Abstract
We describe a method for ratio estimations in 18O-water labeling experiments acquired from low resolution isotopically resolved data. The method is implemented in a software package specifically designed for use in experiments making use of zoom-scan mode data acquisition. Zoom-scan mode data allows commonly used ion trap mass spectrometers to attain isotopic resolution, which make them amenable to use in labeling schemes such as 18O-water labeling, but algorithms and software developed for high resolution instruments may not be appropriate for the lower resolution data acquired in zoom-scan mode. The use of power spectrum analysis is proposed as a general approach which may be uniquely suited to these data types. The software implementation uses power spectrum to remove high-frequency noise, and band-filter contributions from co-eluting species of differing charge states. From the elemental composition of a peptide sequence we generate theoretical isotope envelopes of heavy-light peptide pairs in five different ratios; these theoretical envelopes are correlated with the filtered experimental zoom scans. To automate peptide quantification in high-throughput experiments, we have implemented our approach in a computer program, MassXplorer. We demonstrate the application of MassXplorer to two model mixtures of known proteins, and to a complex mixture of mouse kidney cortical extract. Comparison with another algorithm for ratio estimations demonstrates the increased precision and automation of MassXplorer.
Keywords: power spectral analysis, low-pass and band filtering, correlation of filtered spectrum with a theoretical isotope distribution, mass spectrometry, quantification, ratio estimation, 18O-water labeling, bioinformatics
Introduction
18O-water labeling is a versatile quantitative proteomics approach1 wherein two atoms of heavy oxygen are enzymatically incorporated into the C-termini of peptides2-6. The incorporation changes the molecular mass, but not the physico-chemical and chromatographic properties of the labeled peptides. Labeled and unlabeled samples are mixed together and analyzed in a single liquid chromatography - mass spectrometry (LC-MS) run. This approach limits variations related to chromatography, sample handling and mass spectral acquisition. Advantages of 18O-labeling are that it is not dependent on metabolism, and is not amino acid-specific6. This labeling technique has been commonly used in high resolution instruments6. In this paper we describe new algorithmic approach based on power spectral and correlation analyses to automate 18O-labelingquantification platforms using medium resolution zoom.
Although useful, the use of 18O-water labeling platform requires the use of instrumentation that can resolve isotopes, which can be achieved in a cost effective manner in many ion trap mass spectrometers using zoom-scan mode. In a zoom-scan mode, ion trap mass spectrometers produce, compared to the more high resolution instruments, only moderate resolution mass spectra of selected ions over limited mass-to-charge ratio (m/z) ranges. Zoom scans allow for separation of isotopic peaks of medium mass peptides with up to +4 charge. Because of their lower resolution and special mode of data acquisition, the large body of signal processing methods and software currently available for hybrid instruments such as the LTQ-Orbitrap are not optimal or appropriate. Several algorithmic approaches to estimate the ratios of heavy and light peptide pairs in zoom-scan experiments have previously been reported7-13. Two programs, Matching12 and ZoomQuant7, are publicly available. However, the use of 18O-water labeling will require algorithms capable of coping with overlapping isotopes due to the 4 Da separation in the heavy and light pairs. Only the Matching algorithm allows interpretation of these overlapping mass spectra. In Matching, the mass spectral profiles are modeled as Gaussians. The proportions of the components are estimated by minimizing the differences between the model and experimental profiles. The program is web-accessible, but requires every spectrum to be exported to the host computer manually.
ZoomQuant7 uses a threshold approach to determine isotopic peak positions. The peptide pair ratios are computed via expressions similar to those reported by Yao2 et al. and Johnson10 et al., based on the assumption that all peptides in the labeled sample have incorporated at least one 18O atom. ZoomQuant is publicly available; it can be installed on a local computer and run in a high-throughput mode. Other algorithms have been developed to estimate relative ratios of heavy and light peptide pairs using high mass-resolution data8;14-16. A recent comprehensive review described software solutions for quantitative proteomics17.
Our approach is to make use of power spectrum-based signal processing methodology which may be particularly useful for data from these low resolution instruments. Specifically, contaminant species often co-elute with target peptide pairs, and due to low mass resolution in zoom scans these contaminants cannot be separated from target peptide pairs in the m/z domain. However, a power spectrum based signal processing filters signal from co-eluting species. We apply low-pass and band filtering to clean the zoom scan of high-frequency noise, and of contributions from co-eluting peptides with differing charge states. From the elemental composition of peptides, we generate model isotopic envelopes of heavy-light peptide pairs in five mixing ratios. The isotopic distributions are generated via a convolution of elemental isotope distributions in an approach similar to that in Mercury18. The model isotopic envelopes are then separately correlated with the filtered zoom scan. The maximum from all five correlation functions identifies the position of the monoisotopic (16O2) peak. Zoom scans are fitted to a mixture of Gaussians via the Levenberg-Marquardt non-linear least squares model19. Only peak shapes and peak heights are fitted; the peak position is kept as determined from the cross-correlation. The peptide ratios are estimated from peak heights, much as in the previous applications2;7;10;20.
We present the results of applying our algorithm to two model mixtures of known proteins and a real biological sample. One model sample was comprised of labeled and unlabeled peptides from bovine serum albumin (BSA) mixed in five different concentrations. The second model sample was a mixture of labeled and unlabeled peptides of BSA, bovine alpha and beta caseins and horse cytochrome C, mixed in three different concentrations.
As an example of applications in high-throughput mode on a biological sample, we used our software to analyze data from a mouse kidney cortical extract. For this dataset we present an improved performance compared to ZoomQuant, the only software currently in existence that can operate in high throughput mode. We also show the value of low-pass and band filtering for co-eluting peptides with different charge states and applications to high-mass peptides.
Materials and Methods
Materials
Bovine serum albumin, bovine alpha casein, bovine beta casein, and horse cytochrome C were purchased from Sigma (St. Louis, MO). Modified sequencing grade trypsin was purchased from Promega (Madison, WI). Immobilized trypsin was purchased from Applied Biosystems (South San Francisco, CA). H218O (97% isotopic purity) was purchased from Cambridge Isotope Laboratories (Andover, MA). HPLC grade water and acetonitrile were purchased from Burdick and Jackson (Morristown, NJ). Acetic acid was purchased from Sigma (St. Louis, MO).
Tryptic digestion and post-proteolysis 18O-labeling
Two samples, each containing 1 mg of BSA, were reduced with 10 mM DTT for 30 min at room temperature. Protein cysteinyl residues were alkylated with 30 mM iodoacetamide for 2 h at 37°C. The sample was diluted 10-fold with 100 mM ammonium bicarbonate, and digested with 40 μg of trypsin overnight at 37°C. The tryptic peptide mixture was desalted with a Sep-Pak® C18 cartridge (Waters, Milford, MA) per the manufacturer's instructions. Peptides were eluted from the cartridge with 80% acetonitrile (ACN) and completely dried using a Speedvac. The post-proteolysis 18O-labeling was performed as described21 previously. The labeled and unlabeled peptides were mixed in 1:5, 1:3, 1:1, 3:1 and 5:1 ratios.
In the second model sample (four-protein mix), two protein mixtures, each containing 200 nmole each of BSA, alpha casein and beta casein, and cytochrome C were digested with trypsin, followed by 18O-water labeling. The labeled and unlabeled peptides were mixed with 18O/16O ratios of 5:1, 3:1 and 1:1.
Animal protocols and tissue processing
These procedures were identical to those we previously described22;23.
Liquid chromatography and tandem mass spectrometry
LC-MS/MS experiments were performed with an LTQ linear ion trap mass spectrometer (ThermoFisher, San Jose, CA) equipped with a nanospray source; the mass spectrometer was coupled online to a ProteomX® nano-HPLC system (ThermoFisher, San Jose, CA). Two μL of each peptide solution were manually injected and separated on a reversed-phase nano-HPLC column (PicoFrit™, 75 μm × 10 cm; tip ID 15 μm) with a linear gradient of 0-50% mobile phase B (0.1% acetic acid-90% ACN) in mobile phase A (0.1% acetic acid) over 60 min at 200 nL/min. The mass spectrometer was operated in the data-dependent triple-play mode. In this mode, three most intense ions in each MS survey scan were automatically selected for moderate resolution zoom scans which were followed by MS/MS. Each of the peptide mixtures was repetitively analyzed by nano-HPLC-MS/MS three times.
Data Processing
Power spectrum analysis of zoom scans
In the power spectrum analysis we apply low-pass and band filtering to eliminate high-frequency noise and contributions from other co-eluting species of differing charge state (from the target peptide's charge). The target peptides are read in from database search results. The power spectrum is computed via a periodogram19 estimator. At a frequency fk the corresponding value of the power spectrum is:
where, Ck is a discrete Fourier transform at the frequency fk, k = 1, 2,…(N/2 -1), and N is the number of data points. For f0 and fN/2 the above formula translates into a single term corresponding to the Fourier transform coefficients at the 0th and (N/2)nd frequencies, respectively.
A power spectrum of a zoom scan is a function of the spectral power in the frequency domain. We examine the power spectrum to determine the maximum power frequency outside of the low-frequency region (less than 10). The maximum frequency corresponds to the peak spacing of the most intense signal in the mass spectrum. The mass-to-charge ratio interval between the peaks is obtained by back-transformation into the mass domain:
where fc is the Nyquist critical frequency, Δ is the mass-to-charge ratio scan rate (for zoom scans it is set to 0.02 Thomson, Th), N = 1024, and k is the value of the frequency. Different charge states have distinct peaks in the frequency domain. For example, for a zoom scan rate of 0.02 Th and N = 1024, +2 charged peptides will have their maximum power peak at frequency 41, and +3 charged peptides at frequency 62. In addition to the main peak, every charge state also has satellite peaks whose frequencies are integral divisors of the main frequency. For example, in addition to the main peak (m/z spacing of 0.5 Th and frequency 41), a +2 charged peptide's power spectrum is expected to have peaks corresponding to m/z spacing of 1.0 Th (frequency 20), and mass-to-charge ratio spacing of 1.5 Th (frequency 14). Similarly, a +3 charged peptide will have satellite peaks at 0.66 Th (frequency 31), 0.99 Th (frequency 21), and 1.33 Th (frequency 16). The number of observable satellite peaks and their power abundances are dependent on the concrete isotopic envelope.
Once we determine the maximum power frequency, we eliminate contributions from all frequencies up to ten units higher than this frequency; we also remove low-frequency satellite peaks from the other charge states whose satellite peaks do not coincide with those of the target charge state. It should be noted that this approach cannot filter out components of a contaminant whose charge state is an integral divisor of that of the target peptide. After filtration, the filtered spectrum is transformed back into the mass domain and used downstream in the correlation analysis for peak picking.
Isotopic envelope generation
The isotopic distribution of a sequence results from the natural convolution of the individual isotopic abundances of its elements. We compute theoretical isotopic distributions from the elemental composition of the sequence, first using self-convolution for every chemical element, and then convolutions between elements. Our program convolves arrays of isotopic distributions directly by the dot product.
Peak picking
To determine the monoisotopic peak position, we use cross-correlation between the (frequency-filtered) experimental spectrum and the model isotope distributions. It is expected that the cross-correlation function will have its maximum value when the overlap between the experimental zoom scan and the model isotope distribution is maximized. The position of the maximum overlap corresponds to the position of the monoisotopic peak of the light peptide. It suffices to determine the monoisotopic peak positions of the unlabeled peptides, as the labeled peptides' peak positions are shifted by 4 Da. To speed up the computations we use fast Fourier transforms (FFT) in correlation analyses.
We use five theoretical profiles to correlate with a frequency-filtered zoom scan. The profiles are generated by assumptions of 4:1, 2:1, 1:1, 1:2 and 1:4 ratios of heavy to light peptides. This is done to allow for different H:L ratios in a peptide mixture. All model distributions are normalized to a value of one before the correlations. The monoisotopic peak position is determined from the maximum of all five correlation functions.
Curve fitting
We assume that the peak shapes in zoom scans are Gaussians. The parameters of the Gaussians (peak heights and variances) are determined via the Levenberg-Marquardt nonlinear least-squares fit19. The peak positions are kept fixed as determined in the correlation analysis.
Peptide ratio calculations
After the monoisotopic peak position is determined we compute peptide pair ratios using a previously proposed formula2. This formula assumes that the portion of the second isotopic peak not accounted for by the monoisotopic peak of the unlabeled peptide is due to a single 18O-labeled peptide. The details of the ratio estimations are explained in the Supplementary Materials section. In this paper, we always present the ratio as that of the labeled (heavy) to unlabeled (light) peptides.
For every ratio estimation we report a signal-to-noise ratio (S/N), defined as the ratio of the smaller of the abundances of labeled and unlabeled peptides to the noise abundance24. The noise abundance is determined as a median of all abundances in the zoom scan24.
Implementation
Our approach has been implemented in a program, MassXplorer, written in the C/C++ language of Visual Studio 9. The program accepts mzML25 format for spectra and pepXML26 format for database search results. The summary output includes ratios, abundances of peptide pairs, signal-to-noise ratios and peptide false discovery rates27 as determined from combined target and reversed databases28;29. The summary is in the csv file format. The program is freely available, and can be obtained by contacting the communicating author.
MS/MS database search
Database search conditions are standard and described in the Supplementary Materials section.
Results and Discussion
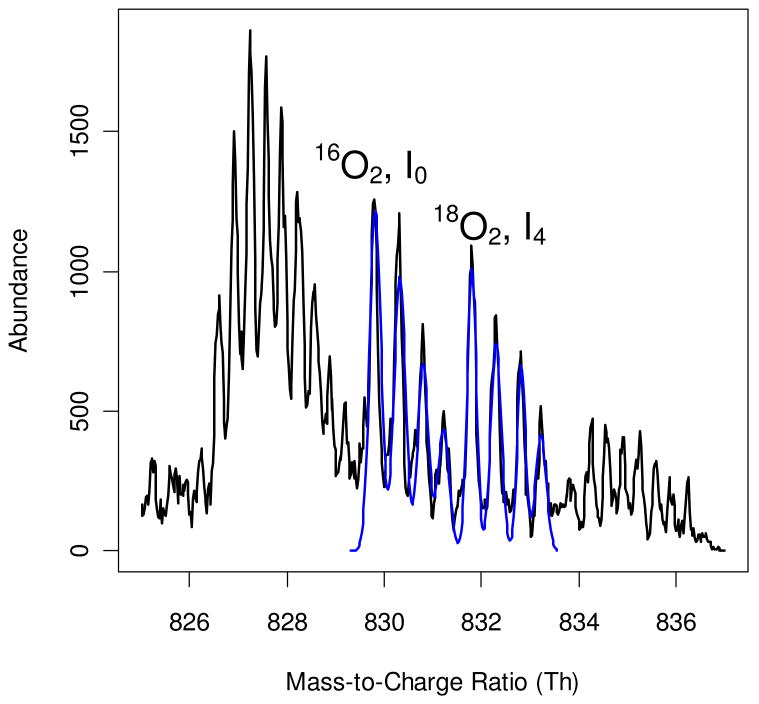
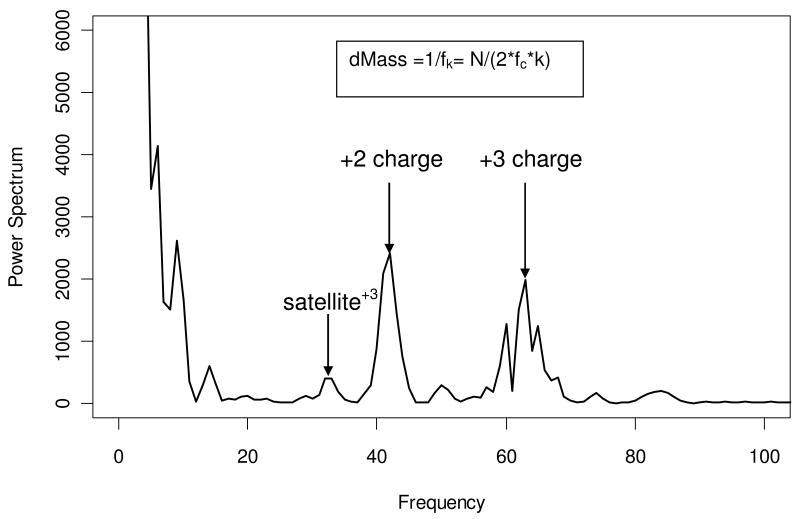
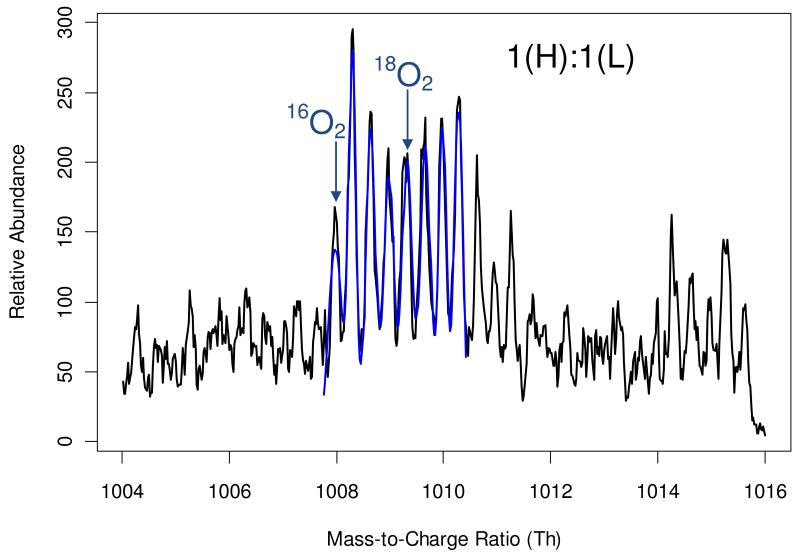
The application of the power spectrum analysis is illustrated in an example of a peptide from the mouse dataset. The zoom scan (black solid line in Figure 1) shows isotopic patterns of co-eluting but non-overlapping +2 and +3 charged species. The subsequent tandem mass spectrum was used to search the mouse protein database twice with the assumed precursor charge states of +2 and +3. The peptide LPDGSEIPLPPILLGK (from cytosolic non-specific dipeptidase, accession numberQ9D1A2 in the SwissProt mouse database) was identified with a +2 charge state at 1% FDR. As is seen from the zoom scan in Figure 1, the signal from the +3 charged species was stronger than that of the +2 peptide. First our algorithm performed power spectrum analysis of this zoom scan (Figure 2). Only a portion of the power spectrum is shown; the rest of the frequency axis contained only high-frequency noise. As expected, the highest value of the power spectrum corresponded to the low frequencies (less than 10). Outside of this region the power spectrum had two major peaks, one at frequency 42 and the other at frequency 63, which corresponded to the +2 charged peptide and +3 charged contaminant, respectively. Note that the peaks are not sharp, but have broad widths. This indicates that the peak spacings are not a single value, but rather a distribution around 0.5 Th and 0.33 Th, respectively.
Figure 1.
A zoom scan (black solid line) of a mixture of +2 and +3 charged species. The +2 charged peptide is a mouse peptide, LPDGSEIPLPPILLGK (from cytosolic non-specific dipeptidase, accession numberQ9D1A2). The signal from the contaminating +3 charged species is stronger than that from the target peptide. Without pre-processing, determining the position of the +2 charged monoisotopic peak is not possible in this case. The blue line is the Levenberg-Marquardt fit to the mass profile of the light and heavy pair of the peptide. Note that only the peak heights and shapes are fitted; the peak positions are fixed as determined by the cross-correlation analysis.
Figure 2.
The power spectrum of the zoom scan from Figure 1. Outside of the low-frequency domain (< 10), the power spectrum has two maxima, at frequencies 42 and 63. When transformed back into the mass domain (dMassk = 1/fk = N/(2* fc *k) = N * Δ/k), these frequencies correspond to mass-to-charge spacings of 0.5 Th and 0.33 Th, respectively. Here N is the array size (in this case 1024), Δ is the mass-to-charge ratio interval (0.02 Th) and k is the frequency. Also shown are the first satellite peaks of +2 and +3 charge species, at the frequencies 21 and 32, respectively.
Besides the major charge peaks at 41 and 63, the power spectrum also showed their satellite peaks. Thus the peak at frequency 32 corresponded to the spacing of 0.66 Th and was a satellite peak of the +3 charged species. To process this zoom scan for a +2 charged peptide, our algorithm removes all high frequencies (in this case, frequencies higher than 52) and band-filters components of the +3 charged species (local maximums around the frequencies 16 and 32). After setting the contaminant frequency components equal to zero, the spectrum was transformed back into the mass domain. The transformed mass spectrum is shown in Figure S1. In this spectrum, the signal from the +3 species was substantially suppressed, and the signal from the +2 species was now stronger. The transformed spectrum was used for peak picking in the correlation analysis.
We generated model isotopic envelopes of the light and heavy forms of the peptide LPDGSEIPLPPILLGK in five different H:L ratios – 4:1, 2:1, 1:1, 1:2 and 1:4. The theoretical isotope distributions were cross-correlated separately with the filtered zoom scan (Figure S1). The cross-correlations between the filtered spectrum and the theoretical spectra had a maximum at an m/z of 829.8 Th (Figure S2). This is the position of the monoisotopic peak; after determining its position we fit the experimental spectrum to a mixture of Gaussians (solid blue line in Figure 1). In the Supplemental Materials section we present the result of processing the zoom scan in Figure 1 for the +3 charged species (filtering out the signal from +2 charged species, Figure S3), and a cross-correlation function (Figure S4) between the theoretical spectrum and the unprocessed, original zoom scan in Figure 1. It is shown that without the filtering, the correlation analysis determined the monoisotopic peak position incorrectly. Also presented in the Supplemental Materials is an example of processing +3 target peptides by suppressing the dominant +2 species (Figures S5, S6, S7). We note here that most of the spectra we worked with are of +2 and +3 charge states. There were a few +4 charged species in our datasets, but no +1 charge species met the 3% FDR cut-off.
We applied MassXplorer to estimate peptide pair ratios from a sample containing a single protein, BSA, in known ratios of labeled and unlabeled peptides. In all we have analyzed five samples with H:L peptide ratios of 5:1, 3:1, 1:1, 1:3 and 1:5. All datasets were collected from triplicate mass spectrometry measurements. For the sample with a 1:1 ratio of labeled and unlabeled peptides, comprehensive results for every identified peptide sequence have been tabulated in Table S1.
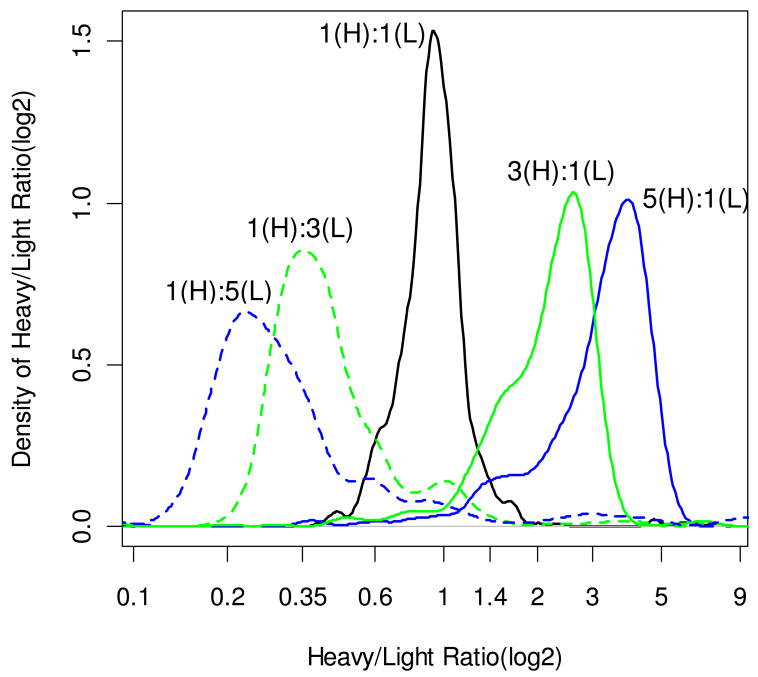
Figure 3 shows the density plots of 2-based logs of computed ratios. The X axis shows actual ratios. The density functions show the regions of highly frequent ratio values. As is seen from the figure, the best results are observed for the sample with a 1:1 ratio of heavy and light peptides (solid black line). The density function of ratios of this sample has a mode at a ratio of 0.92. The ratio at the mode of the density agreed well with the median value, 0.92, and the average ratio of all peptides in this dataset,0.98. Distributions of ratios for samples with higher concentrations of heavy peptides, 3:1 (solid green line), 5:1 (solid blue line)had smaller spreads (compared to the reversed ratios), and more pronounced modes. The modes of the density distributions were at the ratio values 2.6 and 3.8, respectively. The values agreed well with the median and mean values of the ratios from these samples. Thus the mean and median values for 3:1 ratio sample were 2.25 and 2.27, respectively. The corresponding values in the 5:1 ratio sample were 3.25 and 3.42. The datasets contained 499 spectra for 3:1 and 423 spectra for 5:1 ratio samples (at 3% FDR).
Figure 3.
Densities of 2-based logs of relative ratios of labeled to unlabeled peptides from BSA samples as computed by MassXplorer. The densities shown are for 2-based logs of the actual ratios, while the X axis shows the actual ratios. The density figures were drawn using R30. There were 468, 510, 547, 471 and 423 peptide spectrum matches that passed 3% FDR for the samples mixed in 1:5 (broken blue line), 1:3 (broken green line), 1:1 (solid black line), 3:1 (solid green line) and 5:1 (solid blue line) ratios, respectively.
Distributions for smaller ratio samples, 1:5 (broken blue line) and 1:3 (broken green line), had relatively larger spreads, and smaller density values at their corresponding modes. These figures were generated from datasets that contained 468 (1:5) and 510 (1:3) spectra. The density distributions had modes at ratios 0.23 (1:5) and 0.35 (1:3). The corresponding median values were 0.28 and 0.39, respectively. For these samples, the median and mode values were better representations of the actual ratios than the mean values, 0.68 (1:5) and 0.8 (1:3).
In the analyses of model systems the density functions could also help to locate consistent observations that contradict the expectations. Thus, the density function of ratios for the 1:3 sample (green broken line in Figure 3) shows a clear local peak with computed ratios near 1.0. We analyzed this section of the dataset for the identity of sequences and their zoom scans. It turned out that 25 out of 40 peptides with H:L ratios larger than 1 corresponded to the BSA sequence LVNELTEFAK. Examination of the zoom scans showed that the mass profile of the labeled +2 charged peptide LVNELTEFAK overlapped with a +3 charged contaminant species. Since this dataset was so simple, we were also able to determine the identity of the co-eluting contaminant. This was the +3 charged, unlabeled BSA peptide ECCHGDLLECADDR (Figure S8A). The identities of these species were determined from the database searches of the MS/MS spectra. As expected, LVNELTEFAK was in turn a contaminant for the profiles of ECCHGDLLECADDR. However, in this case the effect was not as dramatic, since the contamination was due to the heavy form of LVNELTEFAK, which is only one third of the light peptide in abundance. Another often-repeated peptide in this ratio cluster was the BSA peptide YNGVFQECCQAEDK. In the Supplemental Materials section we provide zoom scans of heavy and light pairs of this peptide, and show that the miscalculations of the ratios were due to overlapping profiles in this case as well (Figure S8B). The Supplemental Materials section also contains results of ratio estimations for the second model dataset comprised of peptides of four known proteins: BSA, alpha and beta caseins, and cytochrome C mixed in three different ratios – 1:1, 3:1 and 5:1, Figure S9.
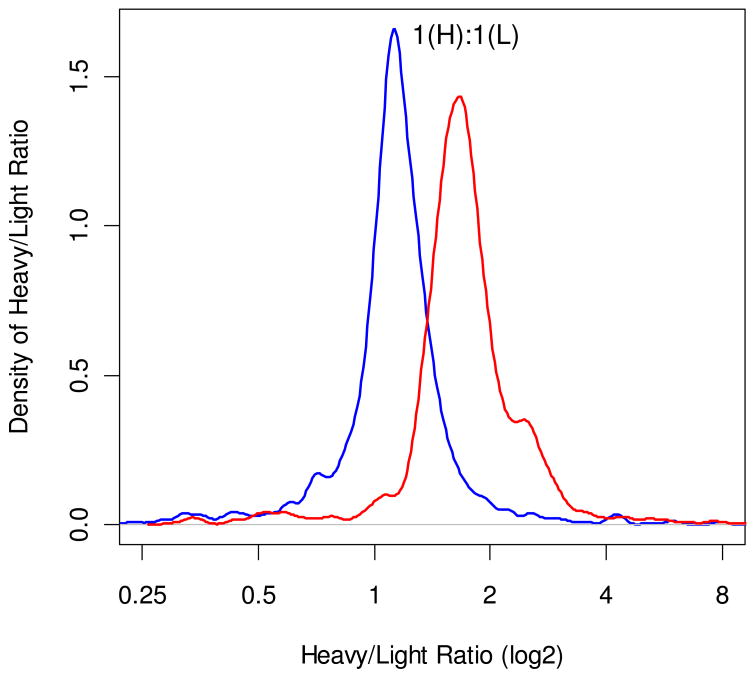
We used ZoomQuant and MassXplorer to compute peptide ratios in the mouse sample. The sample was generated by splitting a mouse tissue extract into two parts. One part was labeled using 18O-water. The labeled and unlabeled species were mixed at a 1:1 (H:L) ratio. Spectra were collected from four mass spectrometry analyses of the mixed sample. At 3% FDR there were 1263 identified spectra that were used for quantification. Figure 4 depicts the density plots of the (log2) ratios for ZoomQuant (red line) and MassXplorer (blue line); the same spectra were used in ratio calculations by MassXplorer and ZoomQuant. Both programs were run in automated mode with no manual curation. For MassXplorer the maximum density mode ratio was 1.12, while for ZoomQuant this value was 1.65. Note that ZoomQuant allows manual assignment of the peaks, and mis-assigned peaks could potentially be correctly re-assigned by manual curation. We only report the results from automated ratio estimations in both ZoomQuant and MassXplorer. Sample colon and zcn files that were generated by ZoomQuant are accessible in the web site indicated in Supplemental Materials section, along with csv files containing results from MassXplorer.
Figure 4.
Density graphs of ratios of abundances of heavy and light peptide pairs in the mouse sample as computed by MassXplorer (blue line) and Zoom Quant (red line). The ratios shown are for 2-based logs of the actual ratios, but the X axis shows the actual ratios. There were a total of 1263 spectra in this dataset that passed the 3% FDR threshold; 1182 spectra of +2 charged, 79 spectra of +3 charged, and 2 spectra of +4 charged peptides.
When using model isotopic distributions for estimating peptide ratios, it is important to point out the limits of application of the model with peptide mass. For high-mass peptides the monoisotopic peak could potentially have a low S/N and mix with the background noise. We estimate the noise abundance as the median abundance in a zoom scan24. The smaller of the monoisotopic peak abundances of unlabeled and labeled peptides is assumed to be the signal abundance24. In both the BSA and the four-protein mixture samples there were no peptides with masses > 2000 Da. Only the mouse sample had large peptides, and we examined them to check the accuracy of our model with increasing peptide mass. The sample had 49 unique peptides with masses > 2000 Da. These peptides have been identified in 113 spectra. In Figure 5 we show the zoom scan of the +3 charged mouse peptide HIADLAGNPEVILPVPAFNVINGGSHAGNK, with the largest mass among all peptides in this dataset. The monoisotopic mass of the peptide was 3021.59 Da, and the S/N for its unlabeled monoisotopic peak was 2.26. As is seen from the figure, MassXplorer correctly identified the position of the monoisotopic peak of the unlabeled species (blue line). The computed H:L peptide ratio was 0.77; in this dataset the expected ratio value is 1.0. The median S/N for the peptides with masses larger than 2000 Da was 4.9, while the median S/N for peptides with masses less than 2000 Da was 10.6. From close examination of the peptides in the mouse sample with masses in the range of 2000 Da to 3000 Da, we noticed that if the monoisotopic peak of the unlabeled peptide is separated from the background signal (relatively high S/N), and there is no overlap of profiles with other co-eluting species, then our algorithm can correctly locate peak positions (Figure S10). However, it is important to note that the peptides in this sample were mixed in a 1:1 (H:L) ratio. As we have seen above with the BSA sample, for other ratios of labeled to unlabeled peptides the accuracy of ratio estimations worsen. This effect is expected to be more pronounced for larger peptides.
Figure 5.
Experimental zoom scan (black line) and fit (blue line) of heavy-light peptide pairs from a +3 charged mouse peptide sequence, HIADLAGNPEVILPVPAFNVINGGSHAGNK, identified at 1% FDR. Note that the peak positions are determined from the correlation analysis, and peak shapes and heights are determined by the Levenberg-Marquardt method. The S/N ratio of the monoisotopic peak of the unlabeled peptide was 2.26. The monoisotopic mass of the unlabeled peptide was 3025.59 Da. The monoisotopic peak position was determined correctly by MassXplorer. The computed heavy to light ratio was 0.77:1, while the expected value was 1:1.
For direct ratio estimations, isotopic profiles of both labeled and unlabeled species must be present in a mass spectrum. However, it is possible that the abundance of one of the species is below the instrument's detection limit. In this case, in moderate mass accuracy instruments it is not possible to determine which form of a peptide (heavy or light) has been detected by the mass spectrometer. Our approach has no rigorous criteria to distinguish labeled and unlabeled species in this case. However, we conditionally assign the peak to the form of the peptide that has been identified in the database search. That is, if the identification has returned a modified peptide then the single isotopic distribution is assumed to be that of the labeled species, and vice versa. In these cases, our algorithm returns a predefined limiting value (20 or 0.05) for the ratios. It has been suggested that one can use the noise signal for the missing peak to estimate the ratio24. We choose to return the predefined value to make it apparent that one of the isotope envelopes is missing in the mass spectrum and the corresponding ratio is out of range of the limits of detection of the instrument.
In our future work, we plan to extend the spectral power processing approach to analyze datasets from other instrument types, high mass accuracy and resolution mass spectrometers, and other labeling platforms where there is isotope profiles of heavy and light peptides overlap (e.g., acrymalide labeling, 3. Da overlap). Also, we will apply machine learning techniques to train a classifier of zoom scans to assign significance to ratio estimations using such features as S/N and differences between experimental and theoretical isotopic profiles.
Conclusion
We have applied signal processing techniques to datasets from an 18O-labeling platform using moderate-resolution ion trap mass spectrometers. Our algorithm uses power spectrum analysis to filter out high-frequency noise and band-filter contaminant peaks from co-eluting peptides with differing charge states. The filtered spectrum is back-transformed into the mass domain and used in a correlation analysis to locate the monoisotopic peak position of unlabeled peptides. After fixing the peak position, peak shapes and peak heights are obtained by a fit using the Levenberg-Marquardt method. The ratios are computed from peak heights using a previously proposed formula2. We observe that the major contributions to erroneous peptide ratio estimations stem from co-eluting contaminants whose profiles overlap with those of the target peptides, low S/N, and high mass values of peptides. This suggests the need to develop a machine learning algorithm to detect noisy spectra and co-eluting peptides to improve automated interpretation of the estimations.
Supplementary Material
Acknowledgments
We thank Prof. Bruce Luxon and Mr. Dennis Obukowicz for discussions on the informatics aspects of the workflow design of quantitative proteomics and Dr. David Konkel for critically editing the manuscript. This work was supported in part by “Clinical Proteomics Centers in Biodefense and Emerging Infectious Diseases” (NIAID contract HHSN272200800048C). The work for generating the experimental data used in this study, was supported by the McCoy Foundation (LD) and, in part, by the Juvenile Diabetes Research Foundation (RGT).
Footnotes
Supporting Information Available. In Supplement 1 we present table S1 (details of ratio estimations in BSA samples) and Figures S1-S10 that show theoretical correlation analysis without frequency filtering, overlapping profiles of co-eluting species, and peak picking for a +4 charged, large-mass peptide.
The raw files used in this study are available at the web site: http://www.scmm.utmb.edu/faculty/rs_software.htm.
Reference List
- 1.Fenselau C, Yao X. J Proteome Res. 2009;8:2140–43. doi: 10.1021/pr8009879. [DOI] [PubMed] [Google Scholar]
- 2.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–42. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 3.Schnolzer M, Jedrzejewski P, Lehmann WD. Electrophoresis. 1996;17:945–53. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 4.Heller M, Menzel C, Mattou H, Yao X. J Am Soc Mass Spectrom. 2003;14:704–18. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 5.Dasari S, Wilmarth PA, Reddy AP, Robertson LJ, Nagalla SR, David LL. J Proteome Res. 2009 doi: 10.1021/pr801054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyagi M, Rao KC. Mass Spectrom Rev. 2007;26:121–36. doi: 10.1002/mas.20116. [DOI] [PubMed] [Google Scholar]
- 7.Halligan BD, Slyper RY, Twigger SN, Hicks W, Olivier M, Greene AS. J Am Soc Mass Spectrom. 2005;16:302–06. doi: 10.1016/j.jasms.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason CJ, Therneau TM, Eckel-Passow JE, Johnson KL, Oberg AL, Olson JE, Nair KS, Muddiman DC, Bergen HR., III Mol Cell Proteomics. 2007;6:305–18. doi: 10.1074/mcp.M600148-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ramos-Fernandez A, Lopez-Ferrer D, Vazquez J. Mol Cell Proteomics. 2007;6:1274–86. doi: 10.1074/mcp.T600029-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson KL, Muddiman DC. J Am Soc Mass Spectrom. 2004;15:437–45. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Hines W, Adamec J, Asara JM, Naylor S, Regnier FE. J Am Soc Mass Spectrom. 2005;16:1181–91. doi: 10.1016/j.jasms.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-de-Cossio J, Gonzalez LJ, Satomi Y, Betancourt L, Ramos Y, Huerta V, Besada V, Padron G, Minamino N, Takao T. Rapid Commun Mass Spectrom. 2004;18:2465–72. doi: 10.1002/rcm.1647. [DOI] [PubMed] [Google Scholar]
- 13.Shinkawa T, Taoka M, Yamauchi Y, Ichimura T, Kaji H, Takahashi N, Isobe T. J Proteome Res. 2005;4:1826–31. doi: 10.1021/pr050167x. [DOI] [PubMed] [Google Scholar]
- 14.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Rapid Commun Mass Spectrom. 2000;14:1226–32. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 15.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, Smith RD. Mol Cell Proteomics. 2005;4:700–09. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin C, Chen J, Goodlett D, Whiteaker J, Paulovich A, McIntosh M. Bioinformatics. 2006;22:1902–09. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- 17.Mueller LN, Brusniak MY, Mani DR, Aebersold R. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 18.Rockwood AL, Van Orden SL. Anal Chem. 1996;68:2027–30. doi: 10.1021/ac951158i. [DOI] [PubMed] [Google Scholar]
- 19.Press William H, Teukolsky Saul A, Vetterling William T, Flamming Brian P. Numerical Recipes: The Art of Scientific Computing. Third Edition. Cambridge University Press; 2007. [Google Scholar]
- 20.Zang L, Palmer TD, Hancock WS, Sgroi DC, Karger BL. J Proteome Res. 2004;3:604–12. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- 21.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, Smith RD. Mol Cell Proteomics. 2005;4:700–09. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilton RG, Haidacher SJ, Lejeune WS, Zhang X, Zhao Y, Kurosky A, Brasier AR, Denner L. Proteomics. 2007;7:1729–42. doi: 10.1002/pmic.200700017. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Denner L, Haidacher SJ, LeJeune WS, Tilton RG. Proteome Science. 2008;6:15. doi: 10.1186/1477-5956-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakalarski CE, Elias JE, Villen J, Haas W, Gerber SA, Everley PA, Gygi SP. J Proteome Res. 2008;7:4756–65. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutsch E. Proteomics. 2008;8:2776–77. doi: 10.1002/pmic.200890049. [DOI] [PubMed] [Google Scholar]
- 26.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Journal of Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 28.Moore RE, Young MK, Lee TD. J Am Soc Mass Spectrom. 2002;13:378–86. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 29.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A Language and Environment for Statistical Computing. 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.