Abstract
Synchronous neurotransmission is triggered when Ca2+ binds to synaptotagmin 1, a synaptic vesicle protein that interacts with SNAREs and membranes. We used single-molecule FRET between synaptotagmin’s two C2 domains to determine that their conformation consists of multiple states with occasional transitions, consistent with domains in random relative motion. SNARE binding results in narrower intra-synaptotagmin FRET distributions and less frequent transitions between states. We obtained an experimentally determined model of the elusive synaptotagmin 1–SNARE complex by using a multi-body docking approach with 34 FRET-derived distances as restraints. The Ca2+-binding loops point away from the SNARE complex, so they could interact with the same membrane. The loop arrangement is similar to that of the crystal structure of SNARE-induced Ca2+ bound synaptotagmin 3, suggesting a common mechanism by which the interaction between synaptotagmins and SNAREs plays a role in Ca2+-triggered fusion.
Keywords: membrane fusion, neurotransmitter release, protein-protein interactions, synaptic vesicle, single molecule FRET
Ca2+-induced membrane fusion of synaptic vesicles at synapses is the central phenomenon that results in triggered inter-neuron signaling. The membrane protein synaptotagmin 1 (Syt1) is the Ca2+ sensor for synchronous neurotransmitter release1,2. Highly coordinated interactions among synaptotagmin, SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins and other neuronal factors are required to create robust and adaptive neural circuits3,4. Syt1 is primarily located on synaptic vesicles and contains two independent C2-type Ca2+-sensing domains5 (termed C2A and C2B, respectively) that are connected by a linker (the fragment containing both domains is designated C2AB). Syt1 interacts with both anionic membranes and SNARE complexes, and both interactions are physiologically relevant1,6.
A general model has emerged where inhibitory and activating interactions among synaptotagmin, complexin, and the SNARE complex (that juxtaposes synaptic vesicles and the plasma membrane) yield a membrane fusion stall that is released by Ca2+ influx following an action potential7,8. Understanding the molecular mechanism underlying the release of the stall requires knowledge of the structures and dynamics of the complexes formed by these proteins. However, the structure of the complex between synaptotagmins and SNAREs has remained elusive.
Here we used single-molecule fluorescence resonance energy transfer (smFRET) between the C2 domains of Syt1 to determine that their configuration consists of multiple states with occasional transitions. Each of these stable FRET states is consistent with the domains being in random relative motion leading to a well-defined average FRET emission at our 100 msec temporal resolution. SNARE binding results in narrowing of the intra-synaptotagmin FRET distributions along with fewer transitions between states, consistent with substantially reduced conformational variability.
Distance-constrained smFRET triangulation methods have been successfully used to localize individual domains within several biomolecular assemblies9–11. Here we generalized this approach to the Syt1–SNARE multi-component system using a multi-body docking strategy guided by 34 FRET-derived distance restraints. We obtained a robust model derived from the major smFRET populations for the Syt1–SNARE complex. This is the first experimentally derived model of a synaptotagmin–SNARE complex, which has resisted crystallization and nuclear magnetic resonance analyses12. The low concentrations of single molecule studies prevented aggregation of the complex and thus enabled us to determine this smFRET-derived model in the presence of 1 mM Ca2+. We observe occasional transitions between different FRET efficiency states and some of the smFRET efficiency distributions have a multimodal appearance, suggesting that the Syt1–SNARE complex is conformationally variable. In our model, the Ca2+-binding loops point away from the SNARE complex and could engage the same membrane. The loop arrangement is similar to that of the structure of the SNARE-induced Ca2+ bound synaptotagmin 3 (Syt3)13, suggesting a common molecular mechanism by which the synaptotagmin–SNARE interaction plays a role in Ca2+-triggered vesicle fusion.
The utility of single molecule experiments is well established for studies of protein folding and enzymatic processes14. Our study of membrane-reconstituted synaptic vesicle proteins demonstrates that smFRET measurements can also determine structural models of weakly bound or flexible multi-component systems.
RESULTS
FRET efficiency distribution of isolated Syt1
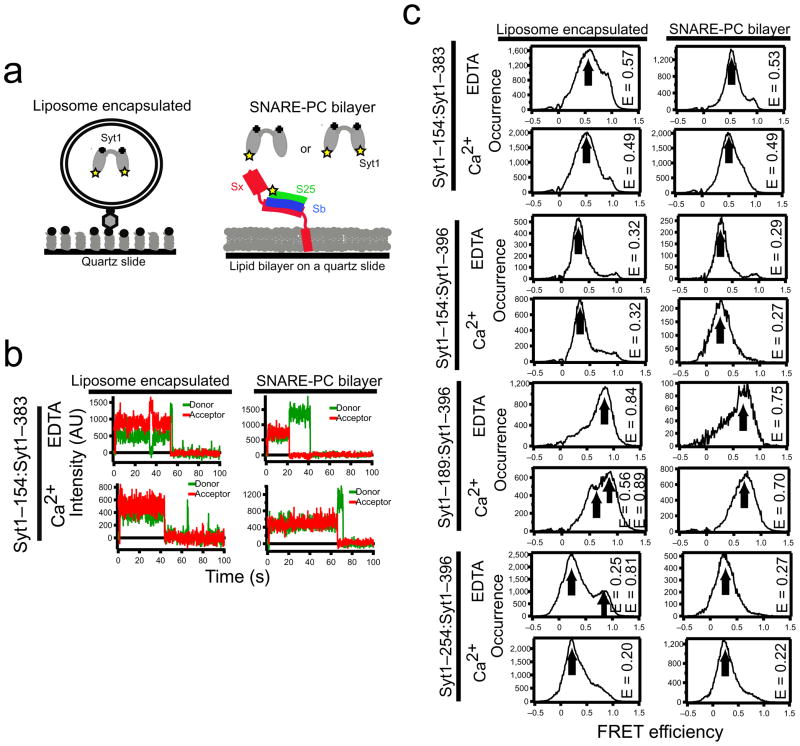
Because isolated C2A and C2B conformations are unchanged upon binding Ca2+, the relative arrangement of the C2 domains largely determines the C2AB configuration15–18, which influences Syt1 function19. We therefore introduced specific dye labeling sites into each of the C2 domains in the Syt1 C2AB fragment (Fig. 1a) to allow FRET to report interactions between C2A and C2B. Individual doubly-labeled C2AB molecules were encapsulated within 100 nm diameter biotinylated liposomes that were tethered to a streptavidin-exposed biotinylated quartz surface or added in solution over supported lipid bilayers containing SNARE complex (Fig. 1a). smFRET efficiencies were measured as IA/(ID + IA) (using background and leakage corrected donor and acceptor intensities: ID and IA, respectively).
Figure 1.
smFRET reveals Syt1 conformations. (a) Surface immobilization schemes using liposome encapsulation (left) or supported lipid bilayers (right). Sx is syntaxin; S25 is SNAP-25; Sb is synaptobrevin; Syt1 is synaptotagmin. 50 mM NaCl and 1 mM Ca2+ or 1 mM EDTA were used for all experiments. (b) Intensity time traces for single molecules of Syt1 with labels attached to residues 154 and 383. Note the transition between mid and high FRET in the upper left graph. The sudden downward transitions to zero acceptor intensity are due to dye photobleaching. (c) Histograms of smFRET measurements using Syt1 with donor in one C2 domain and acceptor in the other (label pairs are indicated on the left edge). Arrows indicate the peak value (designated E in the figure) from Gaussian fits. For comparison, converting Cα separations in the Syt1 crystal structure 22 to FRET using Ro=5.55 yields: 0.99 (residues 154 and 383), 0.77 (residues 154 and 396), 0.95 (residues 189 and 396), 0.75 (residues 254 and 396). See also Supplementary Fig. 9.
First we characterized C2AB conformations in the absence of the SNARE complex. C2AB with dyes attached at residues 154 (C2A) and 383 (C2B) revealed FRET emissions at intermediate levels (0.57) that were stable for many seconds (Fig. 1b). Measurements for 1 mM Ca2+ or 1 mM EDTA exposed molecules were accumulated into histograms (Fig. 1c, left column). For the labeling site pair 254–396 (corresponding to the 410–554 label pair of Syt313) the distribution is similar to that of isolated Syt313 except for a second minor peak at high FRET (Fig. 1c). Small changes in the smFRET efficiency distribution resulted upon exposure to Ca2+. For example, the ~0.08 FRET efficiency decrease when switching from EDTA to Ca2+ for the 154:383 label pair (Fig. 1c) is significant (Fig. 2a). However, FRET emission from yet another label pair, 154–396, was around 0.32 with no dependence on Ca2+ exposure.
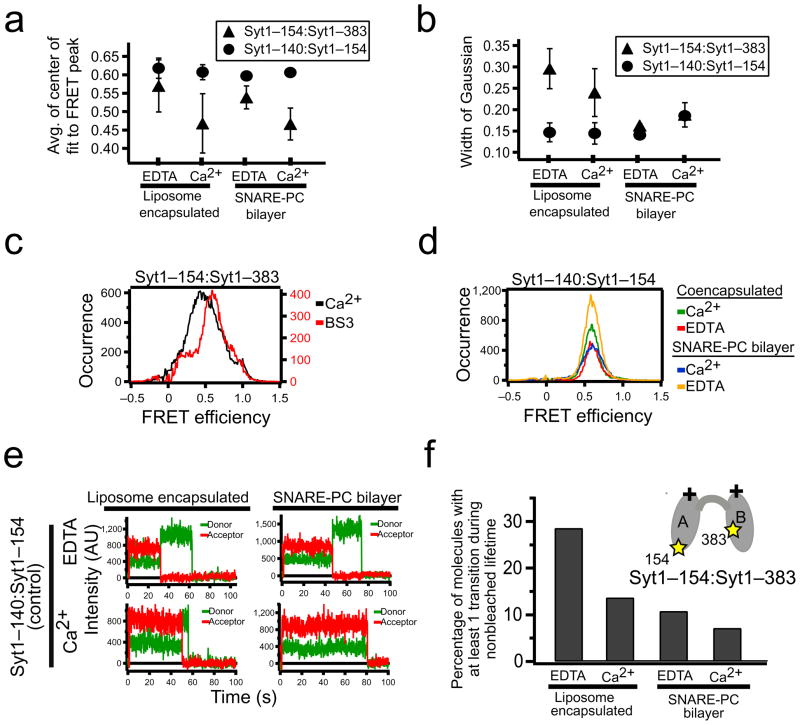
Figure 2.
smFRET shows that Syt1 conformations are dynamic. (a,b) Experiments as in Fig. 1c were repeated three to five times for each indicated condition. The average values of the center and width of the Gaussian fit to the FRET efficiency distribution peak are shown. Error bars indicate two standard deviations. (c) Application of the bi-functional NHS-ester crosslinker (BS3) before liposome encapsulation to Syt1 C2AB with labels attached to residues 154 (C2A) and 383 (C2B) narrows the width of the smFRET efficiency distribution. (d) Control experiments using dye labels that both reside in C2A (residues 140 and 154) for encapsulated or SNARE bilayer immobilized experiments at 50 mM NaCl, and Ca2+ or EDTA, as indicated. (e) smFRET donor and acceptor intensity trajectories in control experiments as in (d) without (top) and with (bottom) Ca2+, using liposome encapsulation (left) and supported bilayers (right). No transitions other than photobleaching were observed. (f) Percent of the molecules with at least one transition between non-zero FRET efficiency levels during the 90 second observation period for Syt1 with labels attached to residues 154 (C2A) and 383 (C2B) under indicated conditions.
FRET efficiency distribution of SNARE-bound Syt1
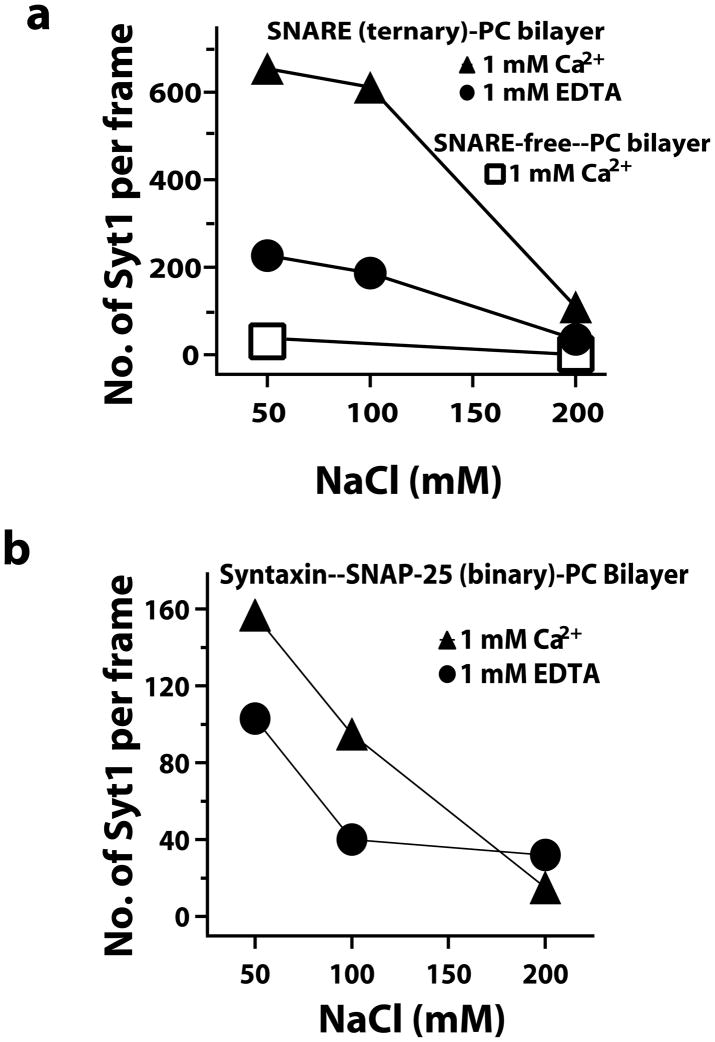
We next used smFRET to characterize the Syt1 conformation when interacting with the assembled SNARE complex (Fig. 1a). Syt1 bound minimally to protein-free, phosphatidylcholine bilayers in both Ca2+ and EDTA buffers (Fig. 3a, squares). Syt1 bound robustly when preassembled, purified SNARE complexes were included in the bilayers via the syntaxin transmembrane domain (Figs. 1a and 3). The binding probability increased in presence of Ca2+ and decreased with increasing NaCl (Fig. 3a), in agreement with other reports20, and we found similar behavior for the interaction with binary SNARE complexes (Fig. 3b). The bound states lasted many seconds in 50 mM NaCl so this condition was used for all subsequent smFRET measurements. smFRET between the dyes in the C2A and C2B domains was measured for the SNARE complex-bound synaptotagmin with the same label sites used for encapsulation studies (Fig. 1c, right column).
Figure 3.
Syt1 binds to SNARE complexes reconstituted in 100% phosphatidylcholine (PC) bilayers more strongly at decreased ionic strength of the buffer. (a) The number of Syt1 bound per 5000 μm2 of PC bilayer containing equal reconstitutions of SNARE complexes is increased at lower ionic strength (circles). Ca2+ further enhances the binding (triangles). Little binding occurred in the absence of SNARE complex (squares). Those few molecules that bound to protein free bilayers diffused freely in the plane of the bilayer. Syt1 bound to SNARE complex containing bilayers exhibit much lower diffusivity. (b) Identical experiments using Syt1 binding to bilayers containing syntaxin– SNAP-25 binary complexes also show increased binding at decreased ionic strength, but the presence of Ca2+ has a smaller effect than for the ternary SNARE complex. The densities of the bilayer-incorporated complexes are 0.4 ternary SNARE complex/μm2 (a) and 0.7 binary SNARE complex/μm2 (b) (Online Methods). Considering the concentration of Syt1 was below 20 nM for all incubations, Syt1 binding to the protein containing surfaces is not saturating.
Comparison of FRET efficiency distributions
The smFRET distribution widths varied for different C2AB-spanning label pairs and generally become narrower upon SNARE complex binding (Figs. 1c and 2b). The large differences of the C2 domain arrangement of available C2AB crystal structures13,21,22 and NMR23 and EPR24 spectroscopic studies of Syt1 C2AB in solution suggest that the two C2 domains are in random relative motion when not bound to the SNARE complex. Attempting to limit possible inter-domain motions, we added bi-functional NHS-ester cross linker to the 154:383 sample before encapsulation without SNARE complex. The width of the smFRET distribution for crosslinked Syt1 was significantly narrower than for the uncrosslinked Syt1 (Fig. 2c). We also performed control experiments in which both the donor and acceptor were positioned within the C2A domain (Figs. 2a,b,d,e and Supplementary Fig. 1a,b). NMR studies have verified that isolated C2A is stable in a nearly identical conformation to that observed in C2AB structures23. smFRET measurements of this control construct (label sites 140–154) revealed a dominant peak centered at 0.60, consistent with the dye–dye separation of 5.4 nm calculated from the Syt1 C2AB crystal structure17,22 (see Supplementary Note for the calculation of the dye dye distances from the crystal structure) and a Förster radius of 5.1 nm for the Alexa555:Alexa647 dye pair, assuming dye parameters measured under standard conditions25 or an empirically determined Förster radius of 5.55 nm for the dyes conjugated to the protein (Supplementary Note). This smFRET efficiency distribution of the control construct did not change upon Ca2+ exposure or upon SNARE complex binding (Fig. 2d). The widths of the crosslinked 154:383 labeled synaptotagmin were close to the widths of the control samples with labels in the same domain, supporting the notion that the wider histograms of unbound Syt1 arise from conformational fluctuations in the relative arrangement of C2A and C2B. This interpretation is consistent with molecular dynamics simulations of C2AB fragments where both C2 domains were modeled as rigid bodies connected by a flexible linker (Supplementary Fig. 2). smFRET measurements of randomly fluctuating single stranded DNA reported similar well defined emission peaks26.
The repeatability of smFRET distribution peak position and width measurements was assessed for the 154:383 label pair (Figs. 2a,b). The smFRET peak location did not significantly shift upon SNARE complex binding in the presence or absence of Ca2+ (Fig. 2a, triangles). A shift due to Ca2+ exposure occurs independent of the SNARE binding. No changes in smFRET distributions are observed for similar experiments using the control pair with both dyes in C2A (140:154) (Fig. 2a, circles and Fig. 2d). In contrast to the peak position, smFRET distribution widths significantly narrowed for SNARE-bound Syt1 compared to isolated Syt1 (Fig. 2b). The SNARE-bound 154:383 smFRET distributions had widths more similar to the 140:154 C2A control labels and to the cross-linked Syt1 construct than to the isolated Syt1, which exhibited larger widths, supporting the notion that SNARE complex binding substantially reduces the motion of the C2 domains.
Conformational states of Syt1
The existence of multiple conformational states for Syt1 is supported by observations that some molecules spontaneously transitioned between two or more non-zero FRET levels during the observation period (Figs. 1b (top left) and 2f); similar transitions were observed for all label combinations involving both C2 domains. Ca2+-free Syt1 (containing EDTA) exhibited about twice as many transitions between intermediate and high FRET levels during a 100 second observation period compared to the experiment prepared with Ca2+ (Fig. 2f). For control experiments with both dyes in C2A, switching was almost non-existent (<0.1%). Thus, the observed transitions are not an artifact of the labels or of the experimental setup, but are due to intrinsic properties of Syt1 C2AB. Binding of SNARE complex reduces the frequency, but does not eliminate the transitions (Fig. 2f).
The individual smFRET efficiency measurements are averages over a 100 msec integration time. It is thus possible that the stable FRET efficiency states are each themselves dynamic with fast motions (faster than 100 msec) averaging to a single FRET efficiency value. These different dynamic states could represent different conformations of the linker connecting the two flexible C2 domains. Such dynamic states would be consistent with the observed lack of cross-peaks between the C2 domains in NMR HSQC spectra23. Alternatively, if one of the FRET efficiency states is a specific arrangement of the C2 domains (such as the one observed the Syt1 C2AB crystal structure22) that is stabilized by interactions between the domains, then the small population (typically less than 10%) would have prevented detection in ensemble NMR or EPR studies23,24. SNARE complex binding significantly reduced the transitions (Fig. 2f) indicating a stabilizing effect of the SNARE complex on the Syt1 conformational dynamics. However, occasional transitions are still observed in the presence of SNARE complex (Supplementary Fig. 3).
SmFRET determines a model of the Syt1–SNARE complex
We used smFRET between membrane-reconstituted SNARE complexes and Syt1 bound from solution to determine an experimentally derived model of the Syt1–SNARE complex. Acceptor labeled ternary SNARE complex was preassembled in solution, purified by denaturant washes to eliminate antiparallel assembly27, and reconstituted into supported lipid bilayers formed from 100% phosphatidylcholine via the transmembrane domain of syntaxin. Donor-labeled, soluble C2AB was then added in solution above the bilayer (Fig. 1a, right panel), leading to FRET emissions upon binding to SNAREs in the bilayer. Our previous studies attempting to determine the conformation of Syt1 binding to membrane tethered SNAREs were limited by the brevity of bound state lifetime in 200 mM NaCl buffers28. In light of the enhanced binding of synaptotagmin to SNARE complex at decreased NaCl concentrations20 (Fig. 3), we conducted FRET experiments in solutions containing 50 mM NaCl. Under these conditions, nonspecific binding of Syt1 to the bilayer was minimal in the absence of SNARE complexes (Fig. 3). The few Syt1 molecules that were bound directly to the bilayer by interactions not involving the SNARE complex could be confidently differentiated from the SNARE-bound Syt1 by their dramatically different mobility as SNARE complexes diffuse much more slowly in these supported bilayers27.
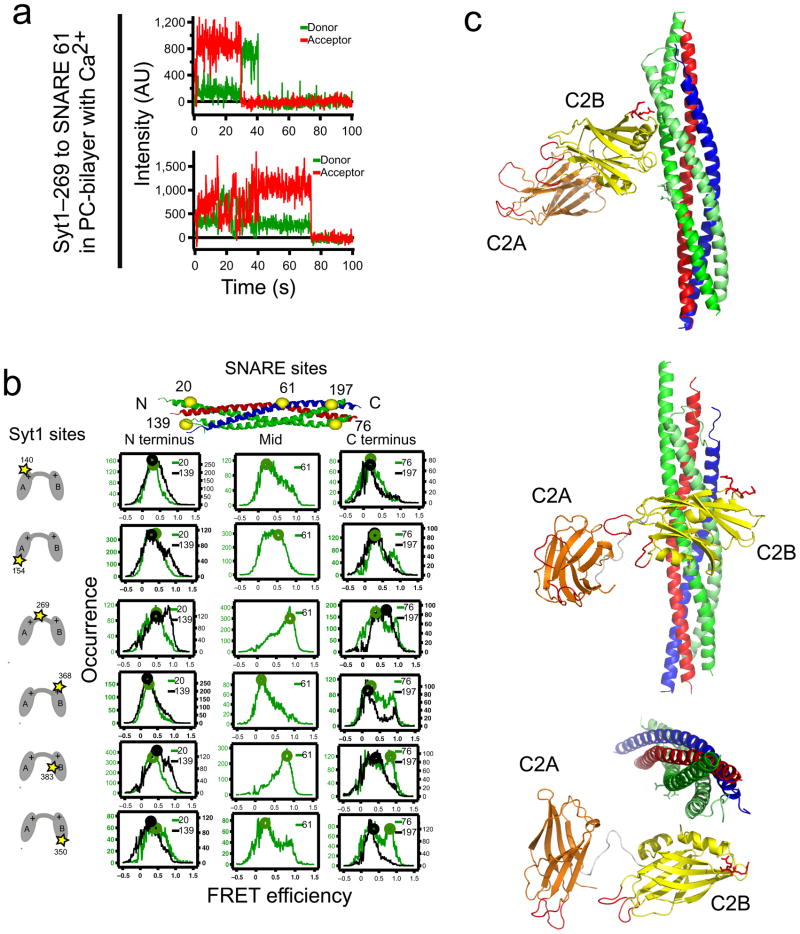
An extensive set of pairs of label attachment sites were used to measure smFRET between C2AB Syt1 fragments and bilayer reconstituted ternary SNARE complexes (Fig. 4a). The SNARE complex was labeled at two N-terminal sites (Fig. 4b, left column), a central site (Fig. 4b, center column), and two C-terminal sites (Fig. 4b, right column). Syt1 was labeled at a variety of sites in C2A, C2B and the linker between C2A and C2B (total of six sites, Fig. 4b, rows). We observed distinct populations of FRET levels that could be fit to sums of Gaussian functions centered at FRET efficiency values ranging from 0.1 to 0.9 that had widely variable widths spanning 0.11 to 0.44 (Supplementary Fig. 4). We used the values at the centers of these Gaussian fits to represent distinct binding states. Fluorescence anisotropy, quantum yield and detection efficiencies must be known for the dyes conjugated to each labeling site to confidently convert smFRET measurements to absolute distances. This is a challenging problem that is a topic of current interest10,29,30. Here we used our control C2A FRET pair to empirically determine the FRET efficiency-to-distance conversion parameters for the Alexa555:Alexa647 dye pair as Ro = 5.55 nm and γ = 1 (Supplementary Note). We verified these conversion parameters by testing three different control labeling site pairs within a single C2 domain (two pairs in C2A and one pair in C2B). Furthermore, there are no significant changes of the γ factor across label site pairs involving all of the label sites used (Supplementary Note, Supplementary Fig. 5 and Supplementary Table 1). Our empirical approach to determine Ro implicitly accounts for orientation averaging effects of the relative alignment of the fluorophores that contribute to the Förster radius so molecular dynamics simulations of orientation effects10 were not performed.
Figure 4.
smFRET-derived model of the Syt1–SNARE complex. (a) smFRET efficiency time traces and (b) smFRET histograms of labeled Syt1–SNARE complex. We used combinations of six different single donor label sites in Syt1 (indicated in the left column) (residues 140 and 154 in C2A; residue 269 in the linker; residues 368, 383, and 350 in C2B) and single acceptor label sites in the SNARE complex (indicated at the top) at the N-terminus (SNAP-25 residues 20 and 139), C-terminus (SNAP-25 residues 76 and 197), and the center (synaptobrevin residues 61) of the SNARE complex. Circles indicate the smFRET efficiency values from which distances were derived for the docking calculations. (c) Shown is the best model (in terms of the rms distance range deviation (rmsdrd) from the borders of the restraining square-well potential, rmsdrd=0.38 nm, see Supplementary Note). Coordinates of the model are available as Supplementary Data. Three rotated views are displayed. C2A is colored orange, C2B yellow, SNAP-25 green, synaptobrevin blue, and syntaxin red. The conserved arginine residues 398 and 399 in C2B are shown as red sticks, and the Ca2+-binding loops are colored red. The docking calculation of the syt1–SNARE complex involves 34 independent distance measurements constraining 12 independent parameters in possible models (3 rigid bodies × (3 rotation + 3 translational degrees of freedom) − (6 over-counting for coupled motions in global translations/rotations) = 12 degrees of freedom).
34 smFRET-derived distances for Syt1 bound to the SNARE complex in the presence of 1 mM Ca2+ (30 label pairs between Syt1 and the SNARE complex and four label pairs within Syt1 C2AB) were used as target restraints for exhaustive docking calculations using simulated annealing where the SNARE complex and the C2 domains were kept rigid, while the torsion angles of the linker connecting the two C2 domains (residues 263–272) were flexible. The dye center positions relative to the corresponding rigid bodies were obtained from separate molecular dynamics simulations and used as pseudoatoms for the distance restraints in the docking calculations (Supplementary Note). FRET histograms for most of the label pairs were characterized by a single dominant state (Fig. 4b), which was converted to the distance between dye center locations using the empirically determined Ro and γ. A few of the label pairs produced FRET efficiency distributions with unusually wide peaks or double peaks that indicate some degree of heterogeneity in the configuration of the Syt1–SNARE complex. We extracted the major population of the FRET efficiency distribution (presumed to be the corresponding most probable configuration of the Syt1–SNARE complex) based on the observation that all histograms were well fit by sums of two Gaussian functions (Supplementary Fig. 4). For most label pairs (26 out of 34) the major Gaussian peak contained >70% of the population. We performed an initial docking calculation using only this subset of the distance restraints, and then compared the top solution of this preliminary docking simulation to the remaining eight more ambiguous label pairs. The distance derived from the two fitted FRET peaks for these remaining eight pairs was selected that was closest to the distance calculated from the preliminary model. We then repeated the docking calculation using all 34 assigned distance restraints (additional details can be found in the Supplementary Note).
A cluster analysis of the ~ 900 models obtained from the docking calculations was performed; each cluster was defined by an root mean square difference (RMSD) of less than 3 Å from a central node (Supplementary Note). The best model (in terms of distance restraint satisfaction) is shown in Fig. 4c (see also Table 1 and Supplementary Fig. 6), and the next best nine models are shown in Supplementary Fig. 7. Out of these nine next best models, seven exhibit unlikely configurations since the Ca2+-binding loops directly contact the C2B domain (note that bound Ca2+ and electrostatics were not used in the docking calculations). Another model is essentially a mirror image of the C2AB configuration with respect to the best structure with similar elements forming the interface between SNARE complex and Syt1.
Table 1.
Comparison of smFRET-derived distances used for the docking calculations and distances calculated from the top model1
| SNARE(acceptor) : syt(donor) | Distance (nm) | ||
|---|---|---|---|
| Calculated from model | smFRET derived | Difference | |
| S25-20:Syt1-140 | 7.0 | 6.4 | 0.6 |
| S25-139:Syt1-140 | 6.0 | 6.5 | −0.5 |
| S25-20:Syt1-154 | 5.6 | 5.9 | −0.3 |
| S25-139:Syt1-154 | 7.3 | 6.5 | 0.8 |
| S25-20:Syt1-269 | 5.1 | 5.5 | −0.4 |
| S25-139:Syt1-269 | 5.8 | 5.6 | 0.2 |
| S25-20:Syt1-350 | 7.3 | 5.7 | 1.6 |
| S25-139:Syt1-350 | 5.4 | 6.4 | −1 |
| S25-20:Syt1-368 | 7.2 | 6.7 | 0.5 |
| S25-139:Syt1-368 | 6.5 | 7.0 | −0.5 |
| S25-20:Syt1-383 | 5.6 | 6.2 | −0.6 |
| S25-139:Syt1-383 | 5.1 | 5.7 | −0.6 |
| S25-76:Syt1-269 | 5.5 | 6.1 | −0.6 |
| S25-197:Syt1-269 | 5.6 | 5.0 | 0.6 |
| S25-76:Syt1-383 | 3.7 | 4.2 | −0.5 |
| S25-197:Syt1-383 | 4.6 | 5.8 | −1.2 |
| S25-76:Syt1-154 | 7.2 | 6.5 | 0.7 |
| S25-197:Syt1-154 | 6.5 | 6.5 | 0 |
| S25-76:Syt1-140 | 6.4 | 7.1 | −0.7 |
| S25-197:Syt1-140 | 7.9 | 7.1 | 0.8 |
| S25-76:Syt1-350 | 4.9 | 4.4 | 0.5 |
| S25-197:Syt1-350 | 6.7 | 6.1 | 0.6 |
| S25-76:Syt1-368 | 6.7 | 6.7 | 0 |
| S25-197:Syt1-368 | 8.1 | 7.3 | 0.8 |
| Sb-61:Syt1-154 | 4.9 | 5.4 | −0.5 |
| Sb-61:Syt1-269 | 4.1 | 4.1 | 0 |
| Sb-61:Syt1-383 | 3.6 | 4.3 | −0.7 |
| Sb-61:Syt1-140 | 7.0 | 6.9 | 0.1 |
| Sb-61:Syt1-368 | 7.2 | 7.6 | −0.4 |
| Sb-61:Syt1-350 | 6.2 | 6.8 | −0.6 |
| SNARE-bound Syt1 | |||
| Syt1-154:Syt1-383 | 5.4 | 5.6 | −0.2 |
| Syt1-154:Syt1-396 | 7.0 | 6.6 | 0.4 |
| Syt1-189:Syt1-396 | 4.4 | 4.8 | −0.4 |
| Syt1-254:Syt1-396 | 6.1 | 6.9 | −0.8 |
| average difference | −0.1 | ||
| RMS difference | 0.6 | ||
The rmsdrd (defined in Supplementary Note) for this model is 0.38 nm. S25 is SNAP-25 and Sb is synaptobrevin.
The best model (Fig. 4c) shows that the bottom of the C2B domain of Syt1 interacts with the middle portion of the SNARE complex on the side that consists of the SNAP-25 helices. This positioning is consistent with the observation that the Syt1-383:Sb-61 label pair displayed the highest FRET of all pairs (Fig. 4b). The other label combinations support this interaction between C2B and the SNARE complex because docking calculations omitting distance assignments involving the Sb-61 or the Syt-383 label sites all resulted in similar models (Supplementary Fig. 8 and data not shown). Furthermore, all of the top ten models from the docking calculations have in common that the “bottom” (a nomenclature commonly used to indicate the face of a C2 fragment opposed to the Ca2+ binding loops) of the C2B domain interacts with the SNAP-25 side of the SNARE complex (Fig. 4c and Supplementary Fig. 7).
The Ca2+-binding loops of Syt1 point away from the SNARE complex (Fig. 4c). The Ca2+-binding region of the C2A domain is more distant from the SNARE complex than that of the C2B domain, which is in agreement with other biochemical studies that find stronger association between the SNARE complex and C2B compared to C2A31 and C2B–SNARE complex binding in the absence of C2A12. This asymmetry of the C2 domain interactions with respect to the SNARE complex is also consistent with the observation that disrupting Ca2+ binding to the C2B domain impairs neurotransmitter release more strongly than with C2A1.
DISCUSSION
Many different approaches have determined a critical role for Syt1 to link Ca2+ influx during neuronal action potentials to triggered neurotransmitter release. Recent work has highlighted the importance of interactions between synaptotagmin, SNARE proteins and other neuronal proteins to establish the robust primed state of a docked vesicle that can be triggered to undergo membrane fusion in less than a millisecond following the arrival of Ca2+. Nevertheless, a precise mechanistic understanding of synaptotagmin’s activity is lacking.
The relative arrangement of the two independent C2 modules in synaptotagmin, C2A and C2B, is a critical feature that any model of the mechanism of triggered neurotransmitter release must consider. Our smFRET studies of Syt1 isolated in solution revealed intermediate FRET signals for all pairs of labeling sites tested. These well-defined FRET states commonly lasted tens of seconds and are consistent with Syt1 existing in a definite relative arrangement of the C2 domains. However, at our time resolution of 0.1 seconds per frame these states are also consistent with C2A and C2B being in rapid relative motion with little interaction. Further experimental advances that allow smFRET interrogations at faster timescales that could probe such rapid molecular motion are required to directly observe this motion. Switching between distinct states within ~1 minute of observation was observed in 10–30% of molecules indicating a capability to shift to different arrangements of Syt1 C2AB (upper left in Fig. 1b), possibly indicating a brief ability of the two domains to stabilize relative each other in a specific conformation. Alternately, the distinct FRET states might all be dynamic with infrequent transitions between them being consistent with the lack of interactions observed in NMR and EPR experiments23,24. The conformation of the C2AB linker may discriminate between these different dynamic states.
Small changes in the FRET efficiency distributions upon binding to the ternary SNARE complex suggest changes in the internal arrangement of Syt1. The widths of the distributions narrowed substantially upon binding the SNARE complexes, which suggests that random motion is stabilized upon SNARE binding. Intermolecular smFRET measurements between Syt1 and the SNARE complex in the bound state revealed broad populations with multiple peaks for many label combinations, suggesting the presence of multiple conformations for the complex.
Rigid body docking calculations using distances derived from the largest FRET populations for each pair of FRET labels led to a robust model of the dominant conformation of the Syt1–SNARE complex. One helix of Syt1 (residues 385–395) is directly positioned at the interface with the SNARE complex (Fig. 4c). The central region of the SNARE complex that mediates Syt1 binding (as predicted by our smFRET-derived model) is essential for function. Mutations of glutamates near this area of SNAP-25 (Glu51, Glu52, Glu55) to lysines eliminated in vitro binding of synaptotagmin to the SNARE complex and greatly reduced Ca2+ stimulated release in PC12 cells32. These same SNAP-25 mutations as well as additional SNAP-25 mutations directly adjacent to this region (Leu50 and Ile171) are critical in the context of docking vesicles in adrenal chromaffin cells.33 Additionally, in our smFRET-derived model the conserved arginine residues at the bottom of C2B34 are close to the interface with the SNARE complex but also are sufficiently exposed to allow potential interactions with membranes. Mutation of these residues results in decreased synchronous neurotransmitter release in hippocampal glutamatergic neurons34. These independent functional assessments of the Syt1–SNARE interactions lend further credence to our smFRET-derived model.
The Syt1–SNARE complex is not a rigid structure since we observe occasional transitions between different FRET efficiency states (Supplementary Fig. 3), and some of the smFRET efficiency distributions have a multimodal appearance (Fig. 4b). This intrinsic flexibility of the Syt1–SNARE complex may allow the complex to adapt to the particular geometry of the interacting membranes in the pre-fusion state.
Our smFRET-derived model leaves the complexin-binding site on the SNARE complex unobstructed. Therefore, the central complexin helix and synaptotagmin could simultaneously bind to the ternary SNARE complex with no apparent clash between them. Indeed, there is some in vitro evidence for simultaneous binding of Syt1 and complexin35,36, although competitive binding has also been observed20. The complexin arrest model20,37 suggests an intermediate state involving trans-SNARE complexes. However, the cis state of the SNARE complex is used in all biophysical and biochemical experiments to date. In any case, our smFRET-derived model suggests that a clamp release may not consist of a displacement reaction but rather a distinct conformational change that is triggered by the increased affinity of the Syt1–SNARE interaction upon Ca2+ binding. The enhanced SNARE interaction and the reduction of C2AB transitions (Fig. 2f) upon Ca2+ exposure may be explained by long-range electrostatic effects caused by the highly bipolar electric potential of the SNARE complex38 (Fig. 6 in ref. 38) and the large change in electric potential of Syt1 upon Ca2+ binding16,17.
Comparisons between the behavior of Syt1 and Syt3 are enabled by the Syt1 labeling sites H254C and N396C, which are aligned in primary protein sequence with amino acids Q410 and N554 in Syt3 used for smFRET measurements elsewhere13. Both Syt1 and Syt3 smFRET distributions for these sites change upon SNARE complex binding, but the changes are different (mostly narrowing and, in few cases, disappearance of minor peaks in the case of Syt1 (Fig. 1c) vs. appearance of a new major peak in the case of Syt3 (Fig. 5 in Ref. 13). Yet, the general arrangement of the Ca2+ binding loops of SNARE-bound Syt1 is strikingly similar to that observed in the SNARE-induced Ca2+-bound crystal structure of Syt313. This Ca2+-bound conformation of Syt3 is also induced by SNARE complex binding13. So, while the configuration of the β-sheets of the C2 domains is different (explaining differences between smFRET measurements of doubly-labeled Syt1 and Syt3), the Ca2+ binding loops converge to a similar arrangement.
This convergent Ca2+ binding loop behavior suggests an evolutionary conserved molecular mechanism: The synaptotagmin–SNARE complex interaction (which exists even in the absence of Ca2+, Fig. 3) recruits synaptotagmin to assembled SNAREs and restricts the conformational variability of the C2 domains by stabilizing a configuration that enables simultaneous membrane binding. This stabilization of synaptotagmin’s dynamic conformations upon SNARE binding that we have observed with smFRET is an example of the importance of conformational flexibility that likely applies to many other biomolecular interactions. The importance of such dynamic equilibria among multiple conformations is increasingly recognized as an integral aspect for the function of many proteins39.
Methods and any associated references are available in the online version of this paper at http://www.nature.com/nsmb/.
METHODS
Proteins
Expression and purification of 6-Histidine fusions of full length syntaxin, SNAP-25 (synaptosome-associated protein of 25 kDa), and synaptobrevin-II (residues 1–96) from pet28a (Novagen) have been described27 and used combinations of metal affinity, ion exchange (Q and/or S), and gel filtration chromatography. Expression and purification of the Syt1 C2AB fragment (residues 96–421) from pGex4T1 (GE Biosciences) have been described28. Briefly, the GST fusion protein was cleaved from glutathione agarose followed by ion exchange and gel filtration chromatography (Mono S and Superdex 200, GE Biosciences). All proteins were examined by SDS-PAGE for purity (>90%) then dialyzed into 20 mM Tris pH 8.0, 200 mM NaCl, 1 mM DTT (TBS).
From cysteine free templates, single cysteine mutations28 of SNAP-25 (Q20C, K76C, N139C, and Q197C), synaptobrevin (S61C) and Syt1 C2AB (E140C, Q154C, K189C, E269C, E350C, G368C, T383C and N396C) and double cysteine mutants of Syt1 C2AB (E140C/Q154C; Q154:G174; Q154:H254; Q154C/T383C; Q154C/N396C; F252C/N396C; H254C/N396C) were generated using Quickchange (Stratagene).
Protein labeling protocols have been described previously27,28,40. Labeling efficiency (assessed by absorbance spectroscopy) was typically >80% for each cysteine in Syt1 and >50% for all proteins. Proteins were labeled with Alexa555 or Alexa647-maleimide (Invitrogen, Carlsbad, CA). One experiment employed Cy3-maleimide (GE Biosciences) and another Alexa488 (Supplementary Figs. 1b and 10). Double cysteine Syt1 mutants were labeled by mixing Alexa555 and Alexa647 simultaneously, both at a ten fold excess over protein during the labeling incubation. Three different populations of doubly labeled synaptotagmin resulted: two-donor dyes, two-acceptor dyes, and synaptotagmin containing exactly one donor and one acceptor.
Syt1 was crosslinked (Fig. 2c) with BS3 (Pierce Biosciences) in 25 mM phosphate (pH 8.0) and 150 mM NaCl at a protein concentration of 50–100 nM with a 1000× molar excess of BS3 for 1 hour at 4 C. These dilute conditions prevented intermolecular crosslinking as assessed by single-molecule fluorescence detection of the number of dyes within each immobilized liposome.
Ternary SNARE complex was formed as described previously27 including the 7.5 M urea wash to minimize antiparallel assembly. Lipids (including phosphatidylcholine (EggPC) and phosphatidylserine (Brain PS) and head group biotinylated 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine (biotin-PE); all from Avanti Polar Lipids), membrane protein reconstitution and supported bilayers formation was performed as described previously40.
Proteins were encapsulated inside liposomes for studies of soluble species41–43. EggPC liposomes were prepared including 0.1% biotin-PE to allow them to be tethered to a quartz surface sequentially treated first with biotinylated BSA (1 mg per ml, 5 minutes) and next with streptavidin (0.1 mg per ml, 5 minutes). Labeled protein solutions diluted to ~50 nM with TBS with indicated final NaCl concentrations were added to glass tubes containing films of dried lipids (1 mg eggPC+0.1% biotin-PE; final lipid concentration, 1 mg per ml). These mixtures were extruded through 100 nm diameter pore filters followed by chromatography (Sepharose CL4B) to remove un-encapsulated protein. This procedure resulted in less than 15% of liposomes containing two or more proteins, similar to the experience of other groups41,42, as verified by counting individual photobleaching events.
Bilayers containing proteins were prepared as previously described.27,40 Concentrations were adjusted before reconstitution into liposomes such that maximum surface density for preassembled ternary SNARE complex with dye labels was 0.04 complexes per μm2 and without dye labels was 0.4 complexes per μm2. For syntaxin bilayers (used to form binary complex upon addition of soluble SNAP-25) (Fig. 3b), the surface protein density was 0.7 syntaxin per μm2. These densities are based upon protein:lipid ratios mixed before liposome reconstitution and purification, causing overestimation.40 Syntaxin density was sufficiently low to minimize the formation of non-productive Sx:S25 2:1 complex.40
Microscopy
Flow cells were constructed between quartz microscope slides and standard microscope coverslips. For experiments with biotinylated BSA/streptavidin surfaces, double sided tape defined the channel edges, whereas for experiments using supported lipid bilayers, UV curing optical adhesive (Norland Products, Cranbury, NJ) sealed the edges of the chamber. The bilayers were formed by 15-minute incubation with SNARE-containing liposomes (3mg per ml) followed by protein-free liposomes (30mg per ml) for an hour. The second liposome application decreased non-specific binding of proteins to the surface. Syt1 was added over these bilayers at 0.1 to 20 nM for 10 minutes before observation.
smFRET measurements used a prism-type, total internal reflection microscope with a Cascade 512B EMCCD (Photometrics) described previously44. Fluorescence emission was split into donor and acceptor channels with a 645dcxr dichoric mirror and filtered with bandpass filters, donor HQ585/70m and acceptor HQ700/75m (all Chroma). Alternating laser illumination (0 to 1 second – 635 nm, 5 mW; 1.5 to 100 seconds – 532 nm, 10 mW; 105 to 110 seconds – 635 nm, 5 mW) allowed us to distinguish the number of acceptor and donor dyes on each molecule by single step photobleaching and quantized intensity levels. This alternating illumination scheme allowed us to exclusively analyze molecular complexes containing exactly one donor and one acceptor for FRET emission [calculated from the measured donor and acceptor intensities that were background subtracted and corrected for donor leakage into the acceptor channel (ID and IA respectively) as FRET=IA/(ID + IA)]. Labeling efficiencies were typically 80–100% (a few samples as low as 50%), which implies little contamination from liposomes containing one donor and one acceptor from two separate C2AB molecules where one has a single donor and the other a single acceptor. All observations were at room temperature in TBS containing 2% glucose (wt/vol), 100 μM cyclooctatetraene, and an enzymatic oxygen scavenger (glucose oxidase 20 units per ml, catalase 1000 units per ml) (Sigma).
Data analysis
We generated all FRET plots by calculating FRET efficiency for each point in intensity traces (at 10 Hz) for which both donor and acceptor are active and combining them in a histogram for many molecules (Supplementary Table 2). This method captures the different FRET efficiency values that sometimes occur within intensity trajectories for dynamic molecules while revealing the distribution of FRET states visited across an ensemble. Histograms were fit to sums of Gaussian functions where one Gaussian function was used for FRET=0 peak, which was removed (Supplementary Fig. 4 and Note).
Supplementary Material
Acknowledgments
We thank M. Bowen, T. Südhof and J. Rizo for comments, and the National Institutes of Health for support to ATB (RO1-MH63105) and the Burroughs Wellcome Fund for a CASI award to KW. A.T.B. presented this work at the first Paul B. Sigler lecture at Yale University on March 23, 2009.
Footnotes
Note: Supplementary Information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS U.B.C. prepared all samples and performed all experimental measurements; P.S., M.V. and A.T.B. performed all simulations; All authors contributed to formulation of experimental design, interpretation of results and preparation of the manuscript.
AUTHOR INFORMATION The coordinates for the model of the Syt1–SNARE complex are available as a supplementary online file. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–9. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 2.Rhee JS, et al. Augmenting neurotransmitter release by enhancing the apparent Ca2+ affinity of synaptotagmin 1. Proc Natl Acad Sci U S A. 2005;102:18664–9. doi: 10.1073/pnas.0509153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Quarterly Reviews of Biophysics. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nature Structural & Molecular Biology. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761:838–49. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Pang ZP, Shin OH, Meyer AC, Rosenmund C, Sudhof TC. A gain-of-function mutation in synaptotagmin-1 reveals a critical role of Ca2+-dependent soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex binding in synaptic exocytosis. J Neurosci. 2006;26:12556–65. doi: 10.1523/JNEUROSCI.3804-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraudo CG, et al. Alternative Zippering as an On-Off Switch for SNARE-Mediated Fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin Controls the Force Transfer from SNARE Complexes to Membranes in Fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Wozniak AK, Schroder GF, Grubmuller H, Seidel CA, Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc Natl Acad Sci U S A. 2008;105:18337–42. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrecka J, et al. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci U S A. 2008;105:135–40. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–63. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrljic M, Strop P, Ernst J, Sutton RB, Chu S, Brunger AT. Molecular mechanism of the synaptotagmin-SNARE interaction in Ca2+-triggered vesicle fusion. 2009 doi: 10.1038/nsmb.1764. submitted. [ we will provide the AOP date and doi number once available] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: at the interface of biology, physics and chemistry. J R Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez I, et al. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–69. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 16.Shao X, et al. Synaptotagmin-syntaxin interaction: the C2 domain as a Ca2+-dependent electrostatic switch. Neuron. 1997;18:133–42. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- 17.Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–15. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 18.Ubach J, Zhang X, Shao X, Sudhof TC, Rizo J. Ca2+ binding to synaptotagmin: how many Ca2+ ions bind to the tip of a C2-domain? Embo J. 1998;17:3921–30. doi: 10.1093/emboj/17.14.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai J, Wang CT, Richards DA, Jackson MB, Chapman ER. Fusion pore dynamics are regulated by synaptotagmin*t-SNARE interactions. Neuron. 2004;41:929–42. doi: 10.1016/s0896-6273(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 21.Sutton RB, Ernst JA, Brunger AT. Crystal structure of the cytosolic C2A-C2B domains of synaptotagmin III. Implications for Ca(+2)-independent snare complex interaction. J Cell Biol. 1999;147:589–98. doi: 10.1083/jcb.147.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–13048. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arac D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–17. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 24.Herrick DZ, et al. Solution and Membrane-Bound Conformations of the Tandem C2A and C2B Domains of Synaptotagmin 1: Evidence for Bilayer Bridging. Journal of Molecular Biology. 2009;390:913–923. doi: 10.1016/j.jmb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haugland RP. The handbook: A guide to fluorescent probes and labeling technologies. Molecular Probes. 2005:1126. [Google Scholar]
- 26.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–27. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 27.Weninger K, Bowen ME, Chu S, Brunger AT. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci U S A. 2003;100:14800–14805. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowen ME, Weninger K, Ernst J, Chu S, Brunger AT. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muschielok A, et al. A nano-positioning system for macromolecular structural analysis. Nat Methods. 2008;5:965–71. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 30.Majumdar DS, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci U S A. 2007;104:12640–5. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annual Review of Biochemistry. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 32.Rickman C, et al. Conserved prefusion protein assembly in regulated exocytosis. Mol Biol Cell. 2006;17:283–94. doi: 10.1091/mbc.E05-07-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wit H, et al. Synaptotagmin-1 Docks Secretory Vesicles to Syntaxin-1/SNAP-25 Acceptor Complexes. Cell. 2009;138:935–946. doi: 10.1016/j.cell.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–8. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokumaru H, Shimizu-Okabe C, Abe T. Direct interaction of SNARE complex binding protein synaphin/complexin with calcium sensor synaptotagmin 1. Brain Cell Biol. 2008;36:173–89. doi: 10.1007/s11068-008-9032-9. [DOI] [PubMed] [Google Scholar]
- 36.Chicka MC, Chapman ER. Concurrent Binding of Complexin and Synaptotagmin to Liposome-Embedded SNARE Complexes (dagger) Biochemistry. 2009 doi: 10.1021/bi801962d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giraudo CG, et al. Distinct domains of complexins bind SNARE complexes and clamp fusion in vitro. Journal of Biological Chemistry. 2008;283:21211–21219. doi: 10.1074/jbc.M803478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–53. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 39.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–72. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 40.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–20. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okumus B, Wilson TJ, Lilley DMJ, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophysical Journal. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boukobza E, Sonnenfeld A, Haran G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. Journal of Physical Chemistry B. 2001;105:12165–12170. [Google Scholar]
- 43.Cisse I, Okumus B, Joo C, Ha TJ. Fueling protein-DNA interactions inside porous nanocontainers. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li YL, Augustine GJ, Weninger K. Kinetics of complexin binding to the SNARE complex: Correcting single molecule FRET measurements for hidden events. Biophysical Journal. 2007;93:2178–2187. doi: 10.1529/biophysj.106.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.