Abstract
Using an Affymetrix GeneChip® Human Mapping 100K Set to study a patient with a late-presenting, right-sided diaphragmatic hernia and microphthalmia, we found a maternally inherited deletion that was 2.7 Mb in size at chromosome 18q22.1. Mapping of this deletion using fluorescence-in-situ hybridization revealed three deleted genes - CDH19, DSEL and TXNDC10, and one gene that contained the deletion breakpoint, CCDC102B. We selected DSEL for further study in 125 patients with diaphragmatic hernias, as it is involved in the synthesis of decorin, a protein that is required for normal collagen formation and that is upregulated during myogenesis. We found p.Met14Ile in an unrelated patient with a late-presenting, anterior diaphragmatic hernia. In the murine diaphragm, Dsel was only weakly expressed at the time of diaphragm closure and its expression in C2C12 myoblast cells did not change significantly during myoblast differentiation, thus reducing the likelihood that the gene is involved in myogenesis of the diaphragm. Although it is possible that the 18q22.1 deletion and haploinsufficiency for DSEL contributed to the diaphragmatic defect in the patient, a definite role for DSEL and decorin in the formation of the collagen-containing, central tendon of the diaphragm has not yet been established.
Keywords: Diaphragmatic hernia, Dsel, decorin, chromosome 18q22.1
INTRODUCTION
Congenital diaphragmatic hernia (CDH) has an estimated frequency of 1 in 2000 to 3000 births and has a high neonatal mortality and morbidity [Torfs et al., 1992]. Several genes that cause diaphragmatic defects as part of multiple congenital anomaly syndromes have been identified [Slavotinek, 2007], but the genes that cause isolated diaphragmatic hernias are almost unknown. To date, one nonsense mutation and several sequence variants of unknown significance in FOG2 have been described in CDH patients [Ackerman et al., 2005; Bleyl et al., 2007]. Chromosome aberrations are found in up to one third of patients with diaphragmatic defects [Howe et al., 1996; Enns et al. 1998; Holder et al., 2007], and array comparative genomic hybridization has therefore been undertaken in CDH patients[Kantarci et al., 2006; Slavotinek et al., 2006; Scott et al., 2007]. These array studies and published reports of patients with CDH and cytogenetic aberrations have led to the identification of numerous chromosome regions that are likely to contain dosage sensitive genes required for normal diaphragm development [Holder et al., 2007]. However, with the exception of FOG2, cloning the causative genes from these chromosome regions has proven difficult, perhaps because hernias show substantial phenotypic heterogeneity, ranging from classical ‘Bochdalek’ diaphragmatic hernias in the posterolateral diaphragm, to anterior retrosternal hernias and hernias of the central part of the diaphragm. These different types of diaphragmatic hernias have been considered to be genetically distinct [Ackerman et al., 2007], and as Bochdalek hernias occur most frequently whilst anterior and central diaphragmatic hernias are rare, case collection for molecular studies can be challenging.
We report on a male who had a late-presenting, right-sided diaphragmatic hernia and unilateral microphthalmia. This child was studied with the Affymetrix GeneChip Human Mapping 100K Set and a 2.7 Mb deletion at chromosome 18q22.1 was identified using this array. The deletion was inherited from his mother, who was reported to be phenotypically normal. We mapped the proband's deletion using fluorescence in-situ hybridization (FISH), and determined that the 18q22.1 deletion contained three genes: CDH19, DSEL, TXNDC10, with the telomeric breakpoint of the deletion likely to interrupt another gene, CCDC102B. Although this deletion was inherited from a normal mother, we chose to further examine the three genes in this small deletion.
We selected DSEL as a candidate gene for the diaphragmatic hernia in the proband, as DSEL is involved in the formation of one of the principal components of collagen, decorin. This paper describes the 18q22.1 deletion, our results re-sequencing DSEL in 125 unrelated diaphragmatic hernia patients, and our evaluation of Dsel as a candidate gene for diaphragm myogenesis.
MATERIALS AND METHODS
Patient Samples and Clinical Details
DNA samples were obtained from probands using two protocols approved by the Committee for Human Subjects Research at the University of California, San Francisco (numbers H41842-22157-06 and H41842-26613-04). We used 23 DNA samples from diaphragmatic hernia patients recruited through UCSF and 96 DNA samples obtained from the newborn blood spots of children with diaphragmatic hernias through the California Birth Defects Monitoring Program. The clinical features of 96 of the diaphragmatic hernia patients have been published [Slavotinek et al., 2006] the remaining 23 patients had non-syndromic diaphragmatic hernias [Slavotinek et al., 2009]. We also sequenced Dsel in DNA from six patients with anterior diaphragmatic hernias kindly provided by Dr Daryl Scott at Baylor.
Array Hybridization
Array hybridization was performed with the GeneChip® Human Mapping 100K Set (http://www.affymetrix.com/products/arrays/specific/100k.affx) using 500 ng genomic DNA according to the manufacturer's instructions. This mapping set has previously been validated for the detection of copy number variants in patients with chromosome aberrations [Rauch et al., 2004; Slater et al., 2005]. The results were analyzed according to the Significance of Mean Difference (SMD) algorithm designed to detect copy number variations and parental studies using the same mapping set [Delaney et al., 2008].
Fluorescence In-situ Hybridization (FISH)
Bacterial artificial chromosome (BAC) probes were purchased from BACPAC resources (http://bacpac.chori.org/), labeled with Cy3dUTP and hybridized as previously described [Slavotinek et al., 2006].
Genomic DNA Sequencing
Genomic sequencing was performed with a BigDye® Terminator v3.1 Cycle Sequencing Kit on an ABI 3730 machine (Applied Biosystems, Foster City, CA) as previously described [Slavotinek et al., 2006]. We sequenced CDH19, DSEL, TXNDC10 and CCDC102B coding exons and intron-exon boundaries in the male patient and the coding exon and the intron-exon boundaries of DSEL in 125 patients with diaphragmatic defects. Control chromosomes were examined by restriction enzyme digestion or by genomic sequencing.
In-situ Hybridization with Murine Embryo Sections
Section in-situ hybridization on murine paraffin sections was performed as previously described using digoxygenin-labeled riboprobes (DIG RNA labeling kit; Roche, Indianapolis, IN) [Chao et al., submitted]. The probe for Dsel was generated using primers: F: 5’gagtgagtgcgtgtgtccag; R: 5’tctcgtttttgtgtgcaagg and Dsel expression was examined in the murine diaphragm at E11.5, E12.5, E13.5, E14.5 and E16.5.
C2C12 Myoblasts and Quantitative Real-Time Polymerase Chain Reaction (RT-PCR) to Examine Dsel Expression during Myoblast Differentiation
Exponentially growing C2C12 myoblasts were plated in 60 × 15 mm Petri dishes at a density of 2.4 × 104 cells per ml. When the cells were 75-90% confluent, growth medium was replaced with differentiation medium containing 2% horse serum (Hyclone, Logan UT) and harvested on days 1, 3 and 6. RNA was obtained by standard methods (RNeasy kit, Qiagen, Valencia, CA). cDNA synthesis was performed from 500-1000 ng of total RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). The expression of Dsel (TaqMan probe Mm01294533_g1; Applied Biosystems, Foster City, CA) and Hprt1 (TaqMan probe Mm03024075_m1; Applied Biosystems, Foster City, CA) were assayed at days 1, 3 and 6 using real-time PCR (Applied Biosystems, Foster City, CA). Quantification analysis was performed with the ΔΔCt method using Hprt as a control.
RESULTS
Clinical Descriptions
The 13-month old male propositus with microphthalmia and diaphragmatic hernia has previously been described [FF264; Pasutto et al., 2007; Chao et al. submitted]. He was ascertained at 13 months of age when he presented with respiratory difficulties and a right diaphragmatic hernia was diagnosed by a chest radiograph. We have no further information regarding the type of hernia, but we assume it to have been relatively small because of the late presentation of the diaphragmatic defect. Surgical repair was successfully performed. He had unilateral right microphthalmia and the diameter of the right globe was reported as 9 mm on computerized tomography imaging at 13 months of age, with an estimated volume of 0.38 cm3 [normal human globe size is 3 cm3 at birth, increasing to 6 cm3 at age 24 months; Galluzzi et al., 2001]. The propositus’ parents were reportedly normal, but a maternal aunt was described as having anophthalmia, although she was unable to be examined. Sequencing of the STRA6 (Stimulated-by-retinoic acid-6; OMIM 610745) gene was negative in the propositus [Pasutto et al., 2007].
The second patient with c.1516G>A, predicting p.Met14Ile, was a female of Hispanic ethnicity. She was born at term with a birth weight of 3033 g (appropriate for gestational age) and an anterior diaphragmatic hernia was surgically corrected at six weeks of age. She presented with respiratory distress after her initial discharge from the nursery and the late presentation also implies a small diaphragmatic defect. She had no other abnormal physical findings.
Array Hybridization and FISH Map a 2.7 Mb Deletion at 18q22.1
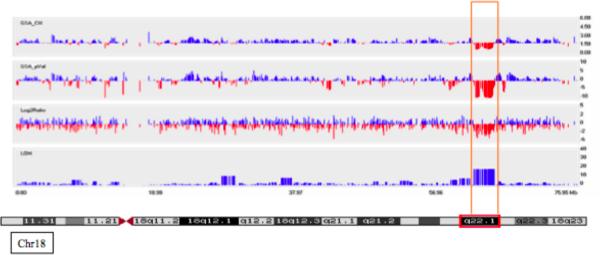
In the propositus, the array results showed loss of heterozygosity and log2ratio results consistent with a deletion of approximately 2.7 Mb in size at chromosome 18q22.1 (Fig. 1). Arrays with the same mapping set were performed on both parents, and the mother shared the same deletion (data not shown), but cells were not available from the mother for verification by FISH. The father did not show any significant copy number variations. We did not find any other significant copy number variations (CNVs) in the propositus or his parents and no other family members were available to be tested. The 18q22.1 deletion in the propositus was verified using FISH, and the centromeric breakpoint was mapped between BAC RP11-1069K2 (not deleted; chr18:61,828,786-62,017,616; numbering according to hg version 18; UCSC Genome Browser) and BAC RP11-246I7 (deleted; chr18:62,306,291-62,490,328) and the telomeric breakpoint was mapped between BAC RP11-105L16 (deleted; chr18:64,478,284-64,627,510) and BAC RP11-22A22 (not deleted; chr18:64,596,566-64,777,467; data not shown). The deleted region (chr18:62,306,291-64,627,510) therefore contains three genes - CDH19 (chr18:62,322,301-62,422196), DSEL (chr18:63,324,799-63,334,947) and TXNDC10 (chr18:64,491,905-64,533,333). CCDC102B is located at the telomeric edge of the deletion at chr18:64,616,550-64,873,406. The sizes, domains and expression pattern of these genes have been summarized in Table I. CDH19 and CCDC102B have been reported to be located in CNV regions (see Database of Genome Variants, http://www.tcag.ca/) with multiple entries for each of these genes. These CNV studies do not definitely eliminate any of the deleted candidate genes from consideration for the etiology of the diaphragmatic defects, although we assessed CDH19 and CCDC102B as less likely to be involved in the pathogenesis of the diaphragmatic hernia due to the frequency of CDH19 and CCDC102B deletions in normal individuals.
Fig. 1.
Array Studies Showed that the Propositus has a 2.7 Mb Deletion at 18q22.1. Chromosome 18, Log2Ratio from an Affymetrix 100K Array in the propositus showing a 2.7 Mb deletion at Chromosome 18q22.1. A chromatogram showing the G-banding pattern for chromsome 18 is located under the figure. The vertical bars of the figure show: GSA_CN (Genome smoothed average, copy number), GSA_pVal (Genome smoother average, p value), log2ratio and LOH (loss of heterozygosity).
Table I.
Genes Contained within the 18q22.1 Deletion
| Gene | Exons | Amino acids | Domains | Expression | Function |
|---|---|---|---|---|---|
| CDH19 | 12 | 772 | 5 extracellular calcium binding repeats; 1 transmembrane domain; cytoplasmic tail | 9.5 kb transcript; Widespread | Atypical cadherin; Cell-cell adhesion |
| DSEL | 2; 1 coding | 1212 | Transcription start site in CpG island; N-terminal signal peptide, 2 transmembrane domains | Widespread in adult tissues, including skeletal muscle | Epimerase and O-sulfotransferase activity in dermatan sulfate synthesis |
| TXNDC10; TMX3 | 16 | 454 | Thioredoxin domain; weak similarity to sulphotransferases | Widespread, including eye | Folding of di-sulfide bonds |
| CCDC102B | 7 | 297 | Coiled-coil domain | Widespread | Unknown |
Re-Sequencing of DSEL shows p.Met14Ile in an Unrelated Patient with Diaphragmatic Hernia
We examined the genes within the deleted interval at 18q22.1 for sequence variants in the propositus. We found no sequence alterations in CDH19, TXNDC10, DSEL or CCDC102B on the non-deleted allele of chromosome 18 (data not shown). We then sequenced DSEL in 125 patients with diaphragmatic hernia. We found one novel missense substitution, c.1516G>A, predicting p.Met14Ile in exon 2 of DSEL, in a patient with an anterior, late-presenting diaphragmatic hernia (Table II; Fig. 2; numbering = initial A of cDNA= 1, DSEL Ensembl transcript ENST00000310045). No other tissues were available for further studies. DNA samples from the parents were not available, but restriction digestion for the p.Met14Ile substitution in 200 Hispanic control chromosomes was negative, although it confirmed the substitution in the propositus (data not shown). Although the Met14 residue was highly conserved, the alteration was predicted to be benign by the PolyPhen website for the prediction of the functional effects of non-synonymous single nucleotide polymorphisms (SNPs). Another sequence alteration, c.4445G>A, predicting p.Asp991Asn, was present in a patient with a diaphragmatic hernia and trisomy 18 (Table II), but was not further studied as the same alteration was present in a healthy parent and our model for the pathogenesis of the hernias predicts haploinsufficiency. We also found c.2301A>G, predicting p.Asn276Ser in CDH patients, but restriction digests showed that this nucleotide substitution was a polymorphism more frequent in panethnic controls (5/84 chromosomes = 6%) than Caucasians (0/180 chromosomes). Comparing allele frequencies between CDH patients and controls from dbSNP, we found that only the A allele from the SNP c.2646A>G, a synonymous SNP resulting in p.Gln390Gln, was significantly reduced in CDH patients compared to controls (p = 0.0038; Table II). However, in view of the difference in ethnicity between the dbSNP population (Caucasian and African American) and our CDH patients (mostly Caucasian and Hispanic), we cannot conclude that this finding has real significance, as the difference in allele frequency may be influenced by ethnicity. In addition, we had no second cohort to replicate this finding.
Table II.
Sequence Alterations in DSEL in Patients with Diaphragmatic Defects
| Nucleotide | Amino acid | Genotype in DH Patients | *AF in DH Patients | Het. score | dbSNP | AF in dbSNP+ | Het.score in dbSNP |
|---|---|---|---|---|---|---|---|
| c.1516G>A | p.Met14Ile |
GG = 124
GA = 1 AA = 0 |
G = 0.996;
A=0.004 |
Het. score = 0.008 | - | - | - |
| c.2301A>G | p.Asn276Ser | AA = 105 AG = 11 GG = 1 |
A = 0.952; G = 0.048 |
Het. Score = 0.09 | - | - | - |
| c.2611T>C | p.Pro379Pro | TT = 124 TC = 1 CC = 0 |
T = 0.996; C = 0.004 |
Het. score = 0.008 | - | - | - |
| c.2646A>G | p.Gln390Gln | AA = 120 AG = 4 GG = 0 |
A = 0.984; G = 0.016 |
Het. score = 0.031* | rs9959648 | A = 0.905 G = 0.095 (74) |
Het. score = 0.171* |
| c.3521C>T | p.Pro683Ser | CC = 21 CT = 73 TT = 31 |
C = 0.46; T = 0.54 |
Het. score = 0.497 | rs2279269 | C = 0.517 T = 0.483 (120) |
Het. score = 0.491 |
| c.3693A>G | p.Tyr740Cys | AA = 124 AG = 1 GG = 0 |
A = 0.996; G =0.004 |
Het. score = 0.008 | rs12953840 | A = 0.986 G = 0.014 (74) |
Het. score = 0.004 |
| c.3998A>T | p.Thr842Ser | AA = 124 AT = 1 TT = 0 |
A = 1.0 | - | rs35479856 | A= 0.944 (74) T = 0.056 |
Het. score = 0.013 |
| c.4445G>A | p.Asp991Asn | GG = 124 GA = 1 AA = 0 |
G = 0.996; A =0.004 |
Het. score = 0.008 | - | - | - |
dbSNP = database of genomic variants (http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp&cmd=search&term=); Nucleotide numbering = A from ATG = 1, Ensembl transcript ENST00000310045; * = allele frequency in 125 diaphragmatic hernia patients. Het. score = heterozygosity score. + = Allele frequency obtained from AGI_ASP population; Coriell cell repositories; Caucasian and African American ethnicity.
Two-tailed P value = 0.0038 using Fisher's exact test.
Fig. 2.
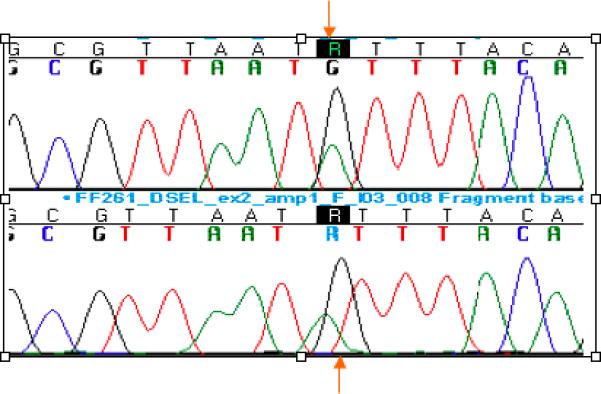
Re-sequencing of DSEL Reveals a Novel Missense Substitution, p.Met14Ile, in an Unrelated Patient with Diaphragmatic Hernia. Chromatogram showing c.42G>A (indicated by arrow) in forward and reverse chromatograms, predicting p.Met14Ile in DSEL.
In-situ Hybridization with Murine Embryo Sections shows Weak Expression of Dsel in the Developing Diaphragm
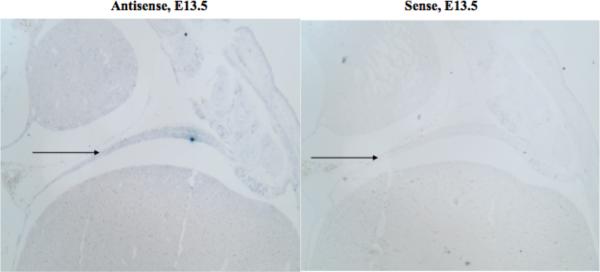
We examined Dsel expression in the murine diaphragm from E11.5 to E16.5. We found only weak expression of Dsel in the murine diaphragm muscle at E13.5 (Fig. 3), and we did not see reliable diaphragmatic expression at later or earlier time periods (data not shown). Our studies continued up to E16.5, several days after diaphragm closure (E12.5 –E13.5 in the mouse), as in some murine models of diaphragmatic defects, hernias have first been noted after the time of diaphragmatic closure as late as E15.5 [Yuan et al., 2003].
Fig. 3.
In-situ Hybridization of the Developing Diaphragm Showed Weak Expression of Dsel at the time of Diaphragm Closure. In-situ hybridization using antisense and sense riboprobes for Dsel, showing weak expression in the murine diaphragm at E13.5.
Quantitative Real Time-Polymerase Chain Reaction (RT-PCR) showed no Increased Dsel Expression during Differentiation of C2C12 cells
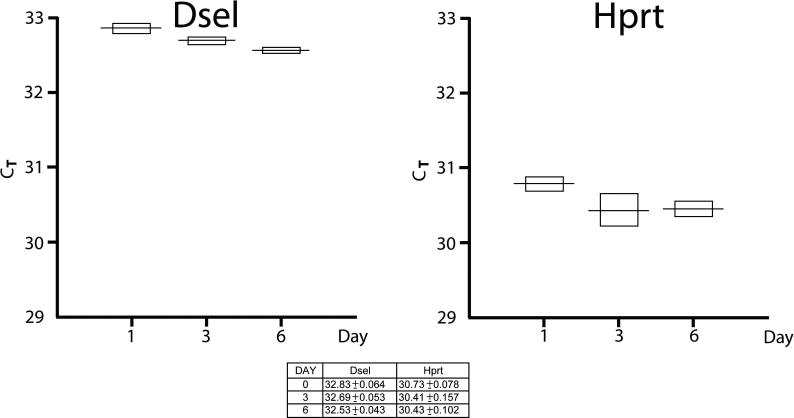
To implicate Dsel in diaphragmatic myogenesis, we tested the hypothesis that Dsel would be upregulated during myoblast differentiation. We examined the expression of Dsel during the differentiation process of C2C12 myoblasts compared to a control gene, Hprt, which showed no increased expression with myoblast differentiation. Our results showed no significant change in the expression of Dsel with myoblast differentiation (Fig. 4)
Fig. 4.
RT-PCR Showed no Significant Change in Dsel Expression During the Differentiation of C2C12 cells. Graph showing mean and standard deviation of Dsel and Hprt Ct at day 0, 3 and 6 of C2C12 cell differentiation, showing minimal change of Dsel expression with C2C12 cell differentiation between day 0 (undifferentiated cells) and day 6 (fully differentiated cells). Each experimental point was repeated in quadruplicate reactions
DISCUSSION
We have demonstrated a maternally inherited, 2.7 Mb 18q22.1 deletion in a male with microphthalmia and a late-presenting diaphragmatic hernia. The deleted genes have been summarized in Table I. CDH19, or Cadherin 19, type 2 preproprotein [OMIM 603016] is a calcium dependent, cell-cell adhesion glycoprotein expressed in neural crest-derived cells that may be important for the development of Schwann cells [Takahashi et al., 2005]. DSEL, or Dermatan Sulfate Epimerase-Like [OMIM 611125], acts as an epimerase and enables the formation of dermatan sulfate from chondroitin sulfate.[Goossens et al., 2003; Maccarana et al., 2006] TXNDC10, or Thioredoxin-domain-containing-10, also known as TMX3, is a thioredoxin that catalyzes the formation and folding of disulfide bonds [Haugstetter et al., 2005; Haugstetter et al., 2007]. Finally, little is known about CCDC102B, or Coiled-coil domain-containing 102B, and both PubMed and OMIM contain no references to this gene.
We chose DSEL for sequencing in a group of 125 patients with diaphragmatic hernia because of the importance of this gene in dermatan sulfate biosynthesis and decorin formation. DSEL acts as a chondroitin-glucuronate C5-epimerase, converting D-glucuronic acid to L-iduronic acid, and catalyzing the formation of dermatan sulfate from chondroitin sulfate. The main dematan sulfate-carrying proteoglycan, decorin, is a leucine-rich proteoglycan located in the extracellular matrix and expressed in the diaphragm of mdx mutant mice [Cáceres et al., 2000]. Decorin binds to collagen types 1 and VI, regulating collagen fibril formation and the stabilization of collagen fibers [Schönherr et al., 1995; Danielson et al., 1997]. Decorin is upregulated during myogenesis and C2C12 myoblast cells that overexpress decorin show increased myoblast proliferation, whereas antisense inhibition of decorin suppresses proliferation and accelerates the differentiation of C2C12 myoblasts due to an enhanced sensitivity to exogenous myostatin [Riquelme et al., 2001; Kishioka et al., 2008].
Our DSEL resequencing results showed one missense substitution, p.Met14Ile (Table I; Fig. 2), in an Hispanic female with a late-presenting, anterior hernia. Parental samples were not available to determine if the substitution was de novo, but it was highly conserved in other species and absent from 200 Hispanic control chromosomes. The missense substitution was also not detected in a paper that sequenced DSEL in 113 patients with bipolar disorder and 160 matched controls [Goossens et al., 2003] although the ethnicity of these subjects was not mentioned. However, the p.Met14Ile sequence alteration was not predicted to be disease-causing by software prediction programs. We also found no evidence for a role for Dsel in myogenesis in C2C12 cells, as expression levels of Dsel were unchanged during myoblast differentiation when examined by quantitative RT-PCR (Fig. 4).
Although we have not shown that the deletion of DSEL is involved in the pathogenesis of the hernias, both the patient with the deletion and the patient with the missense mutation had late-presenting diaphragmatic hernias. Late presenting hernias are defined as diaphragmatic hernias that are detected after the neonatal period and they may constitute a separate phenotypic subdivision of diaphragmatic hernias due to diaphragm weakness.[Numanoglu et al., 1997; Elhalaby et al., 2002; Baglaj, 2004] As decorin is involved in mature collagen formation, a deletion of the entire Dsel gene could result in reduced decorin formation, with a predisposition to later diaphragmatic weakness because of abnormal collagen synthesis. However, the gene appears not to play a major role in the etiology of CDH, as evidenced by the few sequence alterations observed in our patients. It is also possible that another of the deleted genes is responsible for the hernia, or that haploinsufficiency for more than one gene in the deleted interval is needed for the diaphragmatic defect. Re-sequencing of TXNDC10 in 25 patients with diaphragmatic hernias also returned no novel sequence alterations [Chao et al., submitted]. In addition, the occurrence of the deletion in the propositus's phenotypically normal mother must be explained. We hypothesize that the diaphragmatic hernia in the propositus may have been due to an additional contribution of haploinsufficiency for an alternative gene involved in diaphragm formation, unmasking of a recessive allele, or environmental factors.
Finally, this paper did not investigate any of these genes for a role in the microphthalmia in the propositus. However, TXNDC10 is expressed in the eye (Unigene; http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.268041), and therefore this gene may prove to be an attractive candidate for studies into the etiology of the microphthalmia in the propositus.
CONCLUSION
We report on a maternally inherited, 2.7 Mb deletion at chromosome 18q22.1 disrupting four genes in a male with a late-presenting, right-sided diaphragmatic hernia and microphthalmia. We investigated DSEL for a role in the pathogenesis of the diaphragm hernia and identified one amino acid substitution, p.Met14Ile, in an unrelated patient with a late-onset, anterior diaphragmatic hernia. However, a definite role for DSEL in the pathogenesis of diaphragmatic defects has not been established.
ACKNOWLEDGMENTS
Anne Slavotinek was generously funded by an R03 grant 5R03 HDO49411-02 from the National Institute of Child Health and Development (NICHD) and a K08 grant K08HD053476-01A1 from the same institution. Hatem Zayed was supported by a Postdoctoral Training Grant in Medical Genetics, 5T32GM007085-30, to the Division of Medical Genetics, Department of Pediatrics, at UCSF. We are grateful to Dr Daryl Scott for providing patient samples for the project. This publication was supported by NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131. This publication was supported by NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131. The findings and conclusions in this report are are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the California Department of Public of Health. We thank the California Department of Public Health Maternal Child and Adolescent Health Division for providing data for these analyses.
ABBREVIATIONS
- Ensembl
- OMIM
- PolyPhen
- PubMed
- dbSNP
- UCSC
Genome Browser http://genome.ucsc.edu/
- Unigene
http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.268041
REFERENCES
- Ackerman KG, Herron BJ, Vargas SO, Huang H, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, Greer JJ, Beier DR. 2 005. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman KG, Pober BR. Congenital diaphragmatic hernia and pulmonary hypoplasia: new insights from developmental biology and genetics. Am J Med Genet C Semin Med Genet. 2007;145C:105–108. doi: 10.1002/ajmg.c.30133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglaj M. Late-presenting congenital diaphragmatic hernia in children: a clinical spectrum. Pediatr Surg Int. 2004;20:658–669. doi: 10.1007/s00383-004-1269-5. [DOI] [PubMed] [Google Scholar]
- Bleyl SB, Moshrefi A, Shaw GM, Saijoh Y, Schoenwolf GC, Pennacchio LA, Slavotinek AM. Candidate genes for congenital diaphragmatic hernia from animal models: sequencing of FOG2 and PDGFRalpha reveals rare variants in diaphragmatic hernia patients. Eur J Hum Genet. 2007;15:950–987. doi: 10.1038/sj.ejhg.5201872. [DOI] [PubMed] [Google Scholar]
- Cáceres S, Cuellar C, Casar JC, Garrido J, Schaefer L, Kresse H, Brandan E. Synthesis of proteoglycans is augmented in dystrophic mdx mouse skeletal muscle. Eur J Cell Biol. 2000;79:173–178. doi: 10.1078/S0171-9335(04)70020-5. [DOI] [PubMed] [Google Scholar]
- Chao R, Nevin L, Agarwal P, Riemer J, Bai X, Delaney A, Akana M, JiminezLopez N, Bardakjian T, Schneider A, FitzPatrick D, Kwok P-Y, Ellgaard L, Gould D, Zhang Y, Malicki J, Baier H, Slavotinek AM. A Male with Unilateral Microphthalmia Reveals a Role for TXNDC10 in Eye Development. Submitted. [DOI] [PMC free article] [PubMed]
- Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol. 1997;136:729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney AD, Qian H, Friedman JM, Marra MA. Use of Affymetrix mapping arrays in the diagnosis of gene copy number variation. Curr Protoc Hum Genet Chapter. 2008;8:13. doi: 10.1002/0471142905.hg0813s59. [DOI] [PubMed] [Google Scholar]
- Elhalaby EA, Abo Sikeena MH. Delayed presentation of congenital diaphragmatic hernia. Pediatr Surg Int. 2002;18:480–485. doi: 10.1007/s00383-002-0743-1. [DOI] [PubMed] [Google Scholar]
- Enns GM, Cox VA, Goldstein RB, Gibbs DL, Harrison MR, Golabi M. Congenital diaphragmatic defects and associated syndromes, malformations, and chromosome anomalies: a retrospective study of 60 patients and literature review. Am J Med Genet. 1998;79:215–225. [PubMed] [Google Scholar]
- Galluzzi P, Venturi C, Cerase A, Vallone IM, Bracco S, Bardelli AM, Hadjistilianou T, Gennari P, Monti L, Filosomi G. Coats disease: smaller volume of the affected globe. Radiology. 2001;221:64–69. doi: 10.1148/radiol.2211010017. [DOI] [PubMed] [Google Scholar]
- Goossens D, Van Gestel S, Claes S, De Rijk P, Souery D, Massat I, Van den Bossche D, Backhovens H, Mendlewicz J, Van Broeckhoven C, Del-Favero J. A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Mol Psychiatry. 2003;8:83–89. doi: 10.1038/sj.mp.4001190. [DOI] [PubMed] [Google Scholar]
- Haugstetter J, Blicher T, Ellgaard L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J Biol Chem. 2005;280:8371–8016. doi: 10.1074/jbc.M413924200. [DOI] [PubMed] [Google Scholar]
- Haugstetter J, Maurer MA, Blicher T, Pagac M, Wider G, Ellgaard L. Structure-function analysis of the endoplasmic reticulum oxidoreductase TMX3 reveals interdomain stabilization of the N-terminal redox-active domain. J Biol Chem. 282:33859–33867. doi: 10.1074/jbc.M706442200. [DOI] [PubMed] [Google Scholar]
- Holder AM, Klaassens M, Tibboel D, de Klein A, Lee B, Scott DA. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet. 2007;80:825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DT, Kilby MD, Sirry H, Barker GM, Roberts E, Davison EV, McHugo J, Whittle MJ. Structural chromosome anomalies in congenital diaphragmatic hernia. Prenatal Diag. 1996;16:1003–1009. doi: 10.1002/(SICI)1097-0223(199611)16:11<1003::AID-PD995>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Kantarci S, Casavant D, Prada C, Russell M, Byrne J, Haug LW, Jennings R, Manning S, Blaise F, Boyd TK, Fryns JP, Holmes LB, Donahoe PK, Lee C, Kimonis V, Pober BR. Findings from aCGH in patients with congenital diaphragmatic hernia (CDH): a possible locus for Fryns syndrome. Am J Med Genet A. 2006;140:17–23. doi: 10.1002/ajmg.a.31025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishioka Y, Thomas M, Wakamatsu J, Hattori A, Sharma M, Kambadur R, Nishimura T. Decorin enhances the proliferation and differentiation of myogenic cells through suppressing myostatin activity. J Cell Physiol. 2008;215:856–867. doi: 10.1002/jcp.21371. [DOI] [PubMed] [Google Scholar]
- Maccarana M, Olander B, Malmström J, Tiedemann K, Aebersold R, Lindahl U, Li JP, Malmström A. Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J Biol Chem. 2006;281:11560–11568. doi: 10.1074/jbc.M513373200. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, Yang G, Kennedy GC, Webster TA, Cawley S, Walsh PS, Jones KW, Fodor SP, Mei R. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- Numanoglu A, Steiner Z, Millar A, Cywes S. Delayed presentation of congenital diaphragmatic hernia. S Afr J Surg. 1997;35:74–76. [PubMed] [Google Scholar]
- Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nürnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernández-Martínez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nürnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–556. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Rüschendorf F, Huang J, Trautmann U, Becker C, Thiel C, Jones KW, Reis A, Nürnberg P. Molecular karyotyping using an SNP array for genomewide genotyping. J Med Genet. 2004;41:916–922. doi: 10.1136/jmg.2004.022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme C, Larrain J, Schonherr E, Henriquez JP, Kresse H, Brandan E. Antisense inhibition of decorin expression in myoblasts decreases cell responsiveness to transforming growth factor beta and accelerates skeletal muscle differentiation. J Biol Chem. 2001;276:3589–3396. doi: 10.1074/jbc.M004602200. [DOI] [PubMed] [Google Scholar]
- Schönherr E, Hausser H, Beavan L, Kresse H. Decorin-type I collagen interaction. Presence of separate core protein-binding domains. J Biol Chem. 1995;270:8877–8883. doi: 10.1074/jbc.270.15.8877. [DOI] [PubMed] [Google Scholar]
- Scott DA, Klaassens M, Holder AM, Lally KP, Fernandes CJ. Genome-wide oligonucleotide-based array comparative genome hybridization analysis of non-isolated congenital diaphragmatic hernia. Hum Mol Genet. 2007;16:424–430. doi: 10.1093/hmg/ddl475. [DOI] [PubMed] [Google Scholar]
- Slater HR, Bailey DK, Ren H, Cao M, Bell K, Nasioulas S, Henke R, Choo KH, Kennedy GC. High-resolution identification of chromosomal abnormalities using oligonucleotide arrays containing 116,204 SNPs. Am J Hum Genet. 2005;77:709–726. doi: 10.1086/497343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM, Moshrefi A, Davis R, Leeth E, Schaeffer GB, Burchard GE, Shaw GM, James B, Ptacek L, Pennacchio LA. Array comparative genomic hybridization in patients with congenital diaphragmatic hernia: mapping of four CDH-critical regions and sequencing of candidate genes at 15q26.1-15q26.2. Eur J Hum Genet. 2006;14:999–1008. doi: 10.1038/sj.ejhg.5201652. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM. Single gene disorders associated with congenital diaphragmatic hernia. Am J Med Genet C Semin Med Genet. 2007;145C:172–183. doi: 10.1002/ajmg.c.30125. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Moshrefi A, LopezJiminez N, Chao R, Mendell A, Shaw GM, Pennacchio LA, Bates MD. Sequence Variants in the HLX Gene at Chromosome 1q41-1q42 in Patients with Diaphragmatic Hernia. Clin Genet. 2009;75:429–439. doi: 10.1111/j.1399-0004.2009.01182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Osumi N. Identification of a novel type II classical cadherin: rat cadherin19 is expressed in the cranial ganglia and Schwann cell precursors during development. Dev Dyn. 2005;232:200–208. doi: 10.1002/dvdy.20209. [DOI] [PubMed] [Google Scholar]
- Torfs CP, Curry CJ, Bateson TF, Honoré LH. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. 1992. [DOI] [PubMed] [Google Scholar]
- Yuan W, Rao Y, Babiuk RP, Greer JJ, Wu JY, Ornitz DM. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci USA. 2003;100:5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]