Abstract
Motivation: Copy number variation (CNV), a major contributor to human genetic variation, comprises ≥ 1 kb genomic deletions and insertions. Yet, the identification of CNVs from microarray data is still hampered by high false negative and positive prediction rates due to the noisy nature of the raw data. Here, we present CNVineta, an R package for rapid data mining and visualization of CNVs in large case–control datasets genotyped with single nucleotide polymorphism oligonucleotide arrays. CNVineta is compatible with various established CNV prediction algorithms, can be used for genome-wide association analysis of rare and common CNVs and enables rapid and serial display of log2 of raw data ratios as well as B-allele frequencies for visual quality inspection. In summary, CNVineta aides in the interpretation of large-scale CNV datasets and prioritization of target regions for follow-up experiments.
Availability and Implementation: CNVineta is available as an R package and can be downloaded from http://www.ikmb.uni-kiel.de/CNVineta/; the package contains a tutorial outlining a typical workflow. The CNVineta compatible HapMap dataset can also be downloaded from the link above.
Contact: m.wittig@mucosa.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Many recent research findings suggest that copy number variation (CNV) plays a major role in genetic variability and hence human disease (Manolio et al., 2009; Zhang et al., 2009). However, while methodologies for large-scale association studies of single nucleotide polymorphisms (SNPs) are well established, a comprehensive framework for the analysis and interpretation of genome-wide CNV predictions in large case–control datasets is still lacking (Manolio et al., 2009). CNV prediction algorithms for data derived from SNP microarrays have improved considerably over the last few years, but many predicted segments are still false positives (Winchester et al., 2009) and can only be excluded through visual inspection in analogy to the mandatory inspection of genotyping assay scatterplots derived from SNP-based genome-wide association studies (GWAS; WTCCC, 2007). In addition, false negative rates tend to be high (Barnes et al., 2008) which can, for example, cause problems when rare CNV candidates are overlooked in the data. Current visualization tools such as the Affymetrix® Genotyping Console work well with small datasets and are suitable for cancer diagnostics and similarly small-sized sample sets. The difficulties in bridging the gap between packages for CNV prediction, association analysis and visualization for distinguishing genuine signals from false positive/negative predictions in generally noisy datasets have hindered many scientists from conducting genome-wide CNV analysis of their existing large GWAS datasets.
We here present CNVineta, an R package capable of handling CNV data derived from large datasets with implemented analysis tools for the detection and visualization of disease-associated rare (Supplementary Fig. S4) and common CNVs (Supplementary Figs S5, S6). CNVineta was designed to allow researchers to access large datasets from standard desktop computers. We hope that CNVineta will empower scientists to perform genome-wide CNV screening and quickly evaluate raw data of candidate CNVs in existing GWAS datasets.
2 FEATURES OF CNVINETA
2.1 Input and output
In order to allow for rapid access to both predicted CNV data and raw data, as log2 of raw data ratios (LRR) and B-allele frequencies (BAF), CNVineta requires the CNV prediction output from third party tools [e.g. Affymetrix® Power Tools (APT), QuantiSNP (Colella et al., 2007), etc.] as well as the LRR and BAF to be converted to the CNVineta file format. The file format consists of (i) a single large binary data file with binary data on LRR and BAF for all samples; (ii) a SNP array annotation file; (iii) a sample annotation file; (iv) a segment annotation file containing data on predicted CNVs; and (v) a RefGene file with standard annotations for all genes in the human genome [UCSC refGene table (Karolchik et al., 2009)].
A detailed description of the CNVineta input file format and a conversion workflow are available in the online tutorial of CNVineta as well as in the package vignette. As an example for the CNVineta workflow (Fig. 1A).
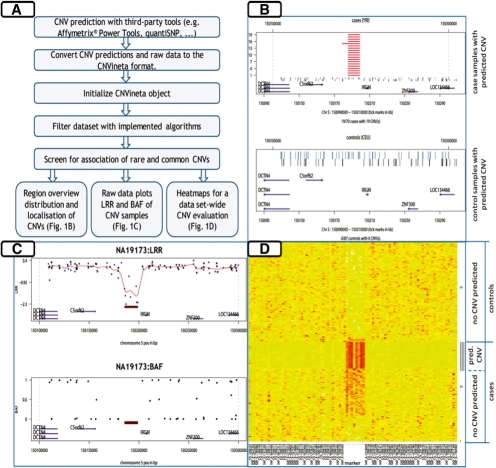
Fig. 1.
Workflow and screenshots from CNVineta. (A) Workflow. Before starting the CNV screening, the SNP array data has to be processed with a third-party CNV prediction algorithm. Subsequent CNV association screening can be performed for rare and/or common CNVs. The functions automatically generate result tables and graphs for all regions that were identified as associated by CNVineta. The visual data mining can be performed in a stepwise fashion. (B–D) Plotting results for a known common deletion at the IRGM gene locus (McCarroll et al., 2008) in the Affymetrix® 6.0 HapMap dataset (International HapMap Consortium, 2003) comprising 180 samples (CEU and YRI). (B) Regional overview plot. From top to bottom the predicted CNVs (deletions highlighted by red horizontal lines), array probe sets within the region (SNP marker in black and non-polymorphic probe sets by blue vertical lines) and annotated genes (purple arrows). (C) Raw data plots. For each sample, the raw data visualization includes LRR (upper panel) and BAF (lower panel). (D) Heat map. To obtain a sample set-wide impression of the particular CNV and in order to identify potential false positive and negative CNVs that should be subjected to further follow-up, heat maps of LRR data can be generated.
Help from the authors is available upon request (Google Group: http://groups.google.com/group/CNVineta).
2.2 Handling and visualization of CNVineta objects
CNVineta combines CNV prediction and raw data of the entire dataset. CNVs can be visualized in user-defined regions of the human genome across all samples using the regional overview option (Fig. 1B). As CNVs often overlap and vary in size, a graphical overview quickly allows for an orientation of the CNV structure at a particular region in the entire sample set. To demonstrate CNVineta's compatibility with large datasets, a regional overview of the proximal part of the chromosome 15q region (13 Mb) in 1587 samples is shown in the Supplementary Figure S1.
2.3 Genome-wide screening of rare and common CNVs
Analysis methods for genome-wide screening of rare and common CNVs are implemented. To reduce the complexity of the CNV predictions, CNVineta minimizes the genomic positions for association analysis. To this end, the array marker set is reduced to those markers, where a predicted CNV in any sample either starts or ends (Supplementary Fig. S2). In CNVineta, these markers are so-called atoms. The collated atoms are then used to scan genome wide for rare CNVs and/or common CNVs using a logistic regression model. The latter analysis method allows for the inclusion of covariates, e.g. gender, age and/or EIGENVECTORs that can help to remove confounding genotyping batch effects and/or population stratification (Price et al., 2006). LRR and BAF plots of samples with CNVs at the candidate regions as well as heat maps can automatically be generated. Primary, temporary and resulting data can all be processed by existing and/or custom R functions.
3 CONCLUSIONS
In this application note, we introduce CNVineta, an R package designed to meet the needs of scientists working with large-scale CNV data derived from SNP arrays. While many recently published CNV analysis programs focus on CNV prediction algorithms, CNVineta aims at handling, visualizing and mining these CNV predictions. The CNVineta workflow, outlined in detail in the online tutorial and Figure 1A, allows for in-depth data mining of a given dataset for associated rare as well as common CNVs for a phenotype of interest. Previous studies on CNVs in large sample sets genotyped with SNP arrays identified a large number of apparently false positive and negative CNV calls, which hampered an accurate interpretation and follow-up. CNVineta aims to overcome these difficulties by combining the visualization of CNV calls and the respective raw data.
Funding: German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN); DFG excellence cluster ‘Inflammation at Interfaces’.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Barnes C, et al. A robust statistical method for case-control association testing with copy number variation. Nat. Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, et al. QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0104s40. Chapter 1, Unit1.4. [DOI] [PubMed] [Google Scholar]
- Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Winchester L, et al. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomic Proteomic. 2009;8:353–356. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, et al. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.