Abstract
Herein, we show that sansalvamide A-amide (San A-amide), a structurally unique molecule, influences a subset of cancer-related pathways involving heat shock protein 90 (Hsp90). We show that San A-amide specifically binds to the N-middle domain of Hsp90 and allosterically disrupts the binding of proteins thought to interact with the Hsp90 C-terminal domain, while having no effect on an N-terminal domain client protein. This unique mechanism suggests that San A-amide is a potential tool for studying C-terminal binding proteins of Hsp90 as well as a promising lead in the development of new cancer therapeutics.
Keywords: Sansalvamide A-amide, Hsp90, cancer therapeutics
There are serious limitations on the molecules currently available to treat drug-resistant cancers. Natural products are a rich source of new therapeutic agents. One such compound is Sansalvamide A (San A), where the peptide derivatives of San A (Figure 1) have shown promise as a scaffold that exhibits anticancer activity.1−5 The cytotoxicity of San A derivatives against pancreatic,5−7 colon,8 and breast and prostate carcinomas as well as melanomas6 suggests that San A-amide derivatives (Figure 1) may have potential as new cancer therapeutics. Although a number of studies have shown San A-amide's structure−activity relationship (SAR) in various cancer cell lines, their apoptotic effect, and their cytotoxicity against numerous cell lines, to date, no mechanistic studies have explained their cytotoxicity in mammalian cells.3,9 Herein, we show that San A-amide binds to heat shock protein 90 (Hsp90) at the N-middle domain, and in doing so, it blocks the binding of Hsp90 to a client protein and a cochaperone that bind to the C terminus of Hsp90: inositol hexakisphosphate kinase-2 (IP6K2) and FKBP52, respectively. Our findings suggest that San A-amide inhibits C-terminal interactions via an allosteric mechanism rather than directly inhibiting their binding to Hsp90.
Figure 1.
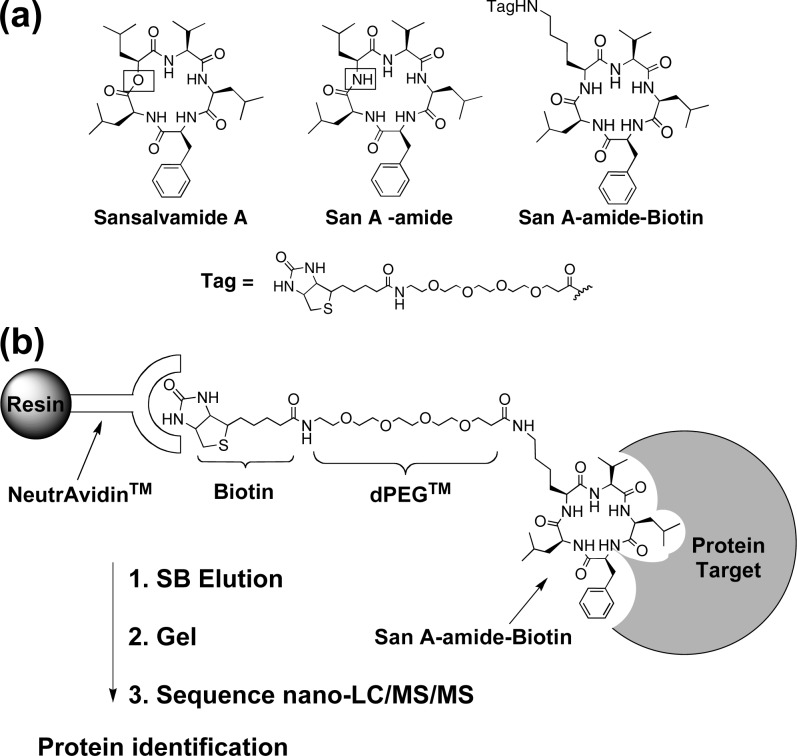
(a) Natural product San A, the derivative San A-amide compound (1), and biotinylated San A-amide (San A-amide-Biotin). (b) Schematic diagram of the pull-down assay.
To investigate the mechanism of action for San A-amide, we identified potential oncogenic protein target(s) that bound to this molecule by employing pull-down assays using a biotinylated derivative, San A-amide-Biotin (Figure 1). Our SAR data indicated that position 4 is not critical for cytotoxicity,4,5 and thus, it represented a logical location for the incorporation of a biotinylated linker into these molecules. San A-amide-Biotin was synthesized and used in pull-down assays with the HCT-116 colon cancer cell line (Figure 2).
Figure 2.
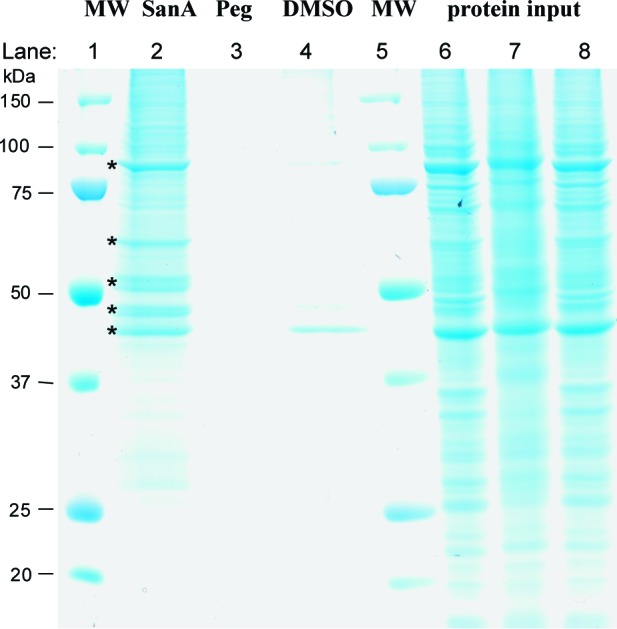
Bands isolated in the pull-down assay using HCT-116 colon cancer cell lyate. Lanes: 1 and 5, MW marker (kDa); 2, San A-amide-Biotin; 3, negative control (PEGlayted biotin linker); 4, DMSO control; and 6−8, protein input for lanes 2−4, respectively. Major proteins are indicated by asterisks with the following order from top to bottom: Hsp90, keratin, α- and β-tubulin, and actin.
The biotinylated compound was incubated with HCT-116 cell lysate, whereupon neutravidin-bound agarose beads were added to immobilize the compound along with the bound target protein(s) (Figure 1b). The beads were washed 10 times to remove nonspecifically bound proteins, followed by elution of proteins using sample buffer. The eluted proteins were run on an sodium dodecyl sulfate−polyacrylamide gel electrophoresis gel and visualized with Coomassie blue (Figure 2). Five major proteins were pulled down by San A-amide-Biotin (asterisks in Figure 2). The most prominent protein bound to San A-amide-Biotin was visualized as a band at approximately 90−95 kDa. Four other major proteins appeared between 40 and 65 kDa. It is possible that other proteins may be related to our mechanism of action that are not seen with Coomassie staining; therefore, protein sequencing of the entire lane 2 was performed using a nano-LC/MS/MS followed by a search of the NCBInr Eukaryotic database and peptide identification using fingerprinting software. No other major proteins were found, and only five major proteins were identified, with one of these being associated with oncogenic pathways. The major bands listed in order from top to bottom were identified as Hsp90, keratin, α- and β-tubulin, and actin. It is well-established that keratin, tubulin, and actin are commonly pulled down due to their hydrophobicity.10−14 Furthermore, comparison of the band at 90−95 kDa in lane 2 to the negative control [lane 3, the PEGylated biotin linker alone, or lane 4, dimethyl sulfoxide (DMSO)] suggests that the interaction between Hsp90 and San A-amide is specific. Given that Hsp90 is the only protein pulled down that is associated with an oncogenic pathway, we examined the interaction between San A-amide and Hsp90 more closely.
Hsp90 is known to play an important role in cancer cell growth because it functions as a molecular chaperone for folding, assembling, and stabilizing proteins and for intracellular cell signaling.15,16 Hsp90 has three distinct regions: the N-terminal, the middle, and the C-terminal domains. It exists as a dimer in which the subunits are constitutively associated via the C-terminal domain.17−19 It has over 100 identified client proteins, many of which are involved in cell signaling.15,20 Interestingly, Hsp90 is up-regulated in the majority of cancers,15,21−25 and inhibiting the function of Hsp90 affects multiple oncogenic pathways that are involved in cancer cell growth and programmed cell death.26−28 Given that the efficacy of target-specific anticancer drugs may decrease or even be lost over time due to the high epigenetic variation within cancer cells, blocking a protein that affects numerous cancer-related pathways, such as Hsp90, can be an effective and efficient means of treating drug-resistant cancers.26−28
There are currently considered to be two classes of Hsp90 inhibitors: the N-terminal inhibitors [e.g., 17-allylamino and 17-demethoxygeldanamycin (17-AAG)] that target the ATP pocket and a relatively weak C-terminal inhibitor, novobiocin (Figures S5 and S6 of the Supporting Information, respectively).29−31 17-AAG is currently in phase I and II clinical trials.26,32,33 There are currently no clinical trials involving molecules that inhibit the C-terminal binding proteins.
Anticipating that the cytotoxic effect of San A-amide might be a direct result of its ability to inhibit the interaction between Hsp90 and cell signaling events, we directly probed the effect of our small molecule on the binding interaction between Hsp90 and three proteins: Her2, IP6K2, and FKBP52. Her2 is a well-established client protein that binds to the N-terminal domain, and it is inhibited from binding to Hsp90 by 17-AAG.35−36 Thus, it was an excellent choice for comparing our molecule's mechanism to that of 17-AAG. San A-amide was shown to cause apoptosis,7,9 and both IP6K229 and FKBPs37 are associated with apoptotic pathways when either is inhibited from binding to Hsp90. Thus, these two proteins were also logical choices for exploring San A-amide's apoptotic mechanism of action.
We monitored the binding between each protein (Her2, IP6K2, and FKBP52) and Hsp90 via an in vitro binding assay. We used native Hsp90, purified from HeLa cells (Stressgen), recombinant 6x-histidine-tagged IP6K2 (His-IP6K2), recombinant GST-tagged Her2, and recombinant GST-tagged FKBP52 (R&D Systems and Abnova, respectively) (Figure 3a). San A-amide (0−10 μM) was added to the reaction; then, Talon metal affinity resin or Immobilized Glutathione agarose was added to the respective reactions, followed by three washes of the beads. The percent of Hsp90 bound to client protein was analyzed using Western blots (Figure 3a). The following procedure was repeated with 17-AAG rather than San A-amide (Figure 3b).
Figure 3.
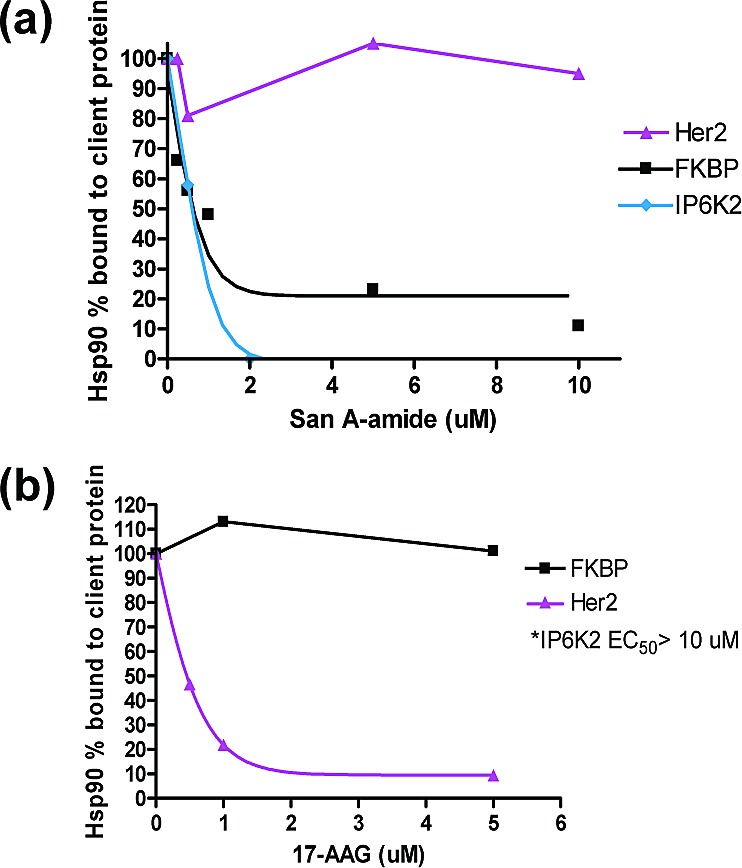
In vitro pull-down binding assay: (a) San A-amide inhibits binding of both IP6K2 (EC50 = ∼1.4 μM) and the FKBP (EC50 = 1.0 μM) to Hsp90. Her2 binding to Hsp90 is not affected. (b) 17AAG inhibits Her2 binding to Hsp90 (EC50 = ∼0.66 μM), while FKBP and IP6K2 binding to Hsp90 are not affected (IP6K2 is not inhibited at 10 μM).29 %Hsp90 bound to client protein was quantified by densitometric scanning of Hsp90 protein on Western blot with normalization to client protein loading using Image J.
We found that San A-amide inhibits the binding of the C-terminal interacting protein, IP6K2, to Hsp90 as well as the C-terminal binding cochaperone FKBP52 but has no effect on the binding of a N-terminal domain client, Her2 (Figure 3a). In contrast, 17-AAG did not affect the binding of IP6K229 and FKBP52 but did affect the binding of Her2 to Hsp90 (Figure 3b). This suggests that our molecule has a mechanism that is unique from that of 17-AAG. Along with demonstrating that Hsp90 binds to IP6K2 in a 17-AAG-independent manner, Snyder's group demonstrated that blocking IP6K2's access to Hsp90's C-terminal domain elicits programmed cell death.29 Furthermore, San A-amide is a significantly more effective binding inhibitor than the Hsp90 C-terminal binder novobiocin, where novobiocin inhibits IP6K2 binding at an EC50 of ∼100 μM29 versus San A-amide's inhibitory EC50 of ∼1 μM. Thus, by disrupting this Hsp90-IP6K2 interaction, San A-amide may provide a new approach to chemotherapeutics.
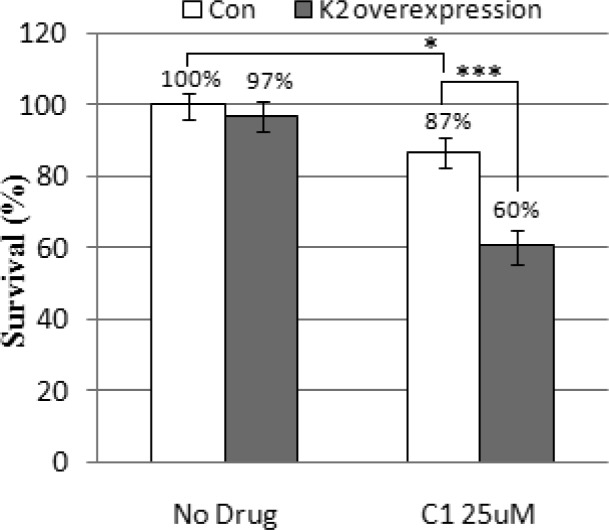
To determine if the in vivo affects of San A-amide were linked to our in vitro observation of the inhibition of Hsp90−IP6K2 interactions, we performed cell-based assays and compared the effect of cell survival in the presence of San A-amide in wild-type (control) and IP6K2 overexpressed (K2-O/E) HEK 293 cell lines. Tet-inducible Myc-IP6K2 constructs were created for IP6K2 overexpression. The control and K2-O/E cells were treated with DMSO (no drug) or 25 μM San A-amide (C1) for 48 h, and viability was determined using a CCK-8 assay. When no drug is present, IP6K2 overexpression only moderately affects cell survival (Figure 4, 3% change when comparing control, white bar, set at 100% to K2-O/E, gray bar, 97%). However, in the presence of San A-amide, cell survival is substantially decreased in cells that overexpress IP6K2 as compared to wild-type cells (∼9-fold, 3 vs 27% change). Thus, San A-amide induces cell death, at least in part, through the Hsp90−IP6K2 pathway (17-AAG did not affect this pathway29).
Figure 4.
Cytotoxicity of San A-amide in IP6K2 overexpressed cell line (K2-O/E, gray bar) as compared to wild-type control (Con, white bar). *p < 0.05; ***p < 0.001.
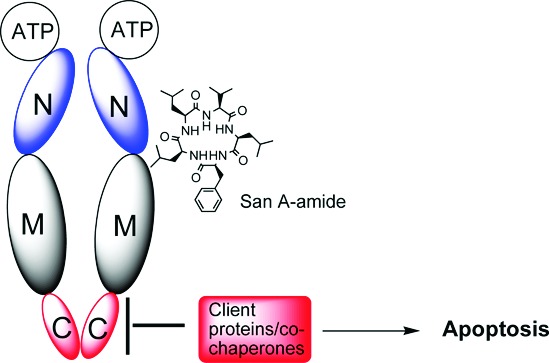
To begin to map the binding site of San A-amide, pull downs were performed against the N-terminal, middle, C-terminal, and N-middle domains of the yeast variant of Hsp90 (Hsc82, Figure 5). The Hsp90 domain variances were expressed in His-fusion vectors, purified, and used in pull-down assays with San A-amide-Biotin (Figure 1a). San A-amide not surprisingly bound to full-length Hsp90 (Figure 5, lane 6). There was only a marginally detectable interaction between the San A-amide and the N-terminal domain (lane 2) and partial affinity for the middle domain (lane 3). Surprisingly, San A-amide has no affinity for the C-terminal domain (lane 4) but has optimal affinity for the N-middle construct (lane 5). These data indicate that although San A-amide binds preferentially to the N-middle domain of Hsp90, its inability to inhibit Her2 from binding to Hsp90 indicates that the Hsp90 binding site of San A-amide is distinct from that of Her2 and 17-AAG. Moreover, unlike 17-AAG, which strongly inhibits the Hsp90 ATPase activity,38 San A-amide has no affect on ATP hydrolysis (Figure S7 of the Supporting Information). Because the macrocyclic peptide does not interact directly with the C-terminal domain, it likely alters the Hsp90 conformational equilibrium by affecting protein binding via an allosteric mechanism.
Figure 5.
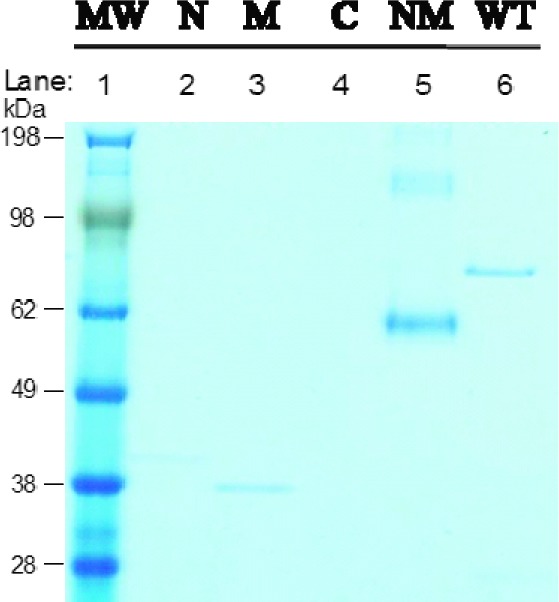
San A-amide pull down of Hsc82 (yeast variant of Hsp90) full length and domains. Lanes: 1, MW marker (kDa); 2, N-terminal domain; 3, middle domain; 4, C-terminal domain; 5, N-terminal and middle domain combined; and 6, full length.
The observed effect of San A-amide on FKBP52 binding provides further support for an allosteric mechanism of inhibition. FKBP cochaperones are tetratricopeptide (TPR) repeat-containing proteins that bind to the MEEVD sequence at the very C terminus of Hsp90. Because the C terminus is disordered in the full-length Hsp90 crystal structures, it is unlikely that San A-amide directly blocks this interaction. Instead, drug-induced alteration of Hsp90 conformational equilibrium likely disrupts FKBP-binding sites remote from the TPR−MEEVD interaction. Importantly, inhibition of FKBP's binding to Hsp90 by perturbing TPR interactions has been linked to programmed cell death.37 Indeed, it is thought that several FKBPs may be involved in multiple cell death mechanisms, one of which acts via the Bcl-2 pathway.39
Thus, San A-amide appears to influence a unique subset of cancer-related pathways by selectively disrupting the binding of C-terminal binding proteins, both clients and cochaperones of Hsp90. By being able to block both Hsp90−IP6K2 and Hsp90−FKBP interactions, where Novobiocin only disrupts Hsp90−IP6K2 and not FKBP binding, San A-amide may provide multiple mechanisms for activating apoptotic pathways, thereby increasing its therapeutic potential. Furthermore, we show that San A-amide is over 100-fold more potent at inhibiting C-terminal client proteins than Novobiocin. Thus, San A-amide offers a potentially efficient means of targeting drug-resistant cancers, where its intriguing multipronged mechanism of action will be further assessed. San A-amide's ability to inhibit other Hsp90−cochaperone and Hsp90−client protein interactions both in vitro and in vivo is under investigation.
Acknowledgments
We thank Prof. Blagg for helpful discussions and Prof. Solomon Snyder and Anutosh Chakraborthy for providing materials used in this study.
Supporting Information Available
Full experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
R.C.V. contributed to the San A-amide-Biotin pull down assay (Figure 2), IP6K2-Hsp90 binding assay in vitro (Figure 3a), IP6K2 over expression and cytotoxicity in vivo (Figure 4), San A-amide-Biotin (inactive compound) pull down (Figure S9); organized and put together the supplemental data; and made minor writing and editing contributions. R.A.R. contributed to the synthesis of San A-amide-Biotin (Figure 1) and the protocol for San A-amide-Biotin pull down assay (Figure 2) and made significant writing and editing contributions. C.N.C. contributed to the San A-amide-Biotin pull down assay (Figure 5), ATPase activity assay (Figure S7), and San A-amide-Biotin pull down E. coli and Yeast Hsp90 (Figure S1). V.C.A. contributed to the Her2 and FKBP binding assay with San A-amide (Figure 3a), Her2 and FKBP binding assay with 17AAG (Figure 3b), and San A-amide-Biotin (inactive compound) pull down (Figure S9) and made significant writing and editing contributions. D.A.A. was the overseeing PI of C.N.C., was a collaborator of the McAlpine lab, and provided significant intellectual input. S.R.M. was the main PI of entire project; was the overseeing PI of R.C.V., R.A.R., and V.C.A.; made significant writing and editing contributions; and provided significant intellectual input.
We thank San Diego State University, the Frasch Foundation (658-HF07), CSUPERB, NIH 1U54CA132379-01A1 and 1R01CA137873 (support of S.R.M. and V.A.), NIH/NIGMS SDSU MARC 5T34GM08303 and NIH MIRT (support of R.A.R.), NIW T90DK07015 (support of R.C.V.), the Howell Foundation (support of R.A.R and R.C.V.) and HHMI (support of C.N.C. and D.A.A.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Cueto M.; Jensen P. R.; Fenical W. N-Methylsansalvamide, a cytotoxic cyclic depsipeptide from a marine fungus of the genus Fusarium. Phytochemistry 2000, 55, 223–226. [DOI] [PubMed] [Google Scholar]
- Belofsky G. N.; Jensen P. R.; Fenical W. Sansalvamide: A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999, 40, 2913–2916. [Google Scholar]
- Hwang Y.; Rowley D.; Rhodes D.; Gertsch J.; Fenical W.; Bushman F. Mechanism of Inhibition of a Poxvirus Topoisomerase by the Marine Natural Product Sansalvamide A. Mol. Pharmacol. 1999, 55, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Otrubova K.; Lushington G. H.; Vander Velde D.; McGuire K. L.; McAlpine S. R. A Comprehensive Study of Sansalvamide A Derivatives and Their Structure-Activity Relationships against Drug-Resistant Colon Cancer Cell Lines. J. Med. Chem. 2008, 51, 530–544. [DOI] [PubMed] [Google Scholar]
- Pan P. S.; Vasko R. C.; Lapera S. A.; Johnson V. A.; Sellers R. P.; Lin C.-C.; Pan C.-M.; Davis M. R.; Ardi V. C.; McAlpine S. R. A comprehensive study of Sansalvamide A derivatives: Their structure-activity relationships and their binding mode to Hsp90. Bioorg. Med. Chem. 2009, 17, 5806–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Gu W.; Lo D.; Ding X.-Z.; Ujiki M.; Adrian T. E.; Soff G. A.; Silverman R. B. N-Methylsansalvamide A peptide Analogues. Potent New Antitumor Agents. J. Med. Chem. 2005, 48, 3630–3638. [DOI] [PubMed] [Google Scholar]
- Ujiki M.; Milam B.; Ding X.-Z.; Roginsky A. B.; Salabat M. R.; Talamonti M. S.; Bell R. H.; Gu W.; Silverman R. B.; Adrian T. E. A novel peptide sansalvamide A analogue inhibits pancreatic cancer cell growth through G0/G1 cell-cycle arrest. Biochem. Biophys. Res. Commun. 2006, 340, 1224–1228. [DOI] [PubMed] [Google Scholar]
- Otrubova K.; Styers T. J.; Pan P.-S.; Rodriguez R.; McGuire K. L.; McAlpine S. R. Synthesis and Novel Structure-Activity Relationships of Potent Sansalvamide A Derivatives. Chem. Commun. 2006, 1033–1034. [DOI] [PubMed] [Google Scholar]
- Rodriguez R. A.; Pan P. S.; Vasko R. C.; Pan C. M.; Disman W.; McAlpine S. R. Structure-activity relationships of Sansalvamide A derivatives and their mechanism of action in the pancreatic cancer cell line PL-45. J. Mex. Chem. Soc. 2008, 52, 201–211. [Google Scholar]
- Sanchez E. R.; Redmond T.; Scherrer L. C.; Bresnick E. H.; Welsh M. J.; Pratt W. B. Evidence that the 90 kDa heat shock protein is associated with tubulin-containing complexes in L cell cytotsol and in intack PtK cells. Mol. Endocrinol. 1988, 756–760. [DOI] [PubMed] [Google Scholar]
- Koyasu S.; Nishida E.; Kadowaki T.; Matsuzaki F.; LIida K.; Harada F.; Kasuga M.; Sakai H.; Yahara I. Two mammalian heat shock proteins, Hsp90 and Hsp100, are actin-binding proteins. Proc Natl. Acad. Sci. 1986, 83, 8054–8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostinis Y.; Theodoropoylos P. A.; Gravanis A.; Stournaras C. Heat shock protein Hsp90 and its association wit the cytoskeleton: A morphological study. Biochem. Cell Biol. 1992, 70, 779–786. [DOI] [PubMed] [Google Scholar]
- Barral J. M.; JHutagalung A. H.; Brinker A.; Hartl F. U.; Epstein H. F. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 2002, 295, 669–671. [DOI] [PubMed] [Google Scholar]
- Pratt W. B.; Toft D. O. Regulation of signaling protein function and trafficking by the Hsp90/Hsp70- based chaperone machinery. Exp. Biol. Med. 2003, 228, 111–133. [DOI] [PubMed] [Google Scholar]
- Neckers L. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol. Med. 2002, 8, S55–S61. [DOI] [PubMed] [Google Scholar]
- Neckers L.; Ivy S. P. Heat shock protein 90. Curr. Opin. Oncol. 2003, 15, 419–424. [DOI] [PubMed] [Google Scholar]
- Maruya M.; Sameshima M.; Nemoto T.; Yahara I. Monomer arrangement in Hsp90 dimer as determined by decoration with N and C-terminal region soecific antibodies. J. Mol. Biol. 1999, 285, 903–907. [DOI] [PubMed] [Google Scholar]
- Nemoto T.; Ohara-Nemoto Y.; Ota M.; Takagi T.; Yokoyama K. Mechanism of dimer formation of the 90-kDa heat shock protein. Eur. J. Biochem. 1995, 233, 1–8. [DOI] [PubMed] [Google Scholar]
- Fang L.; Ricketson D.; Getubig L.; Darimont B. Unliganded and hormone-bound glucocorticoid receptors interact with distinct hydrophobic sites in the Hsp90 C-terminal domain. Proc. Natl. Acad. Sci. 2006, 103, 18487–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C.; Morimoto R. I. Role of the heat shock protein response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [DOI] [PubMed] [Google Scholar]
- Chiosis G.; Huezo H.; Rosen N.; Mimgaugh E.; Whitesell L.; Neckers L. Binding affinity and potent cell activity—Finding an explanation. Mol. Cancer Ther. 2003, 2, 123–129. [PubMed] [Google Scholar]
- Hollingshead M. G.; Alley M.; Burger A. M.; Borgel S.; Pacula-Cox C.; Fiebig H.-H.; Sausville E. A. In vivo antitumor efficacyof17-DMAG(17-dimenthylaminoethylamino-17-demthoxygeldanamycin hydrochloride), a water-soluble geldanamycin derivative. Cancer Chemother. Pharmacol. 2005, 56, 115–125. [DOI] [PubMed] [Google Scholar]
- Senju M.; Sueoka N.; Sato A.; Iwanaga K.; Sakao Y.; Tomimitsu S.; Tominaga M.; Irie K.; Hayashi S.; Sueoka E. Hsp90 inhibitors cause G2/M arrest associated with the reduction of Cdc25C and Cdc2 in lung cancer cell lines. J. Cancer Res. Clin. Oncol. 2006, 132, 150–158. [DOI] [PubMed] [Google Scholar]
- Chang Y.-S.; Lee L.-C.; Sun F.-C.; Chao C.-C.; Fu H.-W.; Lai Y.-K. Involvement of calcium in the differential induction of heat shock protein 70 by heat shock protein 90 inhibitors, Geldanamycin and Radicicol, in human non-small cell lung cancer H460 cells. J. Cell. Biochem. 2006, 97, 156–165. [DOI] [PubMed] [Google Scholar]
- Matei D.; Satpathy M.; Cao L.; Lai Y.-K.; Nakshatri H.; Donner D. B. The platelet-derived growth factor receptor alpha is destabilized by geldanamycins in cancer cells. J. Biol. Chem. 2007, 282, 445–453. [DOI] [PubMed] [Google Scholar]
- Usmani S. Z.; Bona R.; Li Z. H. 17 AAG for HSP90 inhibition in cancer—From bench to bedside. Curr. Mol. Med. 2009, 9, 654–664. [DOI] [PubMed] [Google Scholar]
- Koga F.; Kihara K.; Neckers L. Inhibition of cancer invasion and metastasis by targeting the molecular chaperone heat-shock protein 90. Anticancer Res. 2009, 29, 797–807. [PubMed] [Google Scholar]
- Barginear M. F.; Van Poznak C.; Rosen N.; Miodi S.; Hudis C. A.; Budman D. R. The heat shock protein 90 chaperone complex: An evolving therapeutic target. Curr. Cancer Drug Targets 2008, 8, 522–535. [DOI] [PubMed] [Google Scholar]
- Chakraborty A.; Koldobskiy M. A.; Sixt K. M.; Juluri K.; Mustafa A. K.; Snowman A. M.; van Rossum D. B.; Patterson R. L.; Snyder S. H. Hsp90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. 2008, 105, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly A.; Blagg B. S. J. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr. Med. Chem. 2008, 15, 2702–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu M. G.; Schulte T. W.; Neckers L. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J. Natl. Cancer Inst. 2009, 92, 242–248. [DOI] [PubMed] [Google Scholar]
- Heath E. I.; Hillman D. W.; Vaishampayan U.; Sheng S.; Sarkar F.; Harper F.; Gaskins M.; Pitot H. C.; Tan W.; Ivy S. P.; Pili R.; Carducci M. A.; Erlichman C.; Liu G. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin. Cancer Res. 2008, 14, 7940–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit D. B.; Osman I.; Polsky D.; Panageas K. S.; Daud A.; Goydos J. S.; Teitcher J.; Wolchok J. D.; Germino F. J.; Krown S. E.; Coit D.; Rosen N.; Chapman P. B. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin. Cancer Res. 2008, 14, 8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A.; Gan J.; Mosesson Y.; Vereb G.; Szollosi J.; Yarden Y. Hsp90 restrains ErbB-2/HER2 signalling by limiting heterodimer formation. EMBO J. 2004, 5, 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Mimnaugh E.; Rosser M. F.; Nicchitta C.; Marcu M.; Yarden Y.; Neckers L. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J. Biol. Chem. 2001, 276, 3702–3708. [DOI] [PubMed] [Google Scholar]
- Chen S. Y.; Sullivan W. P.; Toft D. O.; Smith D. F. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones 1998, 3, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A.; Thao L.; Sensintaffar J.; Zhang L.; Boehm M. F.; Fritz L. C.; Burrows F. J. A high- affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407–410. [DOI] [PubMed] [Google Scholar]
- Shames D. S.; Minna J. D. IP6K2 is a client for Hsp90 and a target for cancer therapeutics development. Proc. Natl. Acad. Sci. 2008, 105, 1389–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.