Abstract
The widespread adoption of increasingly sophisticated forms of information technology has paralleled the increase in rapid and far-reaching international travel. The emergence and global spread of the 2009 pandemic influenza A (H1N1) virus illustrated not only the hazards of an interconnected world, but also the powerful role of new methods for detecting, tracking, and responding to infectious diseases.1 Although formal reporting, surveillance, and response structures remain essential to protecting public health,2 a new generation of freely accessible, online, and real-time informatics tools for disease tracking are expanding the ability of public health professionals to detect weak signals across borders and to raise earlier warnings of emerging disease threats.3–5
THE HEALTHMAP H1N1 SYSTEM
HealthMap (http://healthmap.org) is an example of a new effort in transparent, global, public health surveillance.6,7 The HealthMap system combines automated, around-the-clock data collection and processing with expert review and analysis to aggregate reports according to type of disease and geographic location. HealthMap sifts through large volumes of information on events, obtained from a broad range of online sources in multiple languages, to provide a comprehensive view of ongoing global disease activity through a freely available Web site.
To enhance the situational awareness of public health professionals, clinicians, and the general public regarding the global spread of 2009 H1N1 influenza infection, HealthMap partnered with the Journal to create the H1N1 interactive map (available at www.healthmap.org/nejm) as part of the Journal’s H1N1 Influenza Center (http://h1n1.nejm.org). This interactive map used the Health-Map infrastructure to provide information about outbreaks. The information was obtained from both informal sources (e.g., the news media, mailing lists, and contributions from individual users) and formal announcements (primarily from the World Health Organization [WHO], the Centers for Disease Control and Prevention, and the Public Health Agency of Canada).1 Visitors to the site could filter reports according to suspected or confirmed cases or deaths and view a chosen time interval to show the spread of disease. During the two major waves of the H1N1 pandemic, Health-Map collected more than 87,000 reports from both informal and official sources (43,738 reports during the first wave of infection, from April 1 to August 29, 2009, and 43,366 reports during the second wave of infection, from August 30 to December 31, 2009).
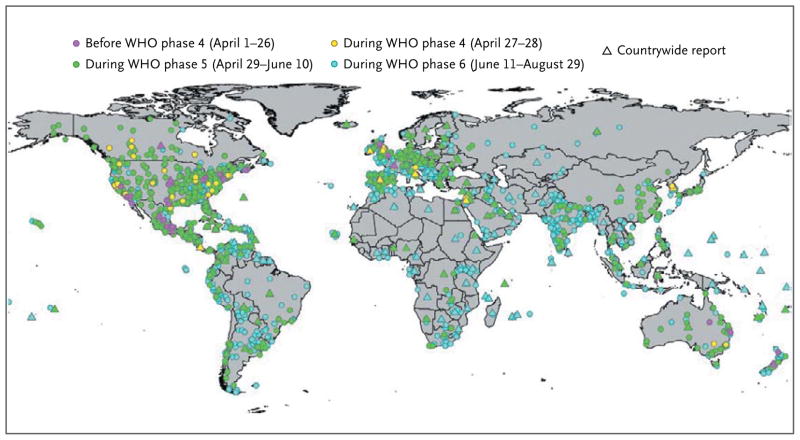
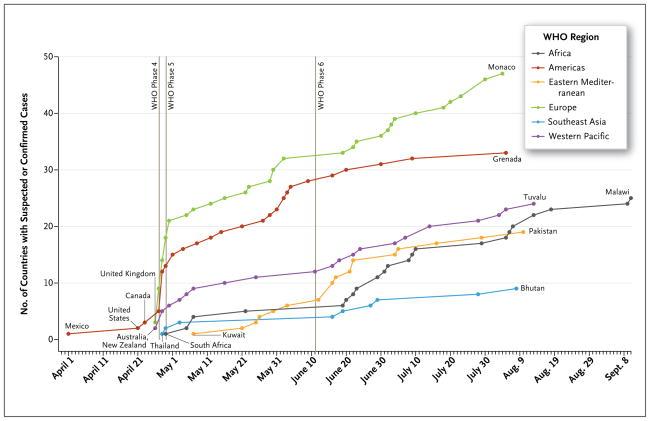
The worldwide spread of the H1N1 virus is shown in Figure 1 and the interactive map available with the full text of this article at NEJM.org. The geographic location was recorded for all reports entered into the HealthMap system, allowing for easy tracking of the regional and global proliferation of H1N1 influenza. HealthMap also identified the increasing number of countries over time with informal reports of suspected or confirmed cases, according to WHO regions and pandemic phases (Fig. 2 and the interactive graphic available at NEJM.org). By the end of WHO pandemic phase 4 (April 28), there had already been reports of either suspected or confirmed cases in 32 countries. During the initial phases of the pandemic, the spread of the virus to new countries was most rapid in Europe and the Americas. This spread gradually slowed over time to about one country per day through the end of phase 5 (June 10), and it further diminished to one country per 2 days in phase 6, possibly because of the implementation of containment measures and the early involvement of countries with a high volume of air travel. Although air travel may have driven the intercontinental spread of the virus, other migration patterns probably influenced the spread of the virus within continents. In Europe, for example, countries that are hubs for international travel (e.g., France and the United Kingdom) reported infections earlier than countries with less international traffic (e.g., Eastern European nations). This trajectory was also affected by public health capacity and the willingness to report cases.
Figure 1. Distribution of Reports of Cases of H1N1 Influenza.
Data were collected by HealthMap during the first wave of the 2009 epidemic (April 1 through August 29, 2009) and were classified according to World Health Organization (WHO) phase.
Figure 2. Timeline of Informal Reporting of Suspected or Confirmed Cases of H1N1 Influenza around the World and in Each of the WHO Regions, 2009.
WHO denotes World Health Organization.
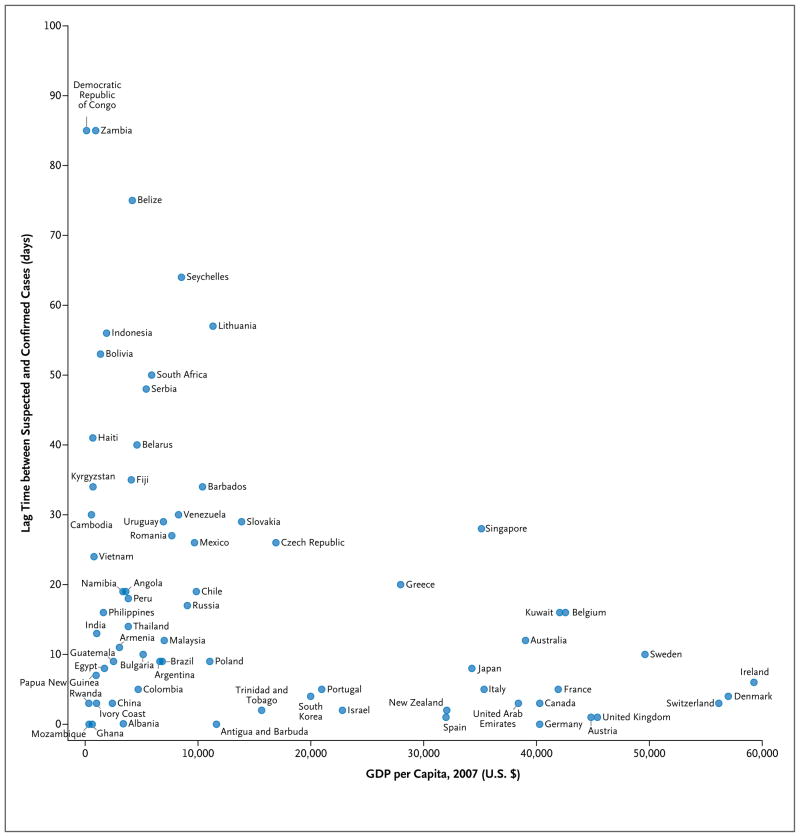
Using informal reports, we explored the time difference between the issue dates of reports of suspected cases and confirmed cases of H1N1 influenza according to country. Overall, there was a median of 12 days (95% confidence interval [CI], 9 to 18) between reports of suspected and confirmed cases among the countries that reported these data. Lag times varied widely according to country. Large delays (up to 85 days) were probably due to a multitude of factors, including the reporting capacity of public health laboratories. We examined the association of this difference with the 2007 national gross domestic product (GDP) per capita (based on data from the United Nations Development Program), an approximate indicator of the strength of the public health infrastructure (Fig. 3 and the interactive graphic available at NEJM.org). Countries with a high GDP per capita tended to have shorter lag times between issue dates of reports of suspected and confirmed cases (Pearson correlation coefficient, −0.4; 95% CI, −0.6 to −0.2), but there was a wide variation in lag times among less affluent nations.
Figure 3. The Relationship between Gross Domestic Product (GDP) per Capita and the Time Lag between the First Informal Report of a Suspected Case of H1N1 Influenza and a Confirmed Case in Each Country.
Only countries in which the first report of a suspected case of infection occurred before or on the same day as the first confirmed case report were included in the analysis. Thus, several countries, including the United States, were excluded.
Many factors can obstruct the timely flow of information; these include deficiencies in the public health infrastructure that may delay case confirmation and political pressures that can restrict the dissemination of information that is likely to affect trade and tourism adversely. The lag may also reflect the intense media coverage that accompanied the emergence of the 2009 H1N1 influenza. In the earliest stages of the pandemic, a proportion of the suspected cases that were reported were probably determined subsequently to be negative; this would create longer lag times between the first report of a suspected case and the first report of a confirmed case. Early reports of suspected cases may also indicate that the country had a reactive and advanced public health system in which almost every patient with suspected H1N1 infection was tested.
IMPLICATIONS, LIMITATIONS, AND FUTURE DIRECTIONS
During the 2009 H1N1 influenza pandemic, non-traditional surveillance sources such as Internet news sources provided new public health data. Collectively, these sources overcame certain limitations of traditional surveillance systems, including reporting delays, inconsistent population coverage, and a poor sensitivity to detect emerging diseases.
Internet news sources, collected and filtered by systems such as HealthMap, can be used effectively to detect sentinel outbreaks that may herald the onset of an epidemic or pandemic and to place them in context. Indeed, unusual outbreaks (e.g., deaths from respiratory disease in the young) attract considerable media coverage. As public health professionals work to avoid panic, they can still capitalize on the media’s tendency to amplify concern through intense reporting of evolving health stories. More broadly, the tools and approaches that we have described demonstrate the ability of “Web 2.0” dissemination systems (i.e., applications for the interactive sharing of information) in concert with online journal resources to communicate real-time public health data. Beyond simply detecting sentinel events, these tools can potentially extract and release information from more isolated areas where traditional public health data remain confined within the country. In 2005, the WHO adopted a revised version of the International Health Regulations to outline more rigorous reporting standards and to encourage strengthened public health surveillance and response capacity. Together with the 2005 International Health Regulations, which went into effect in 2007, these modern information-sharing tools improve information transparency and facilitate a rapid and coordinated global public health response.
Greater interconnectivity between public health stakeholders and the public can broaden the scope of surveillance by encouraging greater participation in reporting activities. Although the bulk of the HealthMap data on H1N1 were collected from online news media, HealthMap is continuing to expand to draw from additional informal sources, including blogs, Twitter, Facebook, and direct reports by users. Persons reporting on their own illnesses can offer important insights into an epidemic, but the lack of confirmation of the diagnosis presents challenges for validation, filtering, and public health interpretation.
Although surveillance activities with the use of innovative Internet-based approaches may overcome some limitations of traditional surveillance systems, the integration of new and traditional approaches offers the greatest promise for future surveillance of influenza and other emerging diseases. These Internet-based data streams as well as new efforts in syndromic surveillance from repurposed clinical data fill critical gaps in traditional approaches by identifying early events (e.g., first cases or early reports of community-level transmission), but they cannot completely describe the epidemiology and global impact of an emerging threat. Traditional surveillance is still necessary to estimate morbidity, mortality, and shifts in the incidence of disease according to age, other demographic factors and shifts, or changes in case fatality rates. A combination of syndromic surveillance and population sampling has been suggested as being important for estimating the burden in real time.8 The grassroots integration of existing local public health efforts is another promising direction.9 The Distribute project (www.isdsdistribute.org), for example, brings together data on visits to emergency departments for influenza-like illness. These data are obtained from more than 30 public health departments that cover more than half the U.S. population.
Syndromic surveillance is the leading edge of what will almost certainly evolve into real-time surveillance of data from electronic medical records. Recent trends in health informatics, including increased adoption of electronic medical records and personally controlled health records, are creating a wealth of electronic data that can be used to monitor population health.10 These types of technology can potentially improve the quality of surveillance data and also enable real-time communication between clinicians, public health departments, and the general population. These efforts have been under way only in leading centers in a limited number of countries, and thus capacity building remains an important global public health priority.
In the future, these broad-reaching, integrated techniques may prove to be valuable for situational awareness in a range of rapidly evolving scenarios beyond epidemics of infectious disease. Overall, the 2009 H1N1 influenza pandemic presented an important test of new disease-surveillance systems. The use of data that were collected, coded, and analyzed through the Journal’s Health-Map system shows how such systems, which were built largely around readily available informal sources, can provide both early warnings and an ongoing operating picture of the patterns of disease spread. Further, by providing a highly accessible picture of the emerging pandemic, the Journal’s H1N1 Influenza Center played a key role in rapidly disseminating information to the public.
Acknowledgments
Supported by grants (G08LM009776-01A2) from the National Library of Medicine, the National Institutes of Health, Google.org, and the Canadian Institutes of Health Research.
Footnotes
Dr. Brownstein and Mr. Freifeld report being cocreators of the HealthMap system. Disclosure forms provided by the authors are available with the full text of this article at NEJM.org
References
- 1.Brownstein JS, Freifeld CC, Madoff LC. Influenza A (H1N1) virus, 2009 — online monitoring. N Engl J Med. 2009;360:2156. doi: 10.1056/NEJMp0904012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novel Swine-Origin Influenza A (H2N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [Erratum, N Engl J Med 2009;361:102.] [DOI] [PubMed] [Google Scholar]
- 3.Heymann DL, Rodier GR. Hot spots in a wired world: WHO surveillance of emerging and re-emerging infectious diseases. Lancet Infect Dis. 2001;1:345–53. doi: 10.1016/S1473-3099(01)00148-7. [DOI] [PubMed] [Google Scholar]
- 4.Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection — harnessing the Web for public health surveillance. N Engl J Med. 2009;360:2153–5. doi: 10.1056/NEJMp0900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartley DM, Nelson NP, Walters R, et al. The landscape of international event-based biosurveillance. Emerging Health Threats J. 2010;3:e3. doi: 10.3134/ehtj.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownstein JS, Freifeld CC, Reis BY, Mandl KD. Surveillance Sans Frontières: Internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med. 2008;5(7):e151. doi: 10.1371/journal.pmed.0050151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freifeld CC, Mandl KD, Reis BY, Brownstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J Am Med Inform Assoc. 2008;15:150–7. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipsitch M, Hayden FG, Cowling BJ, Leung GM. How to maintain surveillance for novel influenza A H1N1 when there are too many cases to count. Lancet. 2009;374:1209–11. doi: 10.1016/S0140-6736(09)61377-5. [DOI] [PubMed] [Google Scholar]
- 9.Diamond CC, Mostashari F, Shirky C. Collecting and sharing data for population health: a new paradigm. Health Aff (Millwood) 2009;28:454–66. doi: 10.1377/hlthaff.28.2.454. [DOI] [PubMed] [Google Scholar]
- 10.Mandl KD, Kohane IS. Tectonic shifts in the health information economy. N Engl J Med. 2008;358:1732–7. doi: 10.1056/NEJMsb0800220. [DOI] [PubMed] [Google Scholar]