sponsor: NIH R01-GM61671
References
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen JA, Chu ST, Amaya E. Maintenance of motor neuron progenitors in Xenopus requires a novel localized cyclin. EMBO Rep. 2007;8:287–292. doi: 10.1038/sj.embor.7400903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Hayes BP, Hunt SP, Roberts A. Sensory physiology, anatomy and immunohistochemistry of Rohon-Beard neurones in embryos of Xenopus laevis. J Physiol. 1984;348:511–525. doi: 10.1113/jphysiol.1984.sp015122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–2882. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129:2639–2648. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev Dyn. 2004;229:289–299. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. Dev Biol. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka M, Sato S, Yajima I, Yamamoto H. Isolation and developmental expression of tyrosinase family genes in Xenopus laevis. Pigment Cell Res. 2003;16:455–462. doi: 10.1034/j.1600-0749.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Imataka H, Morino S, Rogers GW, Jr., Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker C, Bronner-Fraser M, Mayor R. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Dev Biol. 2000;224:215–225. doi: 10.1006/dbio.2000.9723. [DOI] [PubMed] [Google Scholar]
- Martinsen BJ, Frasier AJ, Baker CV, Lohr JL. Cardiac neural crest ablation alters Id2 gene expression in the developing heart. Dev Biol. 2004;272:176–190. doi: 10.1016/j.ydbio.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Owttrim GW, Hofmann S, Kuhlemeier C. Divergent genes for translation initiation factor eIF-4A are coordinately expressed in tobacco. Nucleic Acids Res. 1991;19:5491–5496. doi: 10.1093/nar/19.20.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- Patterson KD, Krieg PA. Hox11-family genes XHox11 and XHox11L2 in xenopus: XHox11L2 expression is restricted to a subset of the primary sensory neurons. Dev Dyn. 1999;214:34–43. doi: 10.1002/(SICI)1097-0177(199901)214:1<34::AID-DVDY4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Roberts A. Early functional organization of spinal neurons in developing lower vertebrates. Brain Res Bull. 2000;53:585–593. doi: 10.1016/s0361-9230(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Steventon B, Araya C, Linker C, Kuriyama S, Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri C, Haremaki T, Weinstein DC. Xema, a foxi-class gene expressed in the gastrula stage Xenopus ectoderm, is required for the suppression of mesendoderm. Development. 2005;132:2733–2742. doi: 10.1242/dev.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Honore E, Hemmati-Brivanlou A. Epidermal induction and inhibition of neural fate by translation initiation factor 4AIII. Development. 1997;124:4235–4242. doi: 10.1242/dev.124.21.4235. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]