Abstract
Motivation: Complex patterns of protein phosphorylation mediate many cellular processes. Tandem mass spectrometry (MS/MS) is a powerful tool for identifying these post-translational modifications. In high-throughput experiments, mass spectrometry database search engines, such as MASCOT provide a ranked list of peptide identifications based on hundreds of thousands of MS/MS spectra obtained in a mass spectrometry experiment. These search results are not in themselves sufficient for confident assignment of phosphorylation sites as identification of characteristic mass differences requires time-consuming manual assessment of the spectra by an experienced analyst. The time required for manual assessment has previously rendered high-throughput confident assignment of phosphorylation sites challenging.
Results: We have developed a knowledge base of criteria, which replicate expert assessment, allowing more than half of cases to be automatically validated and site assignments verified with a high degree of confidence. This was assessed by comparing automated spectral interpretation with careful manual examination of the assignments for 501 peptides above the 1% false discovery rate (FDR) threshold corresponding to 259 putative phosphorylation sites in 74 proteins of the Trypanosoma brucei proteome. Despite this stringent approach, we are able to validate 80 of the 91 phosphorylation sites (88%) positively identified by manual examination of the spectra used for the MASCOT searches with a FDR < 15%.
Conclusions:High-throughput computational analysis can provide a viable second stage validation of primary mass spectrometry database search results. Such validation gives rapid access to a systems level overview of protein phosphorylation in the experiment under investigation.
Availability: A GPL licensed software implementation in Perl for analysis and spectrum annotation is available in the supplementary material and a web server can be assessed online at http://www.compbio.dundee.ac.uk/prophossi
Contact: d.m.a.martin@dundee.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Protein phosphorylation is regarded as a key mechanism for the regulation of many cellular processes including metabolism, cell division and apoptosis (Cohen, 2000). It has been estimated that >50% of expressed proteins are phosphorylated at some point in their life cycle (Hjerrild and Gammeltoft, 2006) though only a small fraction of the potential phosphorylation sites have been identified.
In recent years, the examination of complex protein mixtures by tandem mass spectrometry (MS/MS) has become feasible through advances in instrumentation and computational methodologies (Cox and Mann, 2007). Peptide and protein analysis at the cell extract level has become an almost routine procedure as algorithms such as MASCOT (Perkins et al., 1999) or SEQUEST (Yates et al., 1995) among others allow rapid identification of proteins through matching tandem mass spectra to sequence databases.
Statistical methods have been developed to enable the validity of peptide observations to be assessed. Search strategies such as the inclusion of reversed or scrambled sequences in the database can give an estimate of the likely accuracy, or false discovery rate (FDR), for peptide identifications with respect to the database search engine score for a particular experiment (Elias and Gygi, 2007; Kall et al., 2008). This allows a reasonably robust description of the protein species in a particular sample. Further work has been performed to match peptide fragmentation patterns to peptide sequences through machine learning techniques such as hybrid support vector machines/Bayesian networks (Klammer et al., 2008) and decision trees (Elias et al., 2004) but these are not yet readily applicable to peptides containing post-translational modifications (PTMs).
Although protein and peptide analysis is now a mainstream high-throughput technique, reliable identification of PTMs remains a specialist area. Algorithms designed for peptide database matching can take PTMs into account but are not designed to provide robust identifications. PTMs are often present only in low stoichiometry and ion signals corresponding to phosphopeptides tend to be suppressed in the presence of non-phosphorylated peptides. The low stoichiometry of PTMs requires the use of peptide enrichment approaches such as affinity pull down (Gronborg et al., 2002), immobilized metal affinity chromatography (Andersson and Porath, 1986; Stensballe et al., 2001) or titanium dioxide columns (Pinkse et al., 2004) to enrich the phosphopeptide complement of a complex mixture.
The peptide enrichment strategies required for PTM identification in complex mixtures compromise the scoring assumptions upon which generic database search strategies are based, rendering it unreliable to take the database search scores as a surrogate for PTM identification accuracy. Consequently, confident identification of PTMs requires the manual inspection of the raw MS/MS spectra by a competent mass spectrometrist, a time-consuming step that acts as a major bottleneck in global PTM analyses where many thousands of spectra may require analysis. This examination requires interpretation of the fragmentation pattern in line with the experience of the scientist, and comparison of multiple interpretations of the data, each of which could be correct if the sample contains multiple isobaric isoforms of the phosphopeptide.
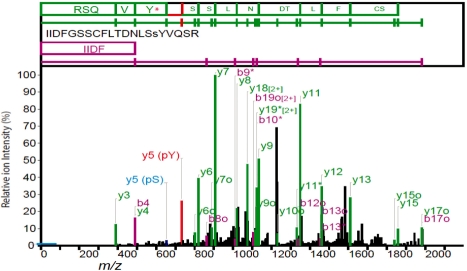
An example is shown in Figure 1 where an experienced mass spectrometrist identifies the phosphoTyrosine (Rank 2 hit) as a better interpretation than the phosphoSerine (top-ranked hit). Full Prophossi reports for these two peptide–spectrum matches (PSMs) are available in the Supplementary Material.
Fig. 1.
An example of misidentification of the correct phosphorylation site. MASCOT identifies the phosphorylation site as pS16 (peptide score 118), though the expected y5-98 ion is much weaker than the weak y5 ion (blue). The second ranked hit (pY17, score 102) is preferred by the experienced analyst with a strong y5 ion match (red) giving a continuous y-ion ladder. The spectrum was annotated with Prophossi and modified. The threshold for ion inclusion is indicated by a blue bar on the y-axis.
This issue has been approached by two other groups, both of whom apply a post-processing step to standard peptide MS/MS search engine results. Beausoleil et al. (2006) report a probabilistic method that calculates an Ascore based on the appearance of phosphorylation sites in multiple candidate solutions to the spectrum-database mapping conundrum. They calculate a probability score based on the appearance of site-determining ions, i.e. those fragment ions that would be specific for a particular phosphopeptide isoform. The method is dependent on the SEQUEST search engine to identify the two top phospho-isoform hits and it reports quality data only for the best phospho-isoform hit. It does not consider the possibility of isobaric phospho-isoform mixtures. The closed source implementation of the software prevents user optimization of the search parameters. Smith et al. (2007) have taken a different approach which is, in some respects, similar to ours. They examine the daughter ion spectrum and assign scores based on a limited range of spectral features. Peptide matches failing to reach a defined score threshold are rejected. Both of the aforementioned methods have some limitations: The Beausoleil method ignores spectral features beyond the site-determining ions and does not consider neutral loss of phosphate from phosphoSerine and phosphoThreonine, resulting in a smaller number of phosphopeptide spectrum matches. The Smith method assigns scores from a limited range of features, which alone may not be sufficient to specifically locate a phosphorylation site, and has not been tested empirically.
Our methodology, described in this article, incorporates a broader range of spectral features and seeks to identify evidence for the specific localization of the putative phosphorylation sites. Thus, every PSM, i.e. every hit from a MASCOT or other search engine search, is assessed on its own merit. The method is, therefore, able to interpret complex spectra derived from multiple isobaric phosphopeptide species. Opinions from three experienced mass spectrometrists were used to derive a set of chemistry-based criteria that could be applied to tandem mass spectra for selection between, and validation of, the database PSM search hits. This method does not perform database searches itself but provides a report on how well the observed spectrum fits to the predicted matches, and whether the predicted match passes these analytical criteria. As such, it can be applied to the results of any such database search.
Typically, relatively few phosphopeptide spectra are observed in proteomics experiments in the absence of specific phosphopeptide enrichment protocols and this low coverage can be treated by hand by an experienced analyst. However, when such enrichment methods are applied, such as in the experiments described here, the proportion of spectra arising from phosphopeptides rapidly expands to a level where automated processing tools are a practical necessity. Our aim was to develop tools that automate rapid processing of large numbers of spectra with few falsely identified phosphorylation sites (high selectivity) and a sufficiently good sensitivity to provide significant coverage. As we are examining proteomes, where little is known about the existing phosphorylation state of the organism, a tool that rapidly and confidently assigns the majority of easy cases is a considerable boost to productivity. All database hits can be assessed, and positive results reported. As all the criteria can be explicitly described in English, marginal hits can be also examined rapidly by an experienced analyst with an appropriate visualization tool. Additionally, a full text report that highlights salient features and annotates the spectrum can be generated. This approach has been validated through assessment of the automated annotation of the Trypanosoma brucei phosphoproteome (Nett et al., 2009b). We manually examined all identified hits for a specific family of proteins (the protein kinases) and examined the error rate and bias in our automated processing. Our method runs rapidly, allows the assessment of more than just the top hit and gives excellent selectivity with good sensitivity. Output can be via an annotated spectrum and report, produced in HTML or PDF, or via a software application programming interface, allowing integration of the analysis in a high-throughput analysis pipeline. Several of our predictions of occupied T.brucei phosphoTyrosine sites have been validated experimentally by both western blot and immunofluorescence microscopy experiments using two well-characterized anti-phosphoTyrosine antibodies (Nett et al., 2009a).
A public web server has been made available at http://www.compbio.dundee.ac.uk/prophossi for individual researchers to examine their peptide spectra online.
2 METHODS
2.1 Phosphopeptide samples and mass spectra
The generation of the T.brucei phosphopeptide mixture and its separation, and analysis have been described previously (Nett et al., 2009b). In summary, the cytosolic fraction of a T.brucei culture was obtained, digested with trypsin and phosphopeptides enriched though a combination of strong cation exchange and titanium dioxide chromatography as described by Olsen et al. (2006). Mass spectrometry was performed using a Q-Star XL mass spectrometer (Applied Biosystems) and an LTQ-Orbitrap (Thermo Electron) both equipped with a nanospray ionisation source. The Q-Star XL mass spectrometer was operated in a data-dependent mode, which consisted of a MS survey scan for 1 s (m/z 400–2000) followed by four 2 s MS/MS scans of the four most intense doubly or triply charged ions (m/z 60–1800) exceeding 10 counts. In the LTQ-Orbitrap mass spectrometer, a survey scan was performed over a split mass range (m/z 300–800 and 800–2000) in the Orbitrap analyser each of them triggering five MS2 LTQ acquisitions of the five most intense ions using multistage activation on the neutral loss of 98, 49 and 32.33 Da. Orbitrap mass spectra were processed with Analyst 1.4 software (Applied Biosystems). Q-star mass spectra were processed with Analyst QS 1.1 software (Applied Biosystems) and centroided and deisotoped peak list files of the SCX–TiO2 experiments were concatenated using the MASCOT daemon engine (Matrix Science, London, UK) for Q-Star XL spectra. Raw files obtained from the LTQ-Orbitrap were converted to MASCOT generic files using Raw2msm software (gift from Prof. Matthias Mann, Max Planck Institute for Biochemistry, Munich) before merging into a single file.
2.2 Database searches
A composite T.brucei database containing all predicted proteins from the genome sequencing projects for T.brucei strains 427 and 497 (downloaded from GeneDB) and all T.brucei peptides in UniProt was created and curated to remove redundant sequences. A decoy dataset containing the reversed sequence of all remaining sequences was generated and appended to the forward dataset. MASCOT (version 2.1, Matrix Science, London, UK) searches were performed on a 4-node cluster using a parent/daughter ion mass accuracy of 0.1 Da (Q-Star) or a parent ion mass accuracy of 10 ppm and daughter ion mass accuracy of 1 Da (Orbitrap). Searches were performed using trypsin as the digestion enzyme, carboxyamidomethylation of cysteine as a fixed modification and with the oxidation of methionine and phosphorylation of serine, threonine or tyrosine classed as variable modifications. The data were searched against the composite and decoy databases described above. MASCOT database search results were processed with custom Perl scripts using the MASCOT developer's toolkit (Matrix Science, London, UK). Large results such as those generated by these kinds of experiments are extremely resource-demanding to view through the MASCOT web interface, so an intermediate relational database (MySQL) was created, the MASCOT Large Results Viewer (MLRV), in which the peptide, protein, modification and search hit information could be stored in a readily queryable manner. MGF files were transformed to DTA files using custom Perl scripts and SEQUEST searches performed on the same sequence database through the TransProteomicsPipeline (TPP) software (Pedrioli, 2010). A maximum of 10 top hits were retained for each spectrum and imported into a custom PostgreSQL database, the SEQUEST Large Results Viewer (SLRV). TPP was also used to post-process each search with PeptideProphet, which provides a score for the top hit for each spectrum query. These data were incorporated into SLRV with custom Perl scripts. Standard SEQUEST parameters were used with peptide mass tolerance = 10 and peptide mass units = 2. PSMs were correlated between SEQUEST/Peptide Prophet and MASCOT output with custom Perl scripts to enable comparison of search methods.
2.3 Spectrum analysis criteria definition
Three experienced mass spectrometrists were observed and interviewed as they manually assessed peptide–spectrum matches. The processes by which they accumulated evidence were noted and formalized as a set of analytical criteria, an assessment of whether it was always applicable, whether it was only applicable to certain peptides or whether it was only applicable to certain types of spectra. Criteria of the first two types were identified and coded as Perl modules for inclusion in the data management infrastructure. It was considered too problematic in this first study to selectively apply criteria of the third type as this would require identifying which spectra these criteria would be applied to. These, typically more difficult cases, remain at present the domain of the mass spectrometry professional. The criteria derived are listed in Table 1. These include an examination of specific proline-directed cleavage products (Breci et al., 2003).
Table 1.
Analysis Criteria for automated validation of PSMs
| Prefilter criteria | ||
| P1 | Forward hit | Only hits against forward, non-redundant sequences are selected |
| P2 | Mass accuracy | Only hits within 0.1 Da (or 0.1 + 1 Da) of the parent ion m/z are selected. |
| P3 | Phospho-PTM | Only hits containing a putative Phospho PTM are selected |
| P4 | Within 20 points | Only hits which are within 20 MASCOT score points of the top ranked hit for that query are selected. |
| P5 | Over FDR threshold | Only peptides with a MASCOT score over the calculated FDR 1% threshold are selected. |
| Validation Criteria—Phosphopeptide assignment | ||
| 1 | 4 in a row | At least four sequential y- or b-series ions are present. This indicates good coverage of the peptide. |
| 2 | 5 of 6 | 5 out of 6 sequential b- or y-series ions are present. This indicates good coverage of the peptide. |
| 3 | 3 desphospho ions | At least three y- or b-series ions with a phosphate loss are present. The phosphate ester bond tends to be more labile than the peptide bond. |
| 4 | Proline-directed fragmentation | The imino bond to the N-terminal side of a proline residue is particularly labile. If the sequence contains a Proline residue then at least one of the imino bonds should give a fragment ion with at least 50% maximum intensity and much stronger than the relatively weakly cleaved amide bond C-terminal to Proline. |
| 5 | 6 of top 10 ions | 6 of the 10 most intense ions should be assigned to y- or b-series ions. |
| Validation criteria—phosphosite assignment | ||
| 6 | Phosphate transitions | To assign the site specifically, at least one ion unique to that peptide species must be observed. This is aided by the high rate of phosphate loss from pSer and pThr residues. |
| 7 | PhosphoTyrosine | Mass differences corresponding to pTyr should be observed between identified peaks. |
With criteria defined, a system for appropriate Boolean combination of the criteria was devised such that an unambiguous validation could be ascertained. The quality assessment criteria listed in Table 1 are combined as follows. All PSMs are subject to a prefilter (a) where they must meet all criteria. Following ion series matching the criteria in section (b) are applied. PSMs must match each of the following set of criteria: either four sequential ions or five ions out of a series of six must match; At least three des-phospho ions must be observed for sequences containing phosphoSerine and phosphoThreonine; if the peptide contains proline, then a strong proline-directed fragmentation ion must be observed; at least 6 of the most intense 10 ions must be positively assigned. For phosphosite identification in section (c), specific mass transitions corresponding to phosphoTyrosine must be observed, and sufficient ions to unambiguously identify the phosphosite. Typically, this would require an ion derived from cleavage between any phosphosite candidates, including residues not identified as potentially phosphorylated by MASCOT.
2.4 Automatic hit validation method
For each database search, a list of all queries, each corresponding to a specific parent ion and with PSMs meeting the prefilter quality criteria, was obtained from the MLRV database. For each dataset, the PSMs were prefiltered to exclude matches with an absolute delta mass >0.1 Da (or 1 ± 0.1 Da for Q-Star data), include only PSMs with phospho modifications and only PSMs that were within 20 MASCOT score points of the highest scoring PSM for that query spectrum. The proportion of reverse sequences identified with respect to forward sequences was plotted against the MASCOT score threshold and the FDR threshold at n% (FDRn) determined as the lowest MASCOT score where reverse sequences comprise <n% of the total (filtered) peptide species identified. FDR results for each dataset are summarized in Supplementary Figure S2. For the purpose of this analysis, we restricted further investigation to peptides with a MASCOT score greater than FDR1.
Each query was then processed as follows: the peak list corresponding to the database search query (parent ion m/z and the related daughter m/z peaks) was read from the peak list data file. This is referred to as the observed spectrum. Each PSM for that query that corresponded to predetermined quality standards was retrieved from the MLRV database. A synthetic spectrum was generated for the peptide in question (the theoretical spectrum) by applying simple fragmentation rules and this was compared to the observed spectrum as a spectrum match. A customizable threshold was used to exclude peaks that may be due to noise. In this study, we excluded any peak with an intensity <5% of the most intense peak observed. This could be examined via a web-based visualization tool with matching ions both labelled on the observed spectrum and tabulated. This process is described in Figure 2. Each rule was then applied to the spectrum match. The results were then combined according to a predetermined Boolean system and the results (pass/fail) stored in a database. In addition, a second layer of criteria was used to determine whether the specific phosphorylation site(s) could be confirmed from the spectral data.
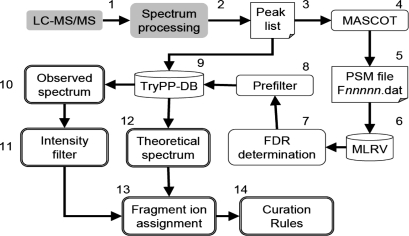
Fig. 2.
Workflow for automated annotation of phosphosites. Experimental LC–MS/MS data is gathered (1) and processed using platform specific software (2) to give a generic peak list file (3). This file, is used as the input to MASCOT (4), which generates a results file (5) containing all the PSMs. This file is parsed into the MLRV relational database (6) and the FDR for the search determined (7). A PSM-quality prefilter is applied (8) and suitable PSMs are exported to the TryPP-DB (9) where they are linked to the source peak list file used for the search. The observed MS/MS spectrum is extracted from the peak list file (10), filtered by an intensity threshold (11) and compared with a calculated fragmentation spectrum (12) for the peptide under examination. Observed ions are assigned to series (13) allowing the curation rules to be applied (14).
2.5 Verification of the validation method
All 501 PSMs from 74 protein kinase sequences, containing 259 putative phosphorylation sites, were examined manually by an experienced mass spectrometrist to provide a ‘gold standard’ reference against which the automated validation method could be evaluated. Detailed statistics for this peptide set are shown in Table 2. All spectra were classified as being either sufficient or insufficient to verify the presence of the phosphopeptide (i.e. pass/fail, the phosphopeptide validation). In addition, the ability to unambiguously identify the precise phosphorylation site was recorded (pass/fail, phosphosite validation), allowing the determination of true positive and false positive rates for the automated analysis.
Table 2.
Kinase dataset statistics
| Site Observations | Unique Sites | Unique Proteins | Peptides Observed | |
|---|---|---|---|---|
| All peptides | 643 | 259 | 74 | 501 |
| Rank 1 and 2 | 449 | 213 | 72 | 355 |
| Rank 1 | 289 | 159 | 72 | 230 |
The subset of peptides from both Orbitrap and Q-Star experiments that were annotated both manually and by the automated system are described. An observed peptide is a single PSM. A PSM may contain more than one observed site. Each protein may contain many PSMs. Each site may be observed in many peptide observations.
3 RESULTS
3.1 Comparing search engine hit score, manual and automated validation
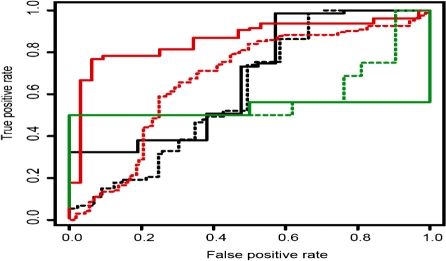
The relationship between search engine score and manual curation was investigated. All PSMs with a score over 1% FDR to a protein from the kinase set which had been evaluated manually were ranked by score. For each search engine score, the false positive rate (PSMs not confirmed after manual curation) and true positive rate (PSMs confirmed after manual curation) were determined. Similar rates were determined for the subset of matches automatically curated as positive by ProPhosSI. ProPhosSI-curated peptides correlate significantly better with manual curation than the search engine-ranked sets (Fig. 3). Performance with the SEQUEST-ranked hits is not as good as with MASCOT, due in part to the differing responses of the search engines and ProPhosSI to phosphate neutral loss. Neutral loss appears to be dependent on local peptide sequence, though no substantial datasets are yet available to model this appropriately. SEQUEST matches are, therefore, biased towards sequences where the phosphate is less labile, and ProPhosSI, in this current implementation, towards those where sites are labile.
Fig. 3.
All manually curated peptide–spectrum matches containing at least one phosphorylated residue were ordered according to their MASCOT (red), SEQUEST (black) or SEQUEST + Peptide Prophet (green) score. Dotted lines indicate the performance of all matches ordered by search engine score. Solid lines indicate the performance of the subset of matches positively curated by ProPhosSI. The increased area under the curve for the solid lines indicates better performance by ProPhosSI.
As SEQUEST only identifies about a third of the phosphopeptides found with MASCOT, and the Ascore software only considers the top SEQUEST hit, it is not possible to rigorously compare Ascore and ProPhosSI. However, using a set of 20 manually assessed PSMs, ProPhosSI and Ascore gave similar results, with 10 PSMs having an Ascore over 16 positively validated by ProPhosSI and 10 PSMs with an Ascore under 9 not confirmed by ProPhosSI.
When the automated validation rate is extrapolated over the whole peptide set, there only appears to be a relation between validation rate and match score at very high scores for MASCOT (ion score >90) and SEQUEST (Xcorr > 6.5 ), and not at all for Peptide Prophet (data not shown). Otherwise the validation rate (proportion curated as positive by ProPhosSI) remains constant at around 60% for scores > 1% FDR.
3.2 Automated validation
Spectra corresponding to 8231 PSMs with MASCOT scores over 1% FDR were assessed. Of these, 3718 were classified automatically as a validated phosphopeptide, with 1236 distinct validated phosphorylation sites. These data are shown broken down by machine type in Table 3.
Table 3.
Automated assignments
| Dataset | Phosphopeptides |
Phosphosites |
||
|---|---|---|---|---|
| Pass | Fail | Pass | Fail | |
| Orbitrap | 1617 | 1992 | 557 | 252 |
| Q-Star | 2101 | 2521 | 939 | 456 |
Automated assignment to the dataset by the methodology described. A single verified site observation at any peptide rank which met the appropriate quality criteria was considered sufficient to call as a phosphosite.
3.3 Verification of validation
Of the ‘gold standard’ manually curated peptide hits, 161/230 (70.0%) were identified with the automated system. Further, 16.1% of peptide hits automatically annotated as positive were not identified as such by manual curation of the spectra (a selectivity of 83.9%). A summary of the validation results broken down by instrument type is shown in Table 4 (Orbitrap and Q-Star, all PSM).
Table 4.
Automated versus manual peptide and phosphosite assignments
| Automated Curation with ProPhosSI |
||||
|---|---|---|---|---|
| Phosphopeptides |
Phosphosites |
|||
| Pass | Fail | Pass | Fail | |
| Orbitrap (all PSM) | ||||
| Manual Curation | ||||
| Pass | 60 | 17 | 41 | 5 |
| Fail | 12 | 79 | 6 | 13 |
| Q-Star (all PSM) | ||||
| Manual Curation | ||||
| Pass | 101 | 52 | 69 | 17 |
| Fail | 19 | 161 | 17 | 33 |
| Orbitrap (Rank 1 PSM) | ||||
| Manual Curation | ||||
| Pass | 32 | 5 | 31 | 3 |
| Fail | 6 | 37 | 2 | 2 |
| Q-star (Rank 1 PSM) | ||||
| Manual curation | ||||
| Pass | 53 | 30 | 53 | 15 |
| Fail | 9 | 58 | 2 | 2 |
The results from independent manual curation were compared with results from the automated validation. Each individual PSM and site observation is considered for each experiment and additionally for the subset of data that only includes top ranked MASCOT PSMs.
Overall the criteria employed appear better suited to the data obtained from the Orbitrap than the Q-Star with a sensitivity of 77.9% versus 66.0% and comparable accuracies with selectivities of 83.3% and 84.2%, respectively. The overall Matthews correlation coefficient (MCC; Matthews, 1975), a metric which considers all elements of the confusion matrix, for the phosphopeptide assignment is 0.600 (0.652 and 0.576 for the Orbitrap and Q-Star, respectively).
Restricting the analysis to just first rank peptides [Table 4, Orbitrap and Q-Star (Rank 1 PSM)] gives little overall change with 85/118 peptides (sensitivity 70.8%) correctly validated and 15/100 incorrect validations (selectivity of 85%). The better performance on Orbitrap data is reflected in the comparison of individual phosphosite assignments by the manual curator and by the automated process. Over all ranked peptides meeting the prefilter quality criteria, the automated process positively validates 47 phosphosite observations in the Orbitrap data [Table 4, Orbitrap (all PSM)]. Manual assignment identifies a further five sites missed by the automated process (a sensitivity of 89.1%) and rejects six validated automatically (selectivity of 87.2%). Both methods reject 13 site observations for an overall MCC of 0.585. In the validation of the Q-Star data [Table 4, Q-Star (all PSM)], 86 site observations are validated automatically of which 17 are rejected by the manual curator (selectivity of 80.2%). The manual curator validates a further 17 sites (sensitivity of 80.2%) and both methods reject 33 site observations giving an MCC of 0.462.
Restricting the analysis to sites observed only in top-ranked peptides from the MASCOT search improves sensitivity and selectivity in both Orbitrap and Q-Star datasets (Selectivity: 93.9% and 96.4%, respectively; Sensitivity: 91.2% and 77.9%, respectively) though the MCC is considerably reduced to 0.374 and 0.151, respectively [Table 4, Orbitrap and Q-Star (Rank 1 PSM)], primarily due to the small number of true negatives.
Taking both Orbitrap and Q-Star data in aggregate, 94 non-redundant phosphorylation sites from the 74 protein kinases were identified automatically. From the 445 peptides examined manually, a further 11 sites were identified (a sensitivity of 88%) and 14 were assigned as negative, giving a selectivity of 85.1%. Twenty-four sites were rejected by both methods giving a MCC of 0.186 (Table 5, all sites). Confidence in a phosphosite assignment can be improved by requiring that at least two automated validated observations are required for verification. Sixty sites were identified with at least two validated observations, of which 8 had no manually validated observation, a selectivity of 86.7%, only a small increase over the single observation level but with a decrease of 35% in sensitivity (Table 5, Sites with two or more positive automatic assignments). Restricting analysis to just those PSMs that are in the top 2 ranks in the MASCOT search results reduces the number of putative phosphosites to 109 from 129. Of these sites, automated assignment validates 72 of the 85 manually validated sites (sensitivity 84.7%) with 8 assignments, which were not validated by any manual curation (selectivity 90.0%). Fourteen sites were rejected by both methods giving a MCC of 0.450.
Table 5.
Aggregated phosphosite assignments (by site)
| Automated assignment |
||
|---|---|---|
| Pass | Fail | |
| All sites (total 129) | ||
| Manual assignment | ||
| Pass | 80 | 11 |
| Fail | 14 | 24 |
| Sites with two or more positive automatic assignments (total 60 sites versus 129) | ||
| Manual assignment | ||
| Pass | 52 | 39 |
| Fail | 8 | 30 |
| Sites with Rank 2 peptides or higher (109 sites) | ||
| Manual assignment | ||
| Pass | 72 | 13 |
| Fail | 8 | 14 |
| Sites with Rank 1 peptides (80 sites) | ||
| Manual assignment | ||
| Pass | 62 | 14 |
| Fail | 3 | 1 |
Each unique site is considered, taking all observations of that site. If any observation of a phosphosite is validated in any experiment then that site is called as validated.
Further restriction to just top-ranked PSMs reduces the number of positively identified sites to 76, 83.5% of those manually curated from all PSMs. The automated validation identifies 62 correctly (sensitivity of 81.6%) and assigns a further 3 sites with no manual curation (selectivity of 95.4%). Only 1 site was rejected by all automated and manual PSM curations giving a MCC of 0.037. Taking into account the sites rejected at the phosphopeptide assignment level (80 sites), the MCC is recalculated as 0.792. This compares favourably with the MCC for all sites (141 rejected at the peptide level) of 0.593 but is almost identical to the MCC for all sites from Rank 2 or better peptides (120 rejected at the peptide level) of 0.793.
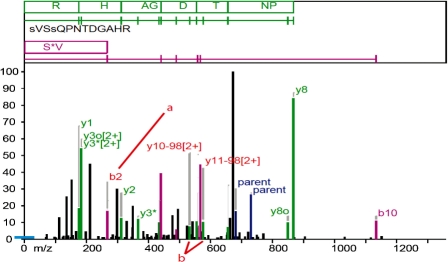
We examined a small number of high-scoring PSMs where ProPhosSI fails to validate an assignment made by an experienced analyst. ProPhosSI appears to be least effective in where the phosphate is not labile, is located on the N-terminal residue or product ions are multiply charged. An example is shown in Figure 4.
Fig. 4.
A manually verified PSM that ProPhosSI fails to validate. Many ion labels are not shown for clarity. Evidence for phosphorylation at S1 arises from the b2 ion (a). ProPhosSI requires ion transitions over a phosphosite and so requires more than one ion. Evidence for phosphorylation at S4 arises from the uniquely assigned des-phospho y10 [2+] ion. ProPhosSI does not consider 2+ ions as they can in many cases be assigned to more than one fragment.
4 CONCLUSIONS
Validation of phosphopeptide identifications by the examination of spectra has historically been a bottleneck in high-throughput phosphoproteomics. It may require many hours of careful cross-checking for each of many thousands of individual PSMs reported by a general database search algorithm such as MASCOT to establish whether it corresponds to a confident identification. We have demonstrated that the search engine score alone is an insufficient parameter for determining whether a phosphorylation site should be accepted. With high-throughput proteomics, this bottleneck becomes critical, precluding the use of phosphosite analysis as a routine screening tool. In this article, we have demonstrated a methodology which, by modelling the analysis and decision process of an experienced scientist, can substantially speed up validation of a large scale phosphoproteomics dataset by processing the data with 80–95% confidence in the positive validations and with a high sensitivity. Not only does it provide a massive speed benefit, allowing the processing of a complete phosphoproteomics experiment overnight, but it provides a report for all considered peptide hits, identifying the features it expects to see and reporting on them. This processing and visualization then provides a framework for an experienced analyst to manually curate the difficult cases or to re-examine those cases that may be interesting from a biological perspective.
We have been conservative in our application of the analysis criteria. Even a small decrease in the number of spectra, which must be validated manually, provides a boost to the researcher in terms of time gained. For this study, we estimate the time saved to be several person–months work on a typical whole cell phosphoproteome screen. It is essential, however, to reduce the number of false assignments to a minimum such that researchers making use of these assignments do not waste time chasing false leads. We have been conservative in our approach, resulting in a low error rate and, including curation at the phosphopeptide level, an exceptionally high MCC. Taking more stringent criteria, such as requiring more than one positive match to automatically call a site, provide a slight but measurable increase in accuracy with the downside that coverage is reduced. Combining multiple experiments from multiple machines should provide a wealth of data that can be combined to improve the overall coverage of phosphoproteome identification with minimal degradation in assignment quality.
In this study, we have demonstrated the utility of a methodology for automatically curating large-scale phosphoproteomics experiments. The principles behind the methods used in the study are simple and easy to comprehend. Access to this and similar methodologies should assist phosphoproteomics as a routine systems biology tool, allowing a deeper understanding of the essential role of phosphorylation in the function of the cell through rapid, global phosphoproteome analysis.
Supplementary Material
ACKNOWLEDGEMENTS
D.M.A.M. conceived the automated analysis system, designed and implemented the analysis framework and curation system. I.R.E.N. provided the experimental dataset and performed the manual curation of phosphopeptides. J.D.B. conceived and constructed the MLRV database. N.A.M. provided guidance on construction of the analytical rules and critical assessment of the software. M.A.J.F. provided guidance for the project and direction in the definition of the criteria used for assessment. F.V. performed the SEQUEST and Peptide Prophet analyses, and provided useful insight into these methods. This report was prepared by D.M.A.M. and refined in collaboration with the other authors. We would like to thank Dougie Lamont and Dr David Campbell for their insights into spectral interpretation, and Dr Tom Walsh for expert systems support.
Funding: Wellcome Trust (programme grant 085622 to M.A.J.F.; PhD studentship to I.R.E.N.); the Medical Research Council (to N.A.M. and F.V.) and the Biotechnology and Biology research Council IRColl RASOR project (to N.A.M. and F.V.).
Conflict of Interest: none declared.
REFERENCES
- Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Breci LA, et al. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal. Chem. 2003;75:1963–1971. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Elias JE, et al. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat. Biotechnol. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Gronborg M, et al. A mass spectrometry-based proteomic approach for identification of serine/threonine-phosphorylated proteins by enrichment with phospho-specific antibodies: identification of a novel protein, Frigg, as a protein kinase A substrate. Mol. Cell. Proteomics. 2002;1:517–527. doi: 10.1074/mcp.m200010-mcp200. [DOI] [PubMed] [Google Scholar]
- Hjerrild M, Gammeltoft S. Phosphoproteomics toolbox: computational biology, protein chemistry and mass spectrometry. FEBS lett. 2006;580:4764–4770. doi: 10.1016/j.febslet.2006.07.068. [DOI] [PubMed] [Google Scholar]
- Kall L, et al. Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- Klammer AA, et al. Modeling peptide fragmentation with dynamic Bayesian networks for peptide identification. Bioinformatics. 2008;24:i348–i356. doi: 10.1093/bioinformatics/btn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Nett IRE, et al. Trypanosoma brucei: identification and specific localization of tyrosine phosphorylated proteins. Eukaryotic Cell. 2009a;8:617–626. doi: 10.1128/EC.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett IRE, et al. The phosphoproteome of bloodstream form Trypanonosoma brucei, causative agent of African Sleeping Sickness. Mol. Cell. Proteomics. 2009b;8:1527–1538. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pedrioli PGA. Trans-proteomic pipeline: a pipeline for proteomic analysis. In: Simon JH, Andrew RJ, editors. Proteome Bioinformatics. Vol. 604. Totowa, NJ: Humana Press; 2010. pp. 213–238. chapter 15. [DOI] [PubMed] [Google Scholar]
- Perkins DN, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pinkse MW, et al. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- Stensballe A, et al. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;2:207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Smith JC, et al. A differential phosphoproteomic analysis of retinoic acid-treated P19 cells. J. Proteome Res. 2007;6:3174–3186. doi: 10.1021/pr070122r. [DOI] [PubMed] [Google Scholar]
- Yates JR, et al. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.