Abstract
Motivation: An observed metabolic response is the result of the coordinated activation and interaction between multiple genetic pathways. However, the complex structure of metabolism has meant that a compete understanding of which pathways are required to produce an observed metabolic response is not fully understood. In this article, we propose an approach that can identify the genetic pathways which dictate the response of metabolic network to specific experimental conditions.
Results: Our approach is a combination of probabilistic models for pathway ranking, clustering and classification. First, we use a non-parametric pathway extraction method to identify the most highly correlated paths through the metabolic network. We then extract the defining structure within these top-ranked pathways using both Markov clustering and classification algorithms. Furthermore, we define detailed node and edge annotations, which enable us to track each pathway, not only with respect to its genetic dependencies, but also allow for an analysis of the interacting reactions, compounds and KEGG sub-networks. We show that our approach identifies biologically meaningful pathways within two microarray expression datasets using entire KEGG metabolic networks.
Availability and implementation: An R package containing a full implementation of our proposed method is currently available from http://www.bic.kyoto-u.ac.jp/pathway/timhancock
Contact: timhancock@kuicr.kyoto-u.ac.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Coordinated gene expression along specific pathways determines which metabolic compounds can be synthesized and, therefore, can be used to infer the function of an entire network. Much of the networked structure of metabolism has already been identified and is readily available through databases such as KEGG (Kanehisa and Goto, 2000). These databases reveal that even for simple organisms, the complete metabolic network is large and highly complex. This mixture of network size and complexity is sufficient to hide the key pathways, which define the response of the metabolic network to external stimuli. Consequently, models of global metabolic networks are required to identify the specific pathways that are driving an observed metabolic response.
Metabolism of specific models such as network expansion (Handorf et al., 2005), flux balance analysis (FBA; Smolke, 2010) and constraints-based modeling (Price et al., 2003) model the chemical properties of metabolic networks. The simplest of these, network expansion, uses the natural limits imposed by the stoichiometry of each reaction to identify the scope of a each compound within the network. The goal of network expansion is to determine a minimum set of compounds required to fully reproduce the entire network. FBA and constraint-based models are explicit physical models of the chemistry that control metabolic networks. Physical models of metabolism explicitly model the flux through the reactions in order to predict the amount of each compound being produced during an experiment. Both physical models and network expansion approaches focus heavily on the chemical properties of the metabolic network and do not directly consider the genetic component of a metabolic network. Extensions to these approaches to include a genetic effect within model have been proposed (Ebenhoh and Liebermeister, 2006; Shlomi et al., 2008). However, these extensions are limited to binary representations of gene expression and do not consider relationships within the gene expression along pathways or within subnetworks.
A considerable amount of research has been undertaken to identify the genetic factors, which dictate the function of metabolic networks (Karp et al., 2010; Mlecnik et al., 2005). Of particular note are methods relating to gene set enrichment analysis (GSEA; Subramanian et al., 2005), which seek to identify known groups of genes that have a non-random response to an external stimulus. GSEA methods identify important groups of genes by first ranking all genes using test statistics such as t-tests or correlation coefficients, and then testing to see if specific groups are at the top or bottom of the ranked list. However, GSEA methods rely on the structure of simple test statistics that do not explicitly use the known networked structure of genes. Furthermore, they can only indicate which large pre-specified groups of genes are important and do not allow for partial responses where only some genes within these known groups are related the observed response.
Machine learning methods such as graph-based kernels (Rapaport et al., 2007) or penalized approaches (Li and Li, 2008; Zhu et al., 2009) overcome the limitations of GSEA and use feature selection to identify functional network components. However, neither graph kernels nor penalized approaches can strictly enforce that the features identified are logically connected within the network, and therefore may produce results that are difficult to justify biologically. Probabilistic network models such as those described in Wei and Li (2007) and Sanguinetti et al. (2008) can enforce these features to be logically connected within the metabolic network. However, these methods require an assumption of a discrete gene expression distribution that may not completely reflect the underlying biology.
Our approach conceptually lies between GSEA and probabilistic models as we assume very little about the structure of the gene expression data but enforce the identified components to be logically connected within the network. We propose a combination of three complementary methods we have previously developed and have proven to be successful in analyzing small metabolic sub-networks. First, we use a non-parametric pathway ranking method (Takigawa and Mamitsuka, 2008), and perform an exhaustive search to identify the top K most coordinated genetic pathways in response to specific experimental conditions. Our path ranking method assumes that the functional components of a metabolic network will possess a highly correlated pathway structure. Then, if any functional components exist the top-ranked pathways will be a clustered list of small pathway variations through these components. Pathway ranking is similar to GSEA; however, it explicitly uses the network structure, does not require the specification of prior groups of genes, and makes no assumption on the distribution of the gene expression.
Pathway ranking has been shown to extract biologically meaningful pathways in small metabolic sub-networks (Takigawa and Mamitsuka, 2008). However, as the network size increases, to ensure we are extracting all biologically relevant structure, we must also increase the number of pathways to be extracted. However, extracting large numbers of pathways, in the order of 1000s, prevents an easy interpretation of the result. Therefore, extending pathway ranking to global metabolism requires further tools to identify the defining structures within the resulting pathway list.
To identify the defining features within the set of top-ranked pathways, we propose both a clustering and a classification algorithm. Both proposed algorithms exploit the natural Markov structure of a pathway. The pathway clustering algorithm is 3M (Mamitsuka et al., 2003), which identifies pathways that possess the same underlying sequence of functional genes. The pathway classifier, HME3M (Hancock and Mamitsuka, 2009) employs the same framework as 3M but restricts the pathway search to identify those pathways that define a specific experimental condition. Both 3M and HME3M have been previously shown to identify biologically significant pathways on metabolic sub-networks (Hancock and Mamitsuka, 2009; Mamitsuka et al., 2003). However, these previous implementations employed a gene activity definition, which required the extraction of all possible pathways between single start and end compounds within the network. Clearly, this approach of extracting all possible pathways through global metabolic networks is infeasible. Therefore, by using the pathways identified by the path ranking method, we are upscaling both 3M and HME3M to analysis of global metabolic networks.
Both 3M and HME3M provide a probability estimate that each edge is a member of an identified functional component within the metabolic network. Furthermore, in this article we define detailed node and edge annotations that allow for an analysis of each pathway with respect to its core genetic dependencies and can scale up to include an analysis of interacting reactions, compounds and KEGG sub-networks. These annotations allow for the analysis of the structure within each identified functional component to be performed at multiple biologically meaningful resolutions. In this article, we show the combination of pathway ranking with Markov clustering and classification models is scalable to the analysis of complete metabolic networks. The results will highlight the flexibility of our combined approach and show that it identifies biologically meaningful pathways within real microarray expression data in both unsupervised and supervised settings.
2 METHODS
2.1 Defining metabolic pathways
In metabolic networks, the same gene can be found in multiple locations within the network, can catalyze different reactions and, therefore, can possess multiple biological functions. When defining a pathway through a metabolic network the location of each gene denotes a specific function and must be precise. We define the specific location of each gene within the metabolic network through node and edge annotations extracted from the KEGG database (Kanehisa and Goto, 2000). Our annotations (1) define each gene as a node within the network. Each gene is annotated by its gene code (G), reaction (R) and KEGG pathway membership (P). Additionally, as the edge between two genes connects two reactions within the metabolic network, it is identified by the first substrate compound (CF); the product compound from the first reaction (CM); final product compound (CT); and the final KEGG pathway membership of CT, (PT). These annotations allow for transparent tracking of each pathway through the entire metabolic network:
| (1) |
Using the annotations in (1), we define a genetic pathway through a metabolic network to be a connected sequence of genes g that extend between specified start (s) and end compounds (t) (2).
| (2) |
In (2), labelk are the edge annotations in (1) and f(gk, gk+1) is a function indicating the strength of the relationship between gk and gk+1.
Our pathway definition requires the specification of start s and end nodes t, which are entry and exit compounds of the metabolic network. In previous analysis, we defined these nodes to be specific compounds of interest within a small metabolic sub-network (Hancock and Mamitsuka, 2009; Takigawa and Mamitsuka, 2008). However, these do not need to be single compounds. Therefore, to extend our original approach to analyze global metabolic networks we define s to be all compounds with edges only leading into the network, and we define t to be all compounds without edges leading into the network. Additionally, we note a slight limitation of our pathway approach for the case where multiple substrates are combined by a single reaction to produce multiple products. In this situation, all possible pairwise combinations of the multiple substrate and products are added to the network, and then treated separately by the path ranking procedure.
Furthermore, each edge is weighted by functions, f(gk, gk+1), which measure the strength of the relationship between gk and gk+1. The specification of the weight function is flexible with the condition that increasing values of the f indicate stronger relationships between gk and gk+1. As the accuracy f is paramount to the path ranking method in this work we define f to be median Pearson's correlation coefficient between gk and gk+1 over 100 bootstrapped replications.
A key assumption of our pathway definition is that correlated gene expression directly relates to the function of a metabolic network. It should be noted here that it is not the mRNA as measured by microarray analysis that performs the metabolic functions but the related proteins. Additionally, it is well established that mRNA expression do not always correlate well with protein abundance as it ignores key biological mechanisms and features such as post-translational modification and sub-cellular location (Gygi et al., 1999). However, in general, highly abundant proteins are also likely to show high mRNA expression levels (Ghaemmaghami et al., 2003). Although the correlation between global gene expression and protein abundance is unclear, by identifying paths of maximum correlated gene expression we are focusing only on regions within the metabolic network where the expression signal is strong and are, therefore, more likely to possess a specific biological function.
2.2 Pathway ranking
Probabilistic pathway ranking identifies the most probable paths through a metabolic network between specified s and t by solving a K-shortest and loop-less path problem on a weighted network (Takigawa and Mamitsuka, 2008). Our path ranking method is a non-parametric method as it does not specify the functional form of the edge weights, f(gk, gk+1) (2), but instead considers the empirical cumulative distribution function (ECDF) over all edge weights within the network. In this work, as we define the edge weights to be the correlation between two genes and the ECDF is simply a probabilistic rank for the most positively correlated genes. Therefore, path ranking is identifying the top K pathways of maximal correlation through the metabolic network. Furthermore, the probabilistic nature of the ECDF edge weights allow for a significance test to determine if a path contains any functional structure or is simply a random walk (Takigawa and Mamitsuka, 2008).
Due to the high levels of redundancy within metabolic networks, it is likely that any pathway ranking procedure will be biased towards short paths consisting of the same gene. Therefore, to ensure that we extract informative paths through global metabolic networks we include two control parameters. First, a parameter to control the minimum number of genes can be set to remove the numerous small and biologically uninteresting pathways from the pathway set. No maximum path length is set. Second, biological redundancy creates chains of reactions that are catalyzed by similar or identical gene sets. Therefore, there are multiple edges within the network where the same gene is connected to itself. As the correlation along these edges will be 1 it is clear that pathways consisting largely of the same gene will dominate the list of extracted pathways. To reduce the effect of the same gene edges, we define a user-specified penalty ρ on all edges, which connect the same gene. We assign the edge correlation, f(gk, gk+1), for all same gene edges to the specified ρ value. Setting ρ allows for explicit control over the diversity of genes selected within the extracted pathway list.
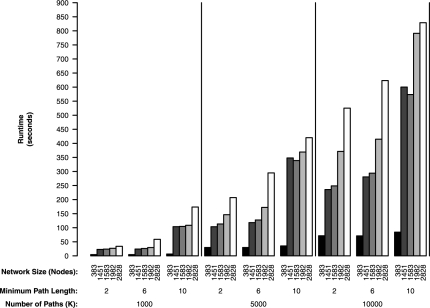
To show the scalability of the path ranking procedure to global metabolism in Figure 1, we present running times for analysis of the entire KEGG yeast metabolic network using the Gasch microarray data (Gasch et al., 2000) and a same gene penalty of ρ = 0. Figure 1 shows the effect on the running time of the path ranking algorithm at various control parameter settings and network sizes. The minimum path length is set to [2, 6, 10], number of paths to be extracted is set to K = [1000, 5000, 10 000] and the network size is increased in terms of number of genes (nodes) until the entire KEGG yeast metabolic network is analyzed. The experiments were performed in R (Ihaka and Gentleman, 1996) running on an desktop PC (Intel Core i7 2.66 GHz CPU,12 GB of RAM).
Fig. 1.
Running times for the path ranking algorithm for various network sizes, minimum path lengths and number of extracted paths.
These experiments clearly show that increasing all parameter values will increase the running time of the path extraction procedure. The maximum run time of ∼850 s (<15 min) for realistic settings, (10 000 paths of ≥10 genes, entire KEGG yeast metabolic network), shows reasonable practical performance of our path ranking method. This performance observed on a desktop PC shows the scalability of our path ranking method to analyze entire metabolic networks.
2.3 3M pathway clustering
The 3M Markov mixture model (Mamitsuka et al., 2003) provides the core framework for both our pathway clustering and classification models. 3M explicitly uses the Markov structure over all extracted pathways to identify the functional components. The 3M model identifies M key functional pathway components through a mixture of first-order Markov chains
| (3) |
where πm is the probability of each component and each component is defined by the transition probabilities θkm, p(si|θ1m) the probability of the start compound si ∈ s and p(gk, labelk|gk−1; θkm) the probability of a path traversing the edge labelk linking genes gk−1 and gk. From the pathway definition (2), every path starts at node s and finishes at t. Therefore, the start compound, s, is the same for all pathways and the probability of the start compound p(s|θ1m) becomes 1 for all components.
The 3M model is simply a mixture model and as such its parameters are conveniently estimated by an expectation maximization (EM) algorithm (Mamitsuka et al., 2003). The result of 3M is M components each defined by θm = {θsm, [θ2m, …, θtm,…, θTm]} where θtm are the probabilities of each gene within each component. The identified components correspond to a cluster of frequently observed pathways that have a similar structure. The parameters θm are probabilities for each gene within each component and, therefore, directly correspond to the importance of each gene within each identified functional component.
2.4 HME3M pathway classification
An extension to the 3M for classification is available through the HME3M model (Hancock and Mamitsuka, 2009). HME3M uses an hierarchical mixture of experts (HME; Jordan and Jacobs, 1994) to create a classification model directly from the 3M model. To supervise 3M an additional term, p(y|X, βm) is added to each functional component within (3) to include information from known experimental groups
| (4) |
where y is a binary response variable and X a binary matrix where each column is a gene, each row is a pathway and a cell value of 1 indicates the inclusion of a particular gene along a specific path. The parameters πm, θkm and βm are estimated simultaneously with an EM algorithm (Hancock and Mamitsuka, 2009). The additional term p(y|X, βm) is simply a classification model, which takes as input the binary pathway matrix X weighted by the EM component probabilities and returns the posterior probabilities for classification of the response variable y. To ensure a scalable and interpretable solution, HME3M uses a penalized logistic regression for each component classifier.
The goal of HME3M is to identify a set of pathways that can be used to classify a particular response label, yl ∈ y. However, as we know the response variable a priori we can optimize performance of the HME3M model by directing the path ranking algorithm through network components that are differently expressed over the response labels. This can be done by normalizing the ECDF edge weights of the path ranking method across all response labels. To do this normalization step, we first compute the ECDF edge weights for each label, yl, independently, and get PEW(gk, gk−1, y = yl). Normalizing exploits the probabilistic nature of the ECDF edge weights, PEW, and divides each PEW(gk, gk−1, y = yl) by the sum over all response labels (5),
| (5) |
We then use PEW as the edge weights and extract the K most likely pathways that are specific to each response label. The process of normalizing the edge weights will highlight edges that display a difference in correlation over response labels. Normalizing the edge weights is useful for supervised analysis where the goal is to identify pathways specific to each response label; however, it is not suitable for unsupervised analyses where the goal is to identify clusters of pathways with a similar structure.
2.5 Software
A complete implementation of our combined approach can be found within our provided R package PathRanker. PathRanker provides all functions to process the KEGG metabolic network, overlay a microarray dataset, extract the K most-correlated metabolic pathways, and analyze the key functional components within these pathways. PathRanker's functions are completely flexible allowing for either an analysis of the complete KEGG network or more tailored analysis of pathways between specific compounds and KEGG pathway groups. Multiple visualization options are also provided allowing the user to interpret the results at each stage of the analysis. PathRanker will soon be submitted to CRAN and is currently available from http://www.bic.kyoto-u.ac.jp/pathway/timhancock.
3 EXPERIMENTS
We evaluate our combined methodology on two microarray datasets constructed with differing experimental objectives. The first microarray is the benchmark Gasch microarray (Gasch et al., 2000), which observes the genetic response of yeast to numerous environmental stress conditions. Our goal is to identify which stress conditions have similar pathway responses. The second is a microarray observing the genetic differences between equally obese patients who are insulin resistant versus those who are insulin sensitive (Yang et al., 2002). Insulin resistance is a known metabolic hallmark of Type II diabetes. Our goal in this analysis is to identify specific pathway differences between insulin resistant and sensitive patients. The microarray datasets were obtained from GEO (Gasch = GSE18, diabetes = GSE121).
3.1 Metabolic profiling of yeast stress responses
Before analysis of the Gasch microarray, pre-processing of the experimental conditions was performed to merge the several original heat shock experiments into one response label ‘Heat Shock’. This merging process did not include the ‘Reverse Heat Shock’. Furthermore, all the experiments concerning cell-cycle factors and strain comparisons were removed due to small numbers of observations. This left 13 stress conditions, and therefore we use 3M to identify 13 functional components, M = 13.
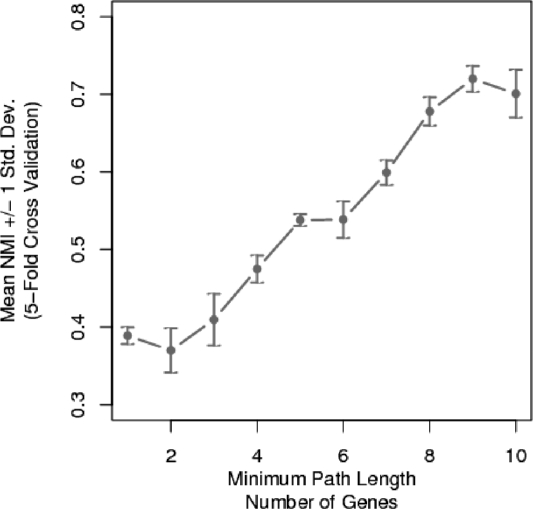
We construct our experiment to highlight path similarities among the stress conditions. We estimate the network edge weights for each stress condition independently and extract the top 1000 paths (K = 1000) with a same gene penalty set to highlight diverse paths (ρ = 0). To find the optimum 3M model, we vary the minimum path length from 1 to 10 genes. The quality of the 3M clustering for each minimum path length setting is evaluated using a 5-fold cross-validated Normalized Mutual Information (NMI).
Figure 2 displays the minimum path optimization curve for 3M clustering of the Gasch yeast stress conditions. Clearly, lengthening the minimum path length yields a pronounced improvement in 3M clustering results. Figure 2 reveals the action of the minimum path parameter to remove small noise pathways that are observed across multiple stress conditions. Only after these small noise pathways have been removed, the functional components of each stress condition can be accurately identified. From Figure 2, the optimal clustering performance is observed to be at a minimum path setting of nine genes. We now set the minimum path parameter to nine genes and analyze the clustering results.
Fig. 2.
The mean of 5-fold cross-validated NMI for clustering the Gasch stress conditions for specified minimum path lengths.
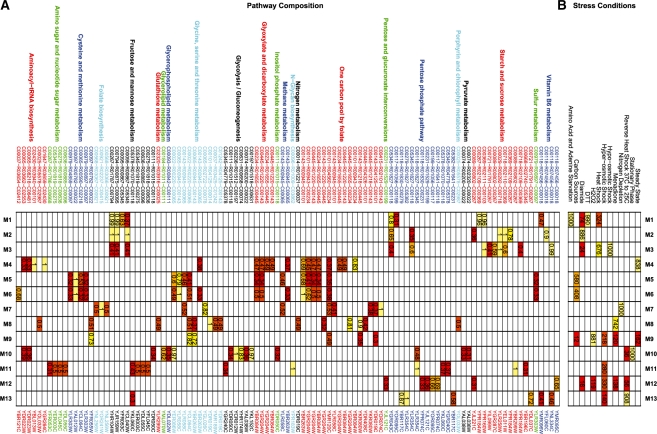
The structure of the 3M model for clustering the Gasch yeast stress conditions is presented in Figure 3. Figure 3 contains two tables where the right table is a heat map of the clustering confusion matrix with the known stress conditions on the top axis and the 3M cluster labels on the right axis. The numbers within each cell are the number of pathways for each stress condition that occur within each 3M cluster. The left table is a heat map of the pathway structure for each 3M component. The top axis shows the KEGG metabolic pathways, compounds and reactions, the bottom axis shows the genes. The number within each cell is probability of each gene within each 3M component. To improve the clarity of the display in Figure 3, all θ parameters less than 0.3 were removed and then only columns with more than one valid θ parameter were retained. The complete image is presented in Figure 1 of the Supplementary Material. From observation of the confusion matrix and the component similarities in Figure 3, we identify six related components with common structure. We now discuss each separately.
Fig. 3.
The 3M pathway clustering results for clustering yeast stress conditions. (A) Represents the cluster structure annotated by KEGG pathway, to compound, reaction, from compound on the top axis and gene on the bottom axis. (B) represents the confusion matrix between stress condition and cluster. Throughout both images yellow represent highly important cluster features and red represents less important features.
3.1.1 (M1, M2, M3): Heat shock, hypo-osmotic shock, DTT, amino-acid starvation, diamide
The stress conditions of heat shock, hypo-osmotic shock, dithiothreitol (DTT), amino-acid starvation and diamide are grouped together as they share common components across starch and sucrose metabolism, fructose and mannose metabolism and the pentose phosphate pathway. The main common reaction path is from C00267 (α-d-glucose) to C00018 (pyridoxal phosphate) through C00794 (Sorbitol), C00095 (d-fructose), C05345 (β-d-fructose 6-phosphate) and C00118 (glyceraldehyde 3-phosphate). Despite this large common element, 3M is able to distinguish three sub-clusters, which group DTT with amino acid starvation, hypo-osmotic stress with heat shock and lastly diamide exposure. These pathway differences occur mainly within starch and sucrose metabolism.
The (DTT, amino acid starvation) (M1) cluster is defined by a edge from α-d-glucose to C00103 (d-glucose 1-phosphate) by the YPR184W gene. YPR184W is induced by Gcn4p, which is a key regulator expressed during amino acid starvation in yeast (Natarajan et al., 2001). Gcn4p is also known to regulate genes relating to the unfolded protein response initiated by DTT exposure (Patil et al., 2004). Therefore, the similarity between the DTT and amino acid starvation stress responses is likely due to Gcn4p regulation.
The diamide response (M2) has considerable overlap with both M1 and M3 but includes key components that allow it to be clustered separately. The similarity between diamide, heat shock, DTT agrees with observation by Gasch et al. (2000). The unique component of the diamide response is largely defined by the conversion from C00369 (starch) to C00267 (alpha-d-Glucose) by YPR184W. YPR184W can be regulated by the Yap1p gene (Garcia et al., 2009) which is known to be activated during diamide exposure (Kuge et al., 2001).
The (heat shock and hypo-osmotic stress) cluster (M3) is defined by the synthesis of α-d-glucose from C00721 (dextrin) or C00089 (sucrose) and not from starch as in M2. In M3, starch is converted to C00103 (α-d-glucose 1-phosphate) by gene YPR160W. The stress response to heat shock and hypo-osmotic shock are known to be similar (Jamieson, 1998) and both can be regulated by the heat shock transcription factors (Cabiscol et al., 2002). Furthermore, YPR160W is known to be regulated by the Hog1p-MAP pathway (Sunnarborg et al., 2001) that is known to be activated under osmotic stress conditions and heat shock (Mollapour and Piper, 2006).
One striking feature common to all M1, M2 and M3 is the use of the gene YPR184W (GDB1). YPR184W is a glycogen debranching enzyme (reaction R02109), which catalyzes the conversion from glycogen (C00718) to starch (C00369) and then from starch to α-d-glucose (C00267). Although all components (M1, M2 and M3) include YPR184W within their components the function of YPR184W is different in each component. M1 uses YPR184W to convert α-d-glucose into starch and then interacts YPR184W with YPR160W (GPH1) to synthesize α-d-glucose 1-phosphate (C00103). M2 is the reverse pathway to M1 and uses YPR184W to convert starch to α-d-glucose, and then interacts YPR184W with YHR104W (GRE3) to produce sorbitol. In contrast, M3 begins with glycogen and uses YBR184W to create starch and then proceeds in a similar path to M1. Therefore, the component differences surrounding the use of YPR184W identifies how starch is metabolized within the network in different stress conditions. The ability to identify multiple functions of single genes and compounds as illustrated by YPR184W is a powerful feature of using pathways to model metabolic networks.
3.1.2 (M4, M5, M6): steady state and carbon sources
The similarity of steady-state growth and alternate carbon sources is expected, as both relate to steady-state growth with either different carbon sources or at different temperatures (Gasch et al., 2000). The backbone of this group is the KEGG one carbon pool by folate pathway from (C00101) tetrahydrofolate (THF) to C00143 (5,10-methylene-THF).
When yeast is grown under steady-state conditions at various constant temperatures (M4) the one carbon pool by folate pathway is entered in through C00101 (THF). THF is a by-product of tRNA metabolism which converts C01647 (tRNA) to C02430 (l-methionyl-tRNA) and then to THF. The observation that genes relating to RNA processing are active during steady-state growth agrees with observations by Li et al. (2000).
Otherwise, when grown on differing carbon sources (M5,M6) the one carbon pool by folate pathway is through C00143 (5,10-methylene-THF) by a reaction series, which converts C00979 (O-acetyl-l-serine), C00097 (l-cysteine), C00022 (pyruvate), C00065 (serine) to 5,10-methylene-THF. The one carbon pool by folate pathway is used to regulate the production of 5,10-methylene-THF in the presence of excess glycine (Piper et al., 2000). As the amount or glycine and serine produced increases when yeast is grown in rich carbon sources (Pasternack et al., 1994), the regulation of the production 5,10-methylene-THF becomes necessary and is identified by 3M in pathway clusters M5 and M6.
3.1.3 (M7): nitrogen depletion
Nitrogen depletion is found to have a very specific response, which is a coordinated chain of reactions from C00568 (4-aminobenzoate) to C00078 (tryptophan) or C00234 (10-formyl-THF) through folate biosynthesis, the one carbon pool by folate pathway, methane metabolism and the glycine, serine and threonine pathway. The importance of the one carbon pool by folate pathway is known in the metabolism of limited nitrogen sources; in particular, relating to the expression of YDR019C (GCV1; Hong et al., 1999; Piper et al., 2002). Furthermore, the identification of glycine, serine and threonine pathway agrees with (Sinclair and Dawes, 1995) who show that glycine and serine levels increase when yeast has limited nitrogen resources and suggest that these compounds are used as a substitute source of nitrogen.
Extrapolation of the M7 component from the beginning compound of the path, 4-aminobenzoate, reveals M7 to be a subset of the larger folate biosythesis pathway. The larger pathway begins with the compound GTP in purine metabolism and flows into the one carbon pool by folate pathway through C00415 dihydrofolate (DHF). This extrapolation agrees with research indicating that nitrogen starvation is characterized by an increase in nucleotide levels as metabolized by purine metabolism (Boer et al., 2010). These nucleotides could then act as a potential nitrogen source required for the synthesis of glycine and serine. However, this required extrapolation is indicative of a potential limitation of the pathway mining approach and its bias towards short path length. The bias of pathway ranking methods towards shorter path lengths results in the identification of only a minimal subset of the full metabolic response. To gain a complete understanding of the full metabolic response requires either extrapolation about the identified paths or the further refinement of the hypothesis through explicit specification of start and end compounds.
3.1.4 (M8, M9): H202 and menadione
The H202 (M9) and menadione (M8) stress have a similar core structure centering around the one carbon pool by folate pathway. The defining feature of menadione exposure (M8) is the cycle from C00014 (ammonia), C00037 (glycine), C01242 (S-aminomethyldihydrolipoylprotein), C00143 (5,10-methylene-THF), C00445 (5,10-methenyl-THF), C00101 (THF) and back to ammonia primarily by the interaction between the genes YDR019C (GCV1) and YMR189W (GCV2). The conversion of ammonia to 5,10-methylene-THF is absent in the H202 component (M9), instead the pathway finishes at 5,10-methenyl-THF or 10-formyl-THF. GCV1 and GCV2 are known to be expressed during menadione exposure (Zhang et al., 2003) and this effect is used to separate the H202 and menadione responses.
3.1.5 (M10): stationary phase
The stationary phase is found to possess a unique response defined by the transition from C00236 (3-phospho-d-glyceroyl phosphate) into C00065 (serine) through glycolysis. The identification of coordinated expression of glycolysis genes agrees with previous research, which indicates that over-expression of the central metabolic pathways is a hallmark of stationary phase metabolism in yeast (Gasch et al., 2000; Martinez et al., 2004).
3.1.6 (M11, M12, M13): common oxidative stress response
Clusters M11, M12 and M13 relate to common oxidative stress response, which is induced by multiple stress conditions . The grouping of the stress conditions, reverse heat shock, menadione, H202 and diamide agree with oxidative stress response being a common component of multiple stress responses (Gasch et al., 2000). Additionally, the identification of the pentose phosphate pathway is expected as it is known to be activated in yeast during oxidative stress (Slekar et al., 1996). Furthermore, the synthesis of C00199 (d-ribulose 5-phosphate) or C00117 (ribose 5-phosphate) within the pentose phosphate pathway in M12 has been shown to be a key marker of oxidative stress in Arabidopsis (Baxter et al., 2007).
3.2 Metabolic profiling of insulin resistance
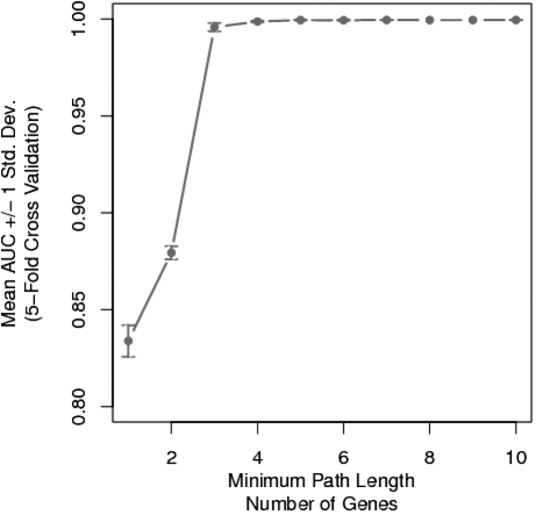
The minimum path analysis for HME3M classifying insulin resistance is presented in Figure 4. From Figure 4, we observe a direct impact between path length and HME3M performance. At small minimum path lengths, the performance is relatively poor and the predictive variance is large. Increasing the minimum path length not only increases accuracy, but also stability of the HME3M model. Optimum performance is reached with a minimum path setting of four genes. This relatively small minimum path setting indicates that insulin resistance is the result of metabolism of only a few critical compounds. We now set the minimum path setting to four and extract the key pathway components for insulin resistance.
Fig. 4.
HME3M 5-fold cross validation (CV) area under a receiver operating characteristic curve (AUC) for classifying insulin resistance.
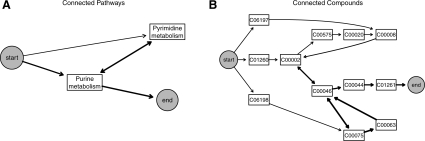
The key component for insulin resistance identified by HME3M is presented in Figure 5. The complete structure of both the insulin resistant and insulin sensitive components is displayed in Figures 2 and 3 of the Supplementary Material. Figure 5 presents the same component in terms of its connected pathways (Fig. 5a) and compounds (Fig. 5b). The line thickness represents highly probable pathways. The connected reaction and gene networks can be found in the Supplementary Material. It is clear from Figure 5a that purine metabolism is the primary driver of insulin resistance. The highly probable edges within compound network (Fig. 5b) relate to conversions between C00002 (ATP) through C00046 (RNA) C00075 (UTP), C00063 (CTP), C00044 (GTP) and C01261 (GppppG). These steps are performed by a common set of reactions and genes (Supplementary Fig. 2). Although this common set of compounds is a major feature of the pathway structure it is not strongly related to insulin resistance but rather serves as the exit point used by all pathways through purine metabolism. The features that relate strongly to insulin resistance are compounds used to create ATP, a key signaling molecule in diabetes and insulin secretion (Koster et al., 2005).
Fig. 5.
The most important HME3M for classifying insulin resistance annotated separately by connected pathways and compounds. The edge thickness represents the number of times each edge has been observed within the selected component.
In Figure 5b, ATP is observed to be synthesized from C01260 (AppppA), C06197 (ApppA), C06198 (UppppU) or converted to C00575 (cAMP), C00020 (AMP) and C00008 (ADP) and then back to ATP. Both AppppA and ApppA are known to have a role in insulin release (Verspohl and Johannwille, 1998) and have more recently been linked to diabetes (Rüsing and Verspohl, 2004; Verspohl et al., 2003). Additionally, cyclic AMP (cAMP) has recently been indicated in the production of GLP-1 that is a known factor in insulin secretion and Type II diabetes (Yu and Jin, 2010). GLP-1 is also target for the drug Byetta that is now being developed to treat Type II diabetes (Yu and Jin, 2010). Furthermore, the conversion from ADP back to ATP is an oxidative phosphorylation step using the nucleoside diphosphate kinase (NDK) enzyme (Lipskaya and Voinova, 2005). NDK has been previously linked to Type II diabetes (Zhu et al., 1999) and abnormal oxidative phosphorylation is a known factor in Type II diabetes (Højlund et al., 2009).
4 CONCLUSIONS
In this article, we have described a complete process for extracting and analyzing functional pathways within global metabolic networks and gene expression data. The main feature of our approach is that it allows for analysis at the sub-network, compound, reaction and gene resolutions, which allows for a complete picture of the metabolic response. We have shown in this article that our combined approach extracts biologically meaningful results by identifying key genetic regulators of environmental stress in yeast and known drug targets for metabolic networks in humans. Furthermore, our approach is flexible allowing complete control over the structure and diversity of the pathways to be found and caters for both unsupervised and supervised styles of analysis. We believe these results highlight our approach to be a powerful framework for the analysis of global metabolic networks.
Funding: Japan Society for the Promotion of Science (JSPS) fellowship (to T.H.); Grant-in-Aid for Scientific Research 20700134 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (in part to I.T.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Baxter C, et al. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant physiol. 2007;143:312. doi: 10.1104/pp.106.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, et al. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E, et al. Mitochondrial Hsp60, resistance to oxidative stress, and the labile iron pool are closely connected in saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44531–8. doi: 10.1074/jbc.M206525200. [DOI] [PubMed] [Google Scholar]
- Ebenhoh O, Liebermeister W. Structural analysis of expressed metabolic subnetworks. Genome Inform. 2006;17:163–72. [PubMed] [Google Scholar]
- Garcia R, et al. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in saccharomyces cerevisiae. J. Biol. Chem. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gygi SP, et al. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock T, Mamitsuka H. Workshop on Algorithms in Bioinformatics (WABI) Philadelphia: 2009. A markov classification model for metabolic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handorf T, et al. Expanding metabolic networks: scopes of compounds, robustness, and evolution. J. Mol. Evol. 2005;61:498–512. doi: 10.1007/s00239-005-0027-1. [DOI] [PubMed] [Google Scholar]
- Højlund K, et al. Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia. 2009:541–551. doi: 10.1007/s00125-009-1624-0. [DOI] [PubMed] [Google Scholar]
- Hong SP, et al. Control of expression of one-carbon metabolism genes of saccharomyces cerevisiae is mediated by a tetrahydrofolate-responsive protein binding to a glycine regulatory region including a core 5′-cttctt-3′ motif. J. Biol. Chem. 1999;274:10523–10532. doi: 10.1074/jbc.274.15.10523. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Jamieson DJ. Oxidative stress responses of the yeast saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Jordan M, Jacobs R. Hierarchical mixtures of experts and the EM algorithm. Neural Comput. 1994;6:181–214. [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. Pathway tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster JC, et al. Diabetes and insulin secretion: the ATP-sensitive k+ channel (k ATP) connection. Diabetes. 2005;54:3065–3072. doi: 10.2337/diabetes.54.11.3065. [DOI] [PubMed] [Google Scholar]
- Kuge S, et al. Regulation of the yeast yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 2001;21:6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24:1175–1182. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Initiation of protein synthesis in saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J. Bacteriol. 2000;182:2886–2892. doi: 10.1128/jb.182.10.2886-2892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipskaya TY, Voinova VV. Functional coupling between nucleoside diphosphate kinase of the outer mitochondrial compartment and oxidative phosphorylation. Biochemistry. 2005;70:1354–1362. doi: 10.1007/s10541-005-0269-y. [DOI] [PubMed] [Google Scholar]
- Mamitsuka H, et al. Mining biologically active patterns in metabolic pathways using microarray expression profiles. SIGKDD Explor. 2003;5:113–121. [Google Scholar]
- Martinez MJ, et al. Genomic analysis of stationary-phase and exit in saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell. 2004;15:5295–5305. doi: 10.1091/mbc.E03-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B, et al. Pathwayexplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper PW. Hog1p mitogen-activated protein kinase determines acetic acid resistance in saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:1274–1280. doi: 10.1111/j.1567-1364.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- Natarajan K, et al. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternack LB, et al. Whole-cell detection by 13C NMR of metabolic flux through the C1-tetrahydrofolate synthase/serine hydroxymethyltransferase enzyme system and effect of antifolate exposure in saccharomyces cerevisiae. Biochemistry. 1994;33:7166–7173. doi: 10.1021/bi00189a020. [DOI] [PubMed] [Google Scholar]
- Patil CK, et al. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, et al. Regulation of the balance of one-carbon metabolism in saccharomyces cerevisiae. J. Biol. Chem. 2000;275:30987–30995. doi: 10.1074/jbc.M004248200. [DOI] [PubMed] [Google Scholar]
- Piper MD, et al. Regulation of the yeast glycine cleavage genes is responsive to the availability of multiple nutrients. FEMS Yeast Res. 2002;2:59–71. doi: 10.1111/j.1567-1364.2002.tb00069.x. [DOI] [PubMed] [Google Scholar]
- Price ND, et al. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 2003;21:162–169. doi: 10.1016/S0167-7799(03)00030-1. [DOI] [PubMed] [Google Scholar]
- Rapaport F, et al. Classification of microarray data using gene networks. BMC Bioinformatics. 2007;8:35. doi: 10.1186/1471-2105-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüsing D, Verspohl EJ. Influence of diadenosine tetraphosphate (Ap4A) on lipid metabolism. Cell. Biochem. Funct. 2004;22:333–338. doi: 10.1002/cbf.1111. [DOI] [PubMed] [Google Scholar]
- Sanguinetti G, et al. MMG: a probabilistic tool to identify submodules of metabolic pathways. Bioinformatics. 2008;24:1078–1084. doi: 10.1093/bioinformatics/btn066. [DOI] [PubMed] [Google Scholar]
- Shlomi T, et al. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Dawes IW. Genetics of the synthesis of serine from glycine and the utilization of glycine as sole nitrogen source by saccharomyces cerevisiae. Genetics. 1995;140:1213–1222. doi: 10.1093/genetics/140.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slekar KH, et al. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- Smolke CD. The metabolic pathway engineering handbook. New York: CRC Press; 2010. [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnarborg SW, et al. Expression of the yeast glycogen phosphorylase gene is regulated by stress-response elements and by the HOG map kinase pathway. Yeast. 2001;18:1505–1514. doi: 10.1002/yea.752. [DOI] [PubMed] [Google Scholar]
- Takigawa I, Mamitsuka H. Probabilistic path ranking based on adjacent pairwise coexpression for metabolic transcripts analysis. Bioinformatics. 2008;24:250–257. doi: 10.1093/bioinformatics/btm575. [DOI] [PubMed] [Google Scholar]
- Verspohl EJ, Johannwille B. Diadenosine polyphosphates in insulin-secreting cells: interaction with specific receptors and degradation. Diabetes. 1998;47:1727–1734. doi: 10.2337/diabetes.47.11.1727. [DOI] [PubMed] [Google Scholar]
- Verspohl EJ, et al. Diadenosine tetraphosphate (Ap4A) induces a diabetogenic situation: its impact on blood glucose, plasma insulin, gluconeogenesis, glucose uptake and GLUT-4 transporters. Pharmazie. 2003;58:910–915. [PubMed] [Google Scholar]
- Wei Z, Li H. A markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia. 2002;45:1584–1593. doi: 10.1007/s00125-002-0905-7. [DOI] [PubMed] [Google Scholar]
- Yu Z, Jin T. New insights into the role of cAMP in the production and function of the incretin hormone glucagon-like peptide-1 (GLP-1) Cell. Signal. 2010;22:1–8. doi: 10.1016/j.cellsig.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Zhang W, et al. Microarray analyses of the metabolic responses of saccharomyces cerevisiae to organic solvent dimethyl sulfoxide. J. Ind. Microbiol. Biotechnol. 2003;30:57–69. doi: 10.1007/s10295-002-0012-2. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. Interaction of the Ras-related protein associated with diabetes Rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc. Natl Acad. Sci. USA. 1999;96:14911–14918. doi: 10.1073/pnas.96.26.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, et al. Network-based support vector machine for classification of microarray samples. BMC Bioinformatics. 2009;10:S21. doi: 10.1186/1471-2105-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.